Abstract
In addition to mediating regulation of endogenous gene expression, RNA interference (RNAi) in plants and invertebrates plays a crucial role in defense against viruses via virus-specific siRNAs. Different studies have demonstrated that the functional diversity of RNAi in animals is linked to the diversification of the Argonaute superfamily, central components of RISCs (RNA induced silencing complexes). The animal Argonaute superfamily is traditionally grouped into AGO and PIWI Argonautes. Yet, by performing phylogenetic analyses and determining the selective evolutionary pressure in the metazoan Argonaute superfamily, we provide evidence for the existence of three conserved Argonaute lineages between basal metazoans and protostomes, namely siRNA-class AGO, miRNA-class AGO and PIWI Argonautes. In addition, it shown that the siRNA-class AGO lineage is characterized by high rates of molecular evolution, suggesting a role in the arms race with viruses, while the miRNA-class AGOs display strong sequence conservation. Interestingly, we also demonstrate that vertebrates lack siRNA-class AGO proteins and that vertebrate AGOs display low rates of molecular evolution. In this way, we provide supportive evidence for the loss of the antiviral siRNA-class AGO group in vertebrates and discuss the consequence hereof on antiviral immunity and the use of RNAi as a loss of function tool in these animals.
Introduction
A large portion of the RNA-species in Metazoans is not translated into proteins, but plays a role in controlling the cellular protein levels1. One of the most important and best-described groups of regulatory RNAs are the small RNAs, which trigger gene silencing through the RNA interference (RNAi) pathway2.
There are three major branches of small RNAs. Whereas genome encoded micro (mi)RNAs play a central role in the regulation of a multitude of cellular and physiological processes by induction of silencing of endogenous transcripts, short interfering (si)RNAs are derived from long exogenous dsRNA-fragments, typically produced during virus replication, and act as the defenders of the cell against invading viruses. A third group are the piwi-interacting (pi)RNAs, which help maintain the genome stability through silencing of selfish nucleic acids, like transposons3–5.
Notwithstanding the enormous complexity of RNAi, only three key protein components are well-conserved in eukaryotes, namely Argonaute proteins, RNA-dependent RNA-polymerases (RdRPs) and Dicer-like enzymes6. The diversity of RNAi appears to be linked to the presence of multiple paralogues of these RNAi-genes and a complex subset of accessory proteins. Argonaute proteins play a central role in binding to small RNAs and silencing of complementary transcripts in the cell by either destroying them or preventing their correct translation. Argonaute proteins found in animals are classified into three major groups, based on their interaction with specific small RNA species. Members of the first group bind to mi- and siRNAs, and are termed the AGO Argonautes, while the second group, the PIWI Argonautes, interact specifically with piRNAs. Nematode specific secondary (S)AGOs constitute the third branch of the Argonaute protein family7, 8. The latter includes Argonautes that interact specifically with newly synthesized (secondary) small RNAs, which are produced by RdRPs9.
The AGO group can be further functionally separated into AGOs specialized in mi- or siRNA-directed silencing. In insects, AGO1 is required for miRNA-directed silencing of endogenous transcripts, while AGO2 is needed for siRNA-based antiviral immunity10. Functional separation of these AGO proteins is further supported by the fact that Drosophila AGO2 was found to be under higher evolutionary pressure than AGO1, which is in accordance with its role in immunity. The situation in the nematode Caenorhabditis elegans is more complex, as 27 different Argonaute proteins are encoded in the genome of this worm, of which 19 are SAGO and 2 are PIWI proteins9. Still, the AGOs ALG1 and ALG2 are identified as the primary AGOs that mediate miRNA-directed regulation of endogenous gene expression, while RDE1 binds to exogenous (exo-)siRNAs to combat viral infections11 and ERGO1 interacts with endogenous (endo-)siRNAs to control expression of duplicating genes9.
Genome defence is presumed to be the ancestral function of RNAi in Eukaryotes6, and it remains a crucial antiviral pathway in plants, fungi and invertebrates12–14. Whether RNAi-based antiviral immunity also operates in vertebrates has been controversial for a long time15–17. Due to the development of alternative innate and adaptive immune systems, vertebrates can effectively combat viruses in the absence of RNAi18. Nevertheless, several studies using deep sequencing technologies identified low amounts of small virus-specific RNAs accumulating in mammals infected with viruses19, 20. Furthermore, virus encoded suppressors of RNAi (VSRs) have been identified in mammalian viruses20, 21. These recent findings reopened the debate on the existence of antiviral RNAi in mammals, and, more broadly, in vertebrates. In fact, it currently remains unclear to what extend viral-derived siRNAs contribute to the antiviral immune response in these animals.
Since an enormous diversity of Argonaute proteins is found in animals, with a clear functional specialization in the regulation of endogenous or exogenous transcripts, studying the phylogenetic relationship of Argonaute proteins can help understand the function of RNAi in different animal phyla. Phylogenetic analyses of the eukaryotic Argonautes superfamily have led to the identification of the AGO, SAGO and PIWI families8. Yet, the evolution of the Argonaute superfamily within animals, and in particular the evolution of the AGO subgroup in animals, has not yet been studied in detail.
Based on the principle of parsimony and the rate of molecular evolution, our analyses demonstrate, for the first time, that there exist two conserved AGO lineages between basal metazoans (sponges and cnidarians) and higher invertebrates, such as arthropods and nematodes, while vertebrates only maintained one type of AGO proteins.
Results
Identification of three conserved Argonaute lineages in metazoans
The mode of action of Argonaute AGO proteins in C. elegans, D. melanogaster and Homo sapiens has been well studied9, 22, 23, but the evolution of the AGO family in animals remains largely undetermined. Therefore, we constructed a ML phylogenetic tree using Argonaute proteins found in different species of nematodes, arthropods and chordates, as well as of flatworms and more primitive metazoans like poriferans (sponges) and cnidarians (jellyfish, corals and sea anemones).
Besides the PIWI and SAGO clades, two groups of AGO proteins can be distinguished (Fig. 1). (I) the first AGO group forms a separate monophyletic group containing sequences found in all groups investigated (Figs 1 and 2). Interestingly, within this group we find all the chordate AGO proteins, as well as the miRNA-interacting AGOs of D. melanogaster (Dme-AGO1) and C. elegans (Cel-ALG1 and 2). These fly and nematode AGOs were experimentally shown to mediate silencing of endogenous transcripts, but not antiviral immunity9, 10. In fact, studies have demonstrated that also AGO1 orthologues of locusts24 and human AGOs25 play a role in miRNA-directed endogenous gene regulation. Therefore, this clade appears to contain miRNA-interacting AGO Argonautes and is for this reason termed the miRNA-class AGO lineage. In addition, the studied poriferan, cnidarian and flatworm species also encode AGO proteins that cluster within this group, which is supported by a high bootstrap value (Figs 1 and 2). (II) The second AGO group does not form a monophyletic group when all Argonautes (AGOs, SAGOs and PIWIs) are included in the dataset (Fig. 1). Yet, when focusing specifically on the AGO subfamily, the existence of two distinct families is supported by high confidence (Fig. 2). This second group contains the Ecdysozoan (nematodes and arthropods) AGOs that mediate siRNA-directed antiviral immunity, as experimentally demonstrated for Cel-RDE1 of C. elegans 11, Dme-AGO2 of D. melanogaster 11 and Isc-AGO2-4 of Ixodes scapularis 26. For this reason, we termed this group the siRNA-class AGOs. In agreement with a role in antiviral defense, the length of the branches, which represent the relative amount of amino acid substitutions, is clearly longer in this group than in the miRNA-class AGO clade (Figs 1 and 2). Amino acid sequence alignment of miRNA-class AGOs of C. elegans (Ce-ALG1), D. melanogaster (Dme-AGO1) and H. sapiens (Hsa-AGO2) revealed ≥60% sequence identity in the entire protein and >80% sequence identity in their PIWI domain. In contrast, amino acid sequence identity between siRNA-class AGOs of C. elegans (Cel-RDE-1 and Cel-ERGO-1) and D. melanogaster (Dme-AGO2) is less than 15% and even AGO2 proteins belonging to different insect species have very low sequence identity. For example, D. melanogaster and B. mori AGO2 possess only 23% sequence identity, while their housekeeping AGO1 counterparts share 81% sequence identity. Also the studied Cnidarian AGO1 proteins, which group in the miRNA class AGO clade, share much higher sequence similarity than their AGO2 counterparts, 75% sequence identity between AGO1s versus 18% between AGO2s of Acropora millepora and Nemastella vectensis (Table 1). It is fascinating that also the most primitive metazoan phyla possess two different AGO-type with clear differences in the rate of molecular evolution. While a direct role in antiviral immunity for the cnidarian and sponge AGO2 Argonautes has not yet been demonstrated, it seems reasonable to assume that the most recent common ancestor of all animals possessed, at least, two types of AGOs, an ancestral siRNA-class and miRNA-class AGO. (III) Nematodes encode an additional Argonaute clade, termed the SAGOs. SAGO proteins were found in all studied nematode species, but were absent in the other animal groups under investigation. Finally, (IV) PIWI proteins were found in all studied animal phyla and cluster as a monophyletic group. Nevertheless, piwi genes were absent in the available genome sequence information of the nematodes Ascaris suum and Brugia malayi and the flatworms S chistosoma japonicum and Schmidtea mediterranea. Making a phylogenetic tree with only the PIWI Argonautes as query resulted in the observation of two well-supported monophyletic subgroups (Supplementary Fig. S1). Both groups contain members of poriferan, cnidarian, flatworm, arthropod and vertebrate species, suggestion that also subfunctionalization of the PIWI Argonaute subgroups accurred early in animal evolution.
Figure 1.
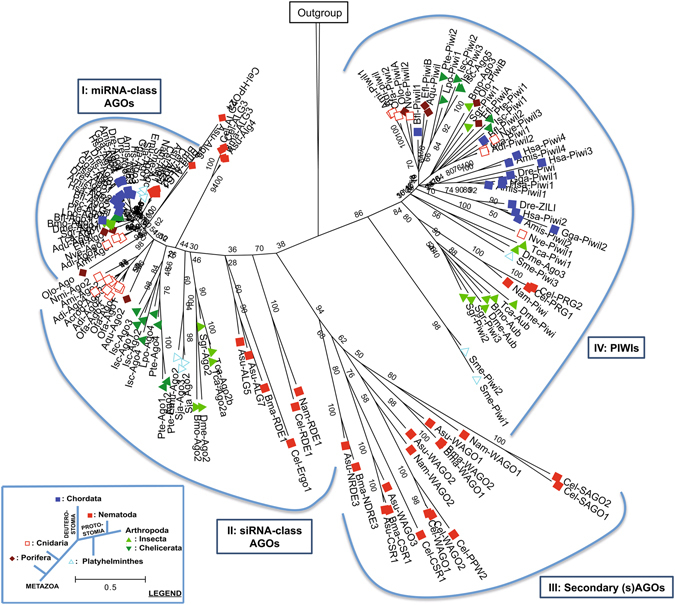
Rooted maximum likelihood phylogenetic tree (100 bootstraps) using the most conserved region between Argonaute superfamily members belonging to Porifera ( : Amphimedon queenslandica; Aqu, Ephydatia fluviatillis; Efl, Oscarella lobularis; Olo), Cnidaria (
: Amphimedon queenslandica; Aqu, Ephydatia fluviatillis; Efl, Oscarella lobularis; Olo), Cnidaria ( : Acropora digitifera; Adi, Acropora millepora; Ami, Orbicella faveolata; Ofa, Nemastella vectensis; Nve), Chordata (
: Acropora digitifera; Adi, Acropora millepora; Ami, Orbicella faveolata; Ofa, Nemastella vectensis; Nve), Chordata ( : Homo sapiens; Hsa, Alligator mississippiensis; Ami, Gallus gallus; Gga, Danio rerio; Dre, Brachiostoma floridae; Bfl), Nematoda (
: Homo sapiens; Hsa, Alligator mississippiensis; Ami, Gallus gallus; Gga, Danio rerio; Dre, Brachiostoma floridae; Bfl), Nematoda ( : Caenorhabditis elegans; Cel, Ascaris suum; Asu, Brugia malayi; Bma, Necator americanus; Nam), Arthropoda (Insecta:
: Caenorhabditis elegans; Cel, Ascaris suum; Asu, Brugia malayi; Bma, Necator americanus; Nam), Arthropoda (Insecta:  : Drosophila melanogaster; Dme, Bombyx mori; Bmo, Tribolium castaneum; Tca, Schistocerca gregaria; Sgr, and Chelicerata:
: Drosophila melanogaster; Dme, Bombyx mori; Bmo, Tribolium castaneum; Tca, Schistocerca gregaria; Sgr, and Chelicerata:  : Ixodes scapularis; Isc, Parasteatoda tepidariorum; Pte, Limulus polyphemus; Lpo) and Platyhelminthes (
: Ixodes scapularis; Isc, Parasteatoda tepidariorum; Pte, Limulus polyphemus; Lpo) and Platyhelminthes ( : Echinococcus multilocularis; Emu, Schistosoma japonicum; Sja, Schmistea mediterranea; Sme). Detailed information on the sequences used is presented in Suppl. Table S2. The Argonaute superfamily can be dissected into four distinct groups: (I) miRNA-class AGOs, (II) siRNA-class AGOs, (III) Secondary (s)AGOs and (IV) PIWI Argonautes.
: Echinococcus multilocularis; Emu, Schistosoma japonicum; Sja, Schmistea mediterranea; Sme). Detailed information on the sequences used is presented in Suppl. Table S2. The Argonaute superfamily can be dissected into four distinct groups: (I) miRNA-class AGOs, (II) siRNA-class AGOs, (III) Secondary (s)AGOs and (IV) PIWI Argonautes.
Figure 2.
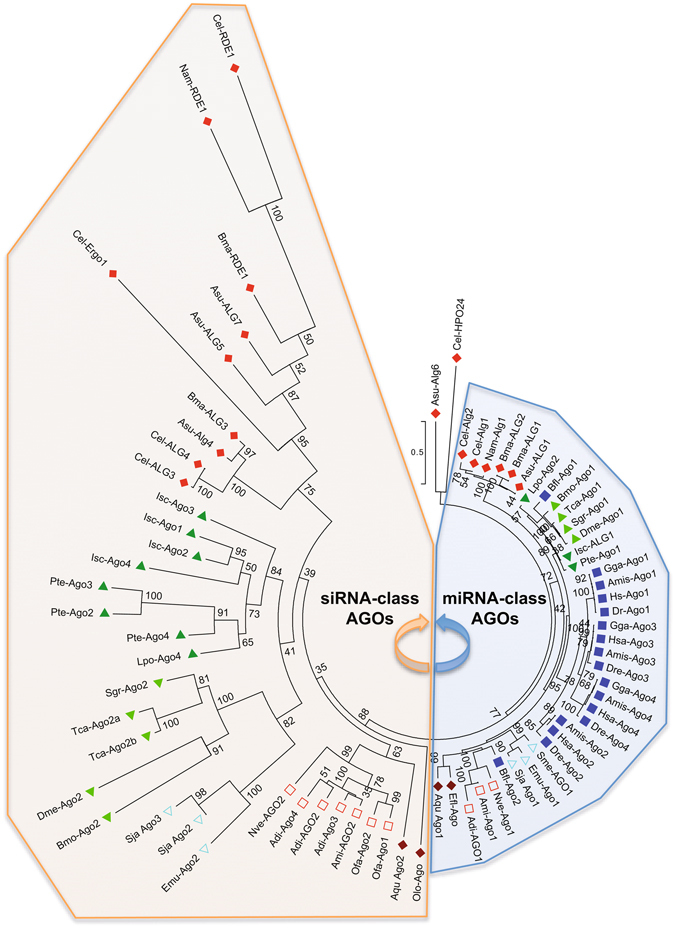
Unrooted maximum likelihood phylogenetic tree (100 bootstraps) using the AGO Argonaute members belonging to Porifera ( : Amphimedon queenslandica; Aqu, Ephydatia fluviatillis; Efl, Oscarella lobularis; Olo), Cnidaria (
: Amphimedon queenslandica; Aqu, Ephydatia fluviatillis; Efl, Oscarella lobularis; Olo), Cnidaria ( : Acropora digitifera; Adi, Acropora millepora; Ami; Orbicella faveolata; Ofa, Nemastella vectensis; Nve), Chordata (
: Acropora digitifera; Adi, Acropora millepora; Ami; Orbicella faveolata; Ofa, Nemastella vectensis; Nve), Chordata ( : Homo sapiens; Hsa, Alligator mississippiensis, Ami; Gallus gallus; Gga; Danio rerio; Dre, Brachiostoma floridae; Bfl), Nematoda (
: Homo sapiens; Hsa, Alligator mississippiensis, Ami; Gallus gallus; Gga; Danio rerio; Dre, Brachiostoma floridae; Bfl), Nematoda ( : Caenorhabditis elegans; Cel, Ascaris suum; Asu, Brugia malayi; Bma, Necator americanus; Nam), Arthropoda (Insecta:
: Caenorhabditis elegans; Cel, Ascaris suum; Asu, Brugia malayi; Bma, Necator americanus; Nam), Arthropoda (Insecta:  : Drosophila melanogaster; Dme, Bombyx mori; Bmo, Tribolium castaneum; Tca, Schistocerca gregaria, Sgr; and Chelicerata:
: Drosophila melanogaster; Dme, Bombyx mori; Bmo, Tribolium castaneum; Tca, Schistocerca gregaria, Sgr; and Chelicerata:  : Ixodes scapularis; Isc, Parasteatoda tepidariorum; Pte, Limulus polyphemus; Lpo) and Platyhelminthes (
: Ixodes scapularis; Isc, Parasteatoda tepidariorum; Pte, Limulus polyphemus; Lpo) and Platyhelminthes ( : Echinococcus multilocularis; Emu, Schistosoma japonicum; Sja, Schmistea mediterranea; Sme). Detailed information on the sequences used is presented in Suppl. Table S2. The AGO Argonaute family can be dissected into two distinct groups: (I) miRNA-class AGOs and (II) siRNA-class AGOs.
: Echinococcus multilocularis; Emu, Schistosoma japonicum; Sja, Schmistea mediterranea; Sme). Detailed information on the sequences used is presented in Suppl. Table S2. The AGO Argonaute family can be dissected into two distinct groups: (I) miRNA-class AGOs and (II) siRNA-class AGOs.
Table 1.
Sequence identity and similarity between different metazoan miRNA-class AGOs (upper table) and siRNA-class AGOs (lower table).
| Identity-Similarity (%) | Hsa-Ago2 | Amis-Ago2 | Cel-ALG1 | Asu-ALG1 | Dme-Ago1 | Bmo-Ago1 | Ami-Ago1 | Nve-Ago1 | Aqu-Ago1 |
|---|---|---|---|---|---|---|---|---|---|
| Hsa-Ago2 | x | ||||||||
| Ami-Ago2 | 94–94 | x | |||||||
| Cel-ALG1 | 60–67 | 57–65 | x | ||||||
| Asu-ALG1 | 60–68 | 58–66 | 79–84 | x | |||||
| Dme-Ago1 | 64–72 | 63–69 | 60–70 | 63–72 | x | ||||
| Bmo-Ago1 | 69–77 | 67–74 | 61–71 | 64–72 | 81–85 | x | |||
| Ami-Ago1 | 67–78 | 65–76 | 55–65 | 56–65 | 58–67 | 63–72 | x | ||
| Nve-Ago1 | 66–77 | 64–75 | 54–66 | 55–66 | 58–68 | 63–73 | 75–84 | x | |
| Aqu-Ago1 | 59–70 | 55–66 | 55–66 | 56–68 | 58–68 | 60–71 | 56–67 | 56–67 | x |
| Identity-Similarity (%) | Dme-Ago2 | Bmo-Ago2 | Cel-RDE1 | Cel-ERGO1 | Asu-ALG5 | Asu-ALG7 | Ami-Ago2 | Nve-Ago2 | Aqu-Ago2 |
| Dme-Ago2 | x | ||||||||
| Bmo-Ago2 | 23–36 | x | |||||||
| Cel-RDE1 | 13–22 | 14–29 | x | ||||||
| Cel-ERGO1 | 14–27 | 14–26 | 13–26 | x | |||||
| Asu-ALG5 | 18–29 | 20–34 | 19–35 | 19–36 | x | ||||
| Asu-ALG7 | 18–29 | 20–34 | 20–35 | 19–33 | 41–56 | x | |||
| Ami-Ago2 | 9–13 | 10–18 | 10–18 | 8–15 | 9–16 | 12–20 | x | ||
| Nve-Ago2 | 21–33 | 22–34 | 15–30 | 14–26 | 21–33 | 20–31 | 18–23 | x | |
| Aqu-Ago2 | 19–29 | 23–35 | 17–31 | 16–31 | 27–41 | 26–39 | 17–25 | 29–40 | x |
The miRNA-class AGO sequences used are of Homo sapiens (chordate, Hsa-Ago2), Alligator mississippiensis (chordate, Ami-Ago2), Caenorhabditis elegans (nematode, Cel-ALG1), Ascaris suum (nematode, Asu-ALG1), Drosophila melanogaster (insect, Dme-Ago1), Bombyx mori (insect, Bmo-Ago1), Acropora millepora (cnidarian, Ami-Ago1), Nemastella vectensis (cnidarian, Nve-Ago1), and Amphimedon queenslandica (sponge, Aqu-Ago1). The siRNA-class AGOs used are of C. elegans (nematode, Ce-RDE1 and Ce-ERGO1), A. suum (nematode, Asu-ALG5 and Asu-ALG7), D. melanogaster (insect, Dme-Ago2), B. mori (insect, Bmo-Ago2), A. millepora (cnidarian, Ami-Ago2), N. vectensis (cnidaria, Nve-Ago2) and A. queenslandica (sponge, Aqu-Ago2). In each cell, the first number corresponds to the percentage of sequence identity, while the second value stands for the percentage of sequence similarity.
Due to the highly expanded AGO-family in nematodes, there are two additional AGO orthologues for which homologues were not found in the other animal groups under investigation. The HPO-24-like sequences, present in C. elegans and A. suum, form a separate cluster closely related to the miRNA-class AGO clade (Figs 1 and 2). In addition, ALG3/4-like sequences cluster within a clade containing the siRNA-class Argonautes Cel-RDE1 and Cel-ERGO1 (Figs 1 and 2). Yet, Conine and coworkers demonstrated that the ALG3/4 play an important role in promoting the transcription of spermatogenesis genes of C. elegans 22. Therefore, it appears that these additional nematode AGO-species are a result of duplication and (sub)functionalization of miRNA-class and siRNA-class ago genes, respectively, in the nematode phyla.
Finally, it appears that all chordates lack siRNA-class AGOs, since none of the available genome sequence information of chordates in Genbank possess siRNA-class AGO sequences. This is exemplified by the fact that the available chordate sequence with highest homology (represented by the lowest E-value) to the D. melanogaster AGO2 protein is B. floridae Bfl-AGO2 (Supplementary Table S1), an Argonaute that clusters within the miRNA-class AGO group (Fig. 2). Yet, due to the limited amount of chordate sequence information available on Genbank, it remains possible that siRNA-class AGOs are present in other primitive chordate species.
Elevated rates of molecular evolution in siRNA-class AGOs and PIWIs, but not in miRNA-class AGOs in insects
Immune proteins are under strong selective pressure, due to the host-pathogen arms race. Directional selective pressure can be identified by testing for an elevated rate of non-synonymous nucleotide substitutions (Ka) compared to synonymous substitutions (Ks)27.
Utilizing tree-based and pairwise comparison for the calculation of Ka/Ks values of Drosophila RNAi genes, elevated rates of molecular evolution were identified in the Drosophila sp. siRNA class-ago (ago2) and piwi genes (ago3, piwi and aub), while not in the miRNA-class ago gene (ago1), i.e. in comparison to the studied housekeeping genes alpha-tubulin1a and gapdh (Fig. 3A and B). However, the ratio averaged over all sites is almost never >1, since positive selection is heterogeneous in nature, unlikely to affect all codons. Therefore, site-specific models in the Codeml package were used to allow the Ka/Ks value to vary among sites. Statistics for codon-specific selection were positive for ago2, ago3 and aub (Table 2).
Figure 3.
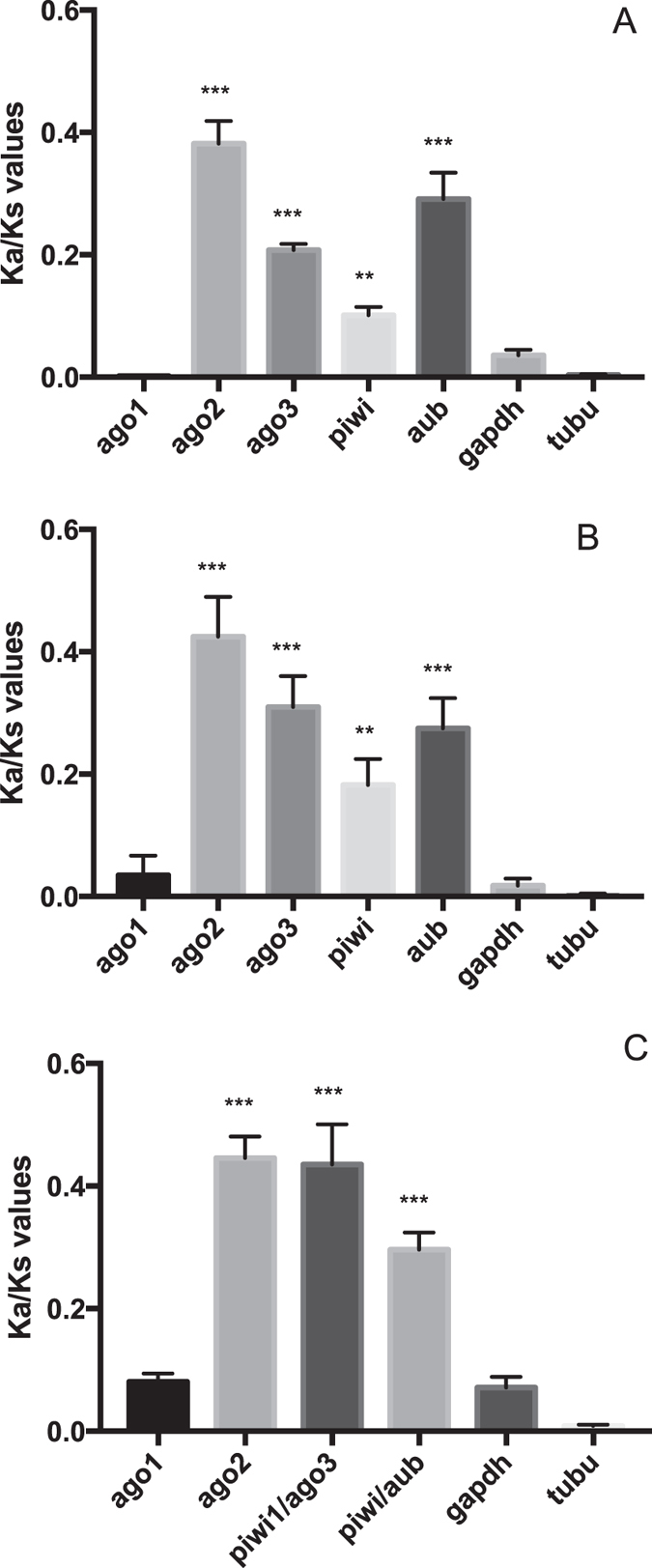
The evolutionary rate of argonaute sequences within the Drosophila genus, estimated by determining the Ka versus Ks substitutions by pairwise (Fig. 3A) and ML-tree based (Fig. 3B) comparison and using sequence information of D. melanogaster, D. simulans, D. sechellia, D. yakuba and D. erecta. Figure 3C: Rate of molecular evolution of argonaute sequences within the Insecta class. For this, sequence information of the D. melanogaster, Bombyx mori, Tribolium castaneum and Schistocerca gregaria argonaute coding regions was used as query. The bars represent the mean +/− SEM of individual Ka/Ks values per gene. Statistics (MWU-test) support difference with the housekeeping genes glyceraldehyde 3-phosphate dehydrogenase (gapdh) and apha-tubulin1a (tubu); **p < 0.01, ***p < 0,0001.
Table 2.
Likelihood ratio tests (LRTs) for codon-specific evolutionary selection in argonaute sequences within Drosophila sp. and primates.
| lnLM7 | lnLM8 | 2ΔlnL (LRT) | p-Value | Tree length | |
|---|---|---|---|---|---|
| Drosophilae | |||||
| ago1 | −4733.497 | −4733.501 | 0.008 | n.s. | 0.232 |
| ago2 | −9056.855 | −9046.185 | 21.341 | 1.16 e-05 | 1.008 |
| ago3 | −5278.170 | −5275.845 | 4.650 | 0.041 | 0.710 |
| aub | −5844.195 | −5840.904 | 6.581 | 0.019 | 0.696 |
| piwi | −5340.199 | −5339.363 | 1.673 | n.s. | 0.593 |
| Primates | |||||
| ago1 | −4733.497 | −4733.501 | −0.008 | n.s. | 0.23791 |
| ago2 | −4167.933 | −4167.937 | −0.008 | n.s. | 0.31090 |
| ago3 | −3698.245 | −3698.246 | −0.001 | n.s. | 0.21646 |
| ago4 | −3601.807 | −3601.809 | −0.003 | n.s. | 0.08879 |
| piwil1 | −5197.149 | −5197.130 | 0.03820 | n.s. | 0.45568 |
| piwil2 | −6167.099 | −6167.045 | 0.10793 | n.s. | 0.49761 |
| piwil3 | −3672.889 | −3668.723 | 8.33137 | 0.0078 | 0.64550 |
| piwil4 | −4812.804 | −4812.653 | −0.00002 | n.s. | 0.15997 |
The log-likelihood scores for model 7 (Neutral, LnLM7) and model 8 (Positive selection, LnLM8) were used to determine significance between these models. Therefore, the Chi-Square Distribution test (df = 2) was performed on the Likelihood ratio test (LRT) values. Sequences information of D. melanogaster, D. simulans, D. sechellia, D. yakuba and D. erecta was used to estimate codon specific evolutionary pressure within Drosophila sp. The primate species that were used for this analysis are Homo sapiens, Pan troglodytes, Pan paniscus, Gorilla gorilla, Pongo abelli, Macaca mulatta and Callithrix jacchus.
To test whether positive evolutionary selection in ago2 and piwi sequences is specific for Drosophila sp. or whether this represents a more general characteristic of these genes in the entire insect class, the Ka/Ks values were estimated for argonautes of insect species belonging to different insect orders, namely of D. melanogaster (Diptera), B. mori (Lepidoptera), T. castaneum (Coleoptera) and S. gregaria (Orthoptera) (Fig. 3C). Elevated Ka/Ks values were detected for the tested siRNA-class ago and piwi genes, but not for ago1. Due to the high sequence divergence of these genes, pairwise comparison, as well as tests for codon-specific selection, could not be performed.
Elevated rates of molecular evolution in PIWI proteins, but not in AGO proteins in vertebrates
To study directional selection in vertebrate argonaute genes, Ka/Ks values were estimated for argonaute genes of different primate species. Elevated Ka/Ks values were found in primate piwi genes (piwil2, 3 and 4), but not in their ago genes (Fig. 4A and B). In agreement with these observations, testing for site-specific directional selection was positive for piwil2 and piwil3 (Table 2), but negative for the ago genes. Furthermore, piwil2 and piwil4 are under elevated rates of selective pressure within vertebrates belonging to different classes (Fig. 4C). Similar as for the insect species with too high sequence divergence, pairwise comparison as well as tests for codon-specific selection could not be performed.
Figure 4.
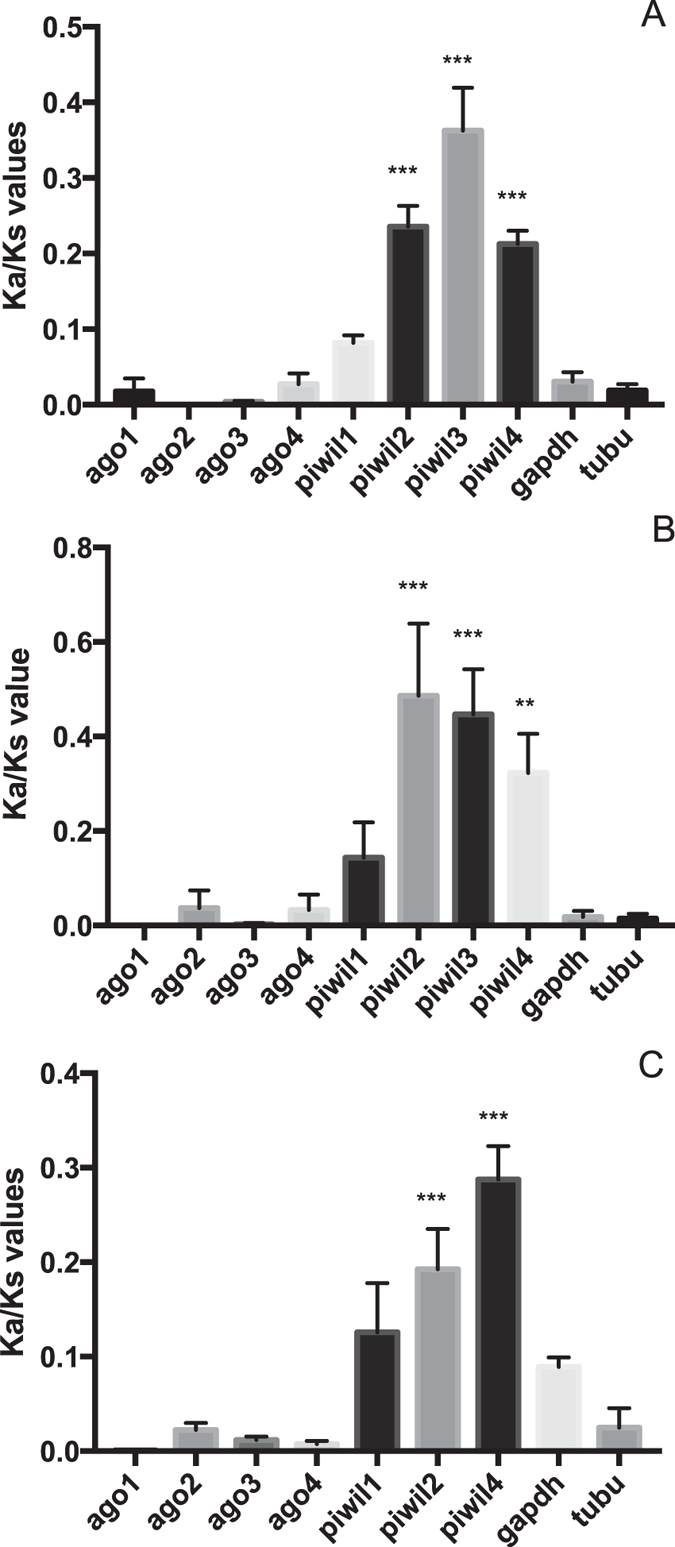
The evolutionary rate of argonaute sequences within the primates, determined by means of pairwise (Fig. 4A) and ML-tree based comparison (Fig. 4B). The nucleic acid sequence of Argonautes of Homo sapiens, Pan troglodytes, Pan paniscus, Gorilla gorilla, Pongo abelli, Macaca mulatta and Callithrix jacchus were used for Ka/Ks estimations. Figure 4C: Rate of molecular evolution of Argonautes within vertebrates. Ka/Ks values are estimated by means of ML-tree based comparison and by using sequence information of H. sapiens, Mus musculus, Alligator mississippiensis, Gallus gallus and Danio rerio. Noteworthy, Piwil4 is absent in the G. gallus and D. rerio genome sequence. The bars represent the mean +/− SEM of individual Ka/Ks values per gene. Statistics (MWU-test) support difference with the housekeeping genes glyceraldehyde 3-phosphate dehydrogenase (gapdh) and apha-tubulin1a (tubu); **p < 0.01, ***p < 0,0001.
Elevated rates of Ka/Ks substitutions in siRNA-class ago and sago genes, but not in miRNA-class ago and piwi genes in nematodes
Elevated rates of molecular evolution were also identified in siRNA-class agos (rde1 and ergo1), hpo24 and the sago genes of different Caenorhabditis species (Fig. 5A). Since C. elegans encodes 19 SAGO proteins, only four sago genes were selected for Ka/Ks estimations. Interestingly, ppw1 and sago1 displayed significantly higher Ka/Ks values than wago1 and csr1. Noteworthy, most studied Caenorhabditis species appear to lack clear ALG-2 orthologues. Furthermore, in contrast to the situation in insects and vertebrates, we did not observe elevated Ka/Ks values for the nematode piwi genes (prg1 and prg2). Unfortunately, we could not estimate the Ka/Ks values by means of pairwise comparison, nor study codon-specific directional selection, due to too low sequence similarity between the siRNA-class ago and sago genes of different Caenorhabditis species. Finally, the rate of molecular evolution between argonautes of species belonging to different nematode classes, namely of C. elegans, A. suum, Necator americanus and B. malayi was determined. Yet, we could only identify orthologues for ALG1 and RDE1 in the studied nematodes. The estimated Ka/Ks values for rde1 were strongly elevated, but not these for the nematode alg1 sequences (Fig. 5B).
Figure 5.
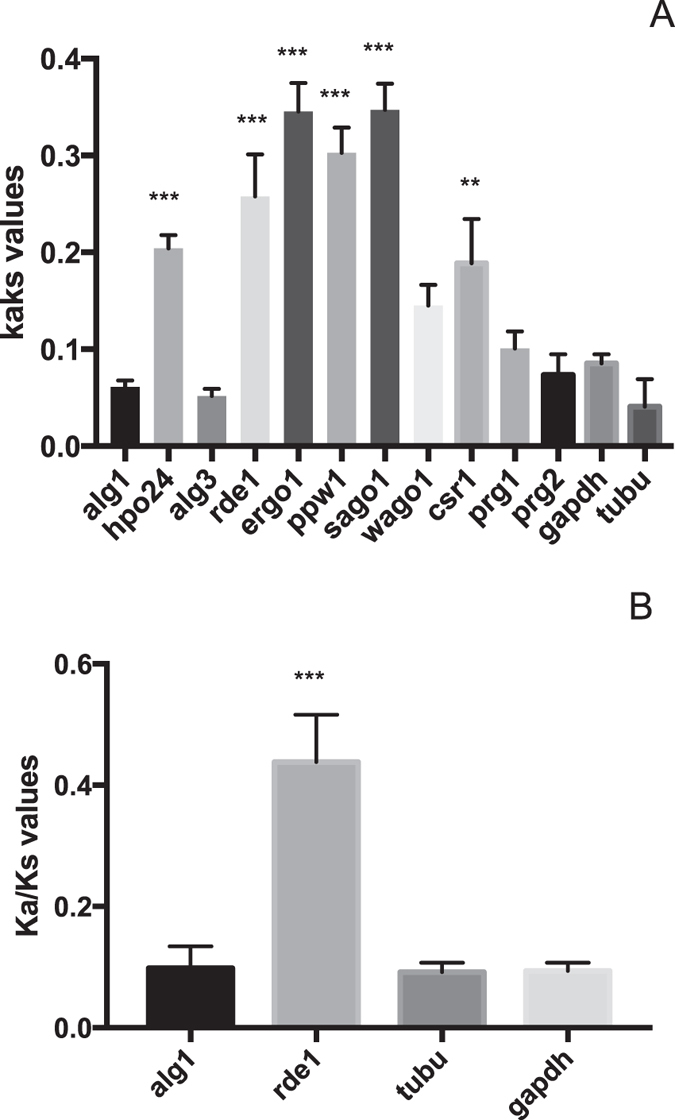
Figure 5A: Evolutionary rate of argonaute transcript sequences within the Caenorhabditis genus. Ka/Ks values are estimated by means of ML-tree based comparison and by using sequence information of C. elegans, C. briggsae, C. remanei and C. brenneri. Figure 5B: The evolutionary rate for Argonaute proteins in nematodes, determined by estimating the Ka/Ks values by means of ML-tree based comparison and by using sequence information of C. elegans, Ascaris suum, Brugia malayi and Necator americanus. Due to the enormous complexity and variation in the Argonaute AGO family present in nematodes, we could only find orthologs of the miRNA-class AGO alg1 and the siRNA-class AGO rde1 in all studied nematode species. The bars represent the mean +/− SEM of individual Ka/Ks values per gene. Statistics (MWU-test) support difference with the housekeeping genes glyceraldehyde 3-phosphate dehydrogenase (gapdh) and apha-tubulin1a (tubu); **p < 0.01, ***p < 0,0001.
Discussion
This article describes the discovery of three conserved Argonaute groups in invertebrates, miRNA-class AGOs with a primary function in endogenous gene regulation, the antiviral siRNA-class AGOs that are under strong selective pressure and PIWI Argonautes that mediate piRNA-directed transposon silencing. The siRNA-class AGOs cluster in a well-supported monophyletic group when focusing on the AGO Argonautes (Fig. 2), but not when including all Argonautes (AGOs, SAGOs and PIWIs). A plausible explanation for this could be found in the high rate of sequence divergence of the siRNA-class AGO group, making it difficult to study their evolutionary relationship. Still, our data support the hypothesis that the genome of the most recent common ancestor of all animals encoded, at least, three different AGOs, an ancestral miRNA-class AGO, siRNA-class AGO and PIWI Argonaute. Furthermore, we demonstrate the exciting finding that vertebrates do not encode siRNA-class AGO Argonautes (Figs 1 and 2).
The miRNA-class AGOs display strong sequence conservation (Table 1) and are not under elevated evolutionary pressure (Figs 3, 4 and 5), which is not in agreement with a direct role in antiviral immunity. This finding corresponds with previously published work, where it was shown that the analogous sequences in D. melanogaster (AGO1) and C. elegans (ALG1/2) function in the control of endogenous gene expression, but not in antiviral immunity9, 11, 28. Since vertebrates only encode miRNA-class AGOs (Figs 1, 2 and 4), our data support the absence of RNAi-based antiviral immunity in vertebrates. This is in accordance to the fact that deep sequencing of infected mammalian cells either failed to detect accumulation of virus-specific siRNAs or only identified extremely low levels19. Nevertheless, mammalian virus-encoded VSR have been identified, which can block RNAi efficiency in insect or plant systems29–31. One plausible explanation could be found in the fact that the majority of mammalian virus-encoded VSRs bind to dsRNA. While shielding the dsRNA might indeed block proper functioning of RNAi, it has also been reported that the VSRs have interferon-antagonistic properties32–35. This is an exciting finding bearing in mind that interferon is considered to be the first line of defense against viruses in mammals36. Interestingly, defence against invading nucleic acids is presumed to be the ancestral function of RNAi in eukaryotes6. Even more, the miRNAs of plants and animals appear to have evolved independently6. Therefore, it seems plausible to assume that the antiviral role of AGOs has been lost in vertebrates (Fig. 6).
Figure 6.
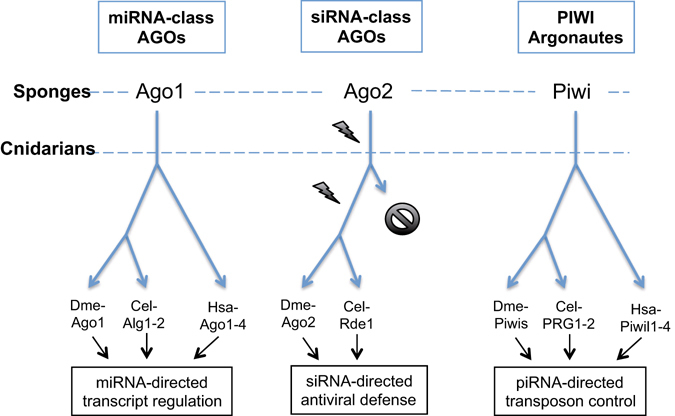
The evolution of the metazoan Argonaute superfamily. The miRNA-class AGO, siRNA-class AGO and PIWI Argonaute lineages are conserved between sponges, cnidarians, nematodes and insects, while chordates only encode miRNA-class AGOs and PIWIs (the absence of the siRNA-class AGOs in vertebrates is depicted by a black stop sign). Several studies have identified that the miRNA-class AGOs in the fruit fly (Dme-Ago1), C. elegans (Cel-ALG1-2) and humans (Hsa-Ago1-4) can interact with miRNAs and mediate regulation of endogenous transcripts (indicated with black arrows), the siRNA-class AGOs of flies (Dme-Ago2) and C. elegans (Cel-RDE-1) play a crucial role in the defense against viruses (black arrows), and finally, the PIWI Argonautes in flies (Dme-Piwis: Ago3, Piwi and Aubergene), C. elegans (Cel-PRG1-2) and humans (Hsa-Piwil1-4) mediate piRNA-directed control of transposons (also depicted by black arrows). The siRNA-class AGOs are characterized by elevated rates of molecular evolution (depicted by lightning signs).
Interestingly, in this paper, it is also demonstrated that several PIWI proteins in vertebrates are under evolutionary pressure (Fig. 4). Also for insects, elevated rates of evolutionary pressure were identified in PIWI proteins (Fig. 3), which is in accordance to the demonstrated antiviral role of the piRNA pathway in mosquitoes37. On the other hand, after deep sequencing of virus-infected D. melanogaster and silencing of the PIWI Argonaute proteins in these flies, Petit and colleagues (2016) concluded that the piRNA-pathway is not required for antiviral defense in the common fruit fly38. Nevertheless, we identified that the Drosophila sp. PIWI proteins are under elevated selective pressure (Fig. 4). This should not necessarily imply that the piRNA pathway plays a role in antiviral immunity in Drosophila flies, since other types of selfish nucleic acids, such as transposons, could be responsible for the identified evolutionary pressure in the PIWI proteins. In fact, it is well established that the PIWI pathway in D. melanogaster plays a crucial role in controlling retrotransposons39. Another interesting observation in the present study is the fact that selective pressure in the Caenorhabditis PIWI sequences is absent (Fig. 5A). Furthermore, piwi genes are absent in the genomes of B. malayi and N. americanus (Figs 1 and 2). In full agreement with this observation, Sarkies et al. (2015) sequenced small RNAs from multiple nematode species and observed that in the majority of studied species piRNAs are absent. In addition, they identified that RdRP-activity helps to compensate for the absence of piRNAs40. We also identified elevated rates of molecular evolution in the studied nematode SAGO proteins. SAGO Argonautes have only been identified in nematodes and are extremely diverse, with 19 SAGO genes in C. elegans. Interestingly, the rate of molecular evolution differs for the different studied SAGOs, with significantly higher rates of Ka versus Ks substitutions in Caenorhabditis ppw1 and sago1 than in wago1 and csr1 (Fig. 5). Remarkably, this corresponds well to the published results of Yigit and colleagues (2006), demonstrating that PPW1 and SAGO1 function primarily in long dsRNA-based RNAi, while CSR1 and WAGO1 act mainly in miRNA-based endogenous gene regulation9.
Besides its important role in the regulation of biological processes, RNAi is also a widely used tool for reverse genetics and holds potential for a vast array of application possibilities, such as the development of therapeutic agents and species-specific insecticides5, 41, 42. In insects and nematodes, long dsRNA, which is an important viral pathogen associated molecular pattern (PAMP), is used to activate the RNAi-pathway. In this way the antiviral siRNA-pathway is activated, which is supported by the observation that dsRNA-derived siRNAs are incorporated into the D. melanogaster AGO2 and the C. elegans RDE1 Argonautes9, 28. In order to purge a viral infection, the host must actively destroy as many viral transcripts as possible, with minimal off-target effects on endogenous transcripts. On the other hand, evidence from microarray studies indicated that a single miRNA can target hundreds of different mRNAs in D. melanogaster 43. Given this discrepancy between the efficiency and specificity upon activation of the siRNA and miRNA pathways, the finding that vertebrates have lost the siRNA-class AGOs, and therefore most likely the entire antiviral siRNA-pathway, is of particular interest for the usage of RNAi as a loss-of-function tool. In general, utilizing the RNAi-technology as a loss-of-function tool in vertebrates is much less user-friendly than in insects and nematodes44. For instance, when long dsRNA, and even siRNAs, is introduced in mammalian cells, other immune pathways are activated, such as the interferon and JAK-STAT pathways, which leads to a non-specific translation stop in the cell45. Furthermore, several species of insects and nematodes possess the remarkable ability to take up dsRNA from the environment and further spread the RNAi-signal to the rest of the body5, 46. Therefore, a single injection or feeding event with long dsRNA can induce potent gene silencing effects in these animals47, 48. Yet, the body does not take up smaller dsRNA-fragments, such as miRNAs, efficiently. In accordance with the absence of RNAi-based antiviral immunity, mammals lack efficient dsRNA-uptake mechanisms. For this reason, delivery of the RNAi signal in vertebrates requires liposome- and virus-based approaches to transport the siRNAs into the cells and tissues. Therefore, the absence of siRNA-based immunity in vertebrates might help explain the relatively insensitive and unspecific RNAi-response observed in most vertebrates.
In conclusion, we describe for the first time that there are three conserved Argonaute lineages in animals and that vertebrates lost the antiviral AGO Argonaute lineage.
Methods
Phylogenetic analyses
Transcript sequence information for Argonaute proteins of different chordate, insect, chelicerate, nematode, flatworm, cnidarian and poriferan species were retrieved from NCBI, flybase and wormbase. Detailed information on the used sequences is available in the Supplementary Table S1. Shortly, regarding the chordates, Argonaute sequence information of Homo sapiens (primates), Alligator mississippiensis (Crocodilla), Gallus gallus (Galliformes), Danio rerio (Cypriniformes), Branchiostoma floridae (Amphioxiformes) was used. The selected nematode species for sequence analyses were C. elegans (Rhabditidae), Ascaris suum (Ascaridia), Brugia malayi (Spiurida) and Necator americanus (Strongylida). Concerning the Arthropoda, D. melanogaster (Diptera), Bombyx mori (Lepidoptera), Tribolium castaneum (Coleoptera), Schistocerca gregaria (Orthoptera), Parasteatoda tepidariorum (Araneae), Ixodes scapularis (Ixodida) and Limulus polyphemus (Xiphosura) were selected for phylogenetic analyses. The flatworm species under investigation are Echinococcus multilocularis (Cestoda), Schistosoma japonicum (Trematoda) and Schmidtea mediterranea (Rhabditophora). For the Cnidaria, Argonaute sequences of Acropora digitifera (Acroporidae), Acropora millepora (Acroporidae), Orbicella faveolata (Merulinidae) and Nemastella vectensis (Actiniaria) were used. Finally, regarding the Porifera, Argonautes were identified in the available genome database of the sponge Amphimedon queenslandica (Haplosclerida) and in the available sequence data of the sponges Ephydatia fluviatilis (Haplosclerida) and Oscarella lobularis (Homosclerophorida). The deduced amino acid sequences, determined by in silico translation using Prosite (ExPASy, ETH Zurich), was used to predict the PIWI protein domains (Pfam, Sanger institute). The amino acid sequences of the PIWI domains were aligned with T-coffee software using the PAM matrix (EMBL-EBI). The region with highest sequence homology, which was approximately 200 amino acids in length, was selected using Jalview software. The latter was used for the construction of a phylogentic tree using the maximum likelihood (ML) method with 100 bootstraps (PhylML). The Jones-Taylor-Thornton (JTT) substitutions model was used, since we determined with MEGA7 that this is the best substitution model for our dataset. The final tree was visualized in MEGA7. All Argonaute proteins found in the studied animal species were used, with the exception of several SAGO proteins of C. elegans. This nematode encodes 19 different SAGO proteins. Previous studies have shown that all C. elegans SAGO proteins cluster in one clade and overrepresentation of one branch can however istort the formation of the correct phylogenetic tree. Therefore, six C. elegans SAGO orthologues were randomly selected for analyses. On the other hand, all SAGO proteins found in the other nematode species under investigation were included in the data set.
Rate of molecular evolution analyses
Codon-based multiple sequence alignments were performed in MEGA7, using the nucleic acid sequence of the entire coding region of argonautes. ML-phylogentic trees were constructed in MEGA7 to confirm the correct identity of the selected Argonaute sequences. The amount of nonsynonymous (Ka) versus synonynomous (Ks) nucleic acid substitutions was estimated by means of pairwise comparisons (runmode = −2) in Codeml (implemented in the PAML package) and by ML-tree-based methods using KaKs Calculator (UiB-CBU). The Drosophila species (sp.) used in these analyses were D. melanogaster, D. simulans, D. sechellia, D. yakuba and D. erecta, retrieved from flybase. The primate species included in the analyses were H. sapiens (human), Pan troglodytes (chimpanzee), Pan paniscus (bonobo), Gorilla gorilla (western gorilla), Pongo abelli (Bornean orangutan), Macaca mulatta (rhesus macaque) and Callithrix jacchus (marmoset). Regarding the Caenorhabditis s, we retrieved argonaute sequence information of C. elegans, C. briggsae, C. remanei and C. brenneri. Besides determining the Ka/Ks values using homologues sequences of closely related species, we also determined the rate of molecular evolution upon comparison of homologous sequences of more distantly related species. Due to high sequence divergence (especially between the siRNA-class AGO and PIWI proteins), only the tree-based method (KaKs calculator, CBU) was used for Ka/Ks calculations. We used sequences of insect species of different insect orders, namely of D. melanogaster (Diptera), B. mori (Lepidoptera), T. castaneum (Coleoptera) and S. gregaria (Orthoptera). For the vertebrates, we used sequence information of H. sapiens (Primates), Mus musculus (Rodentia), A. mississippiensis (Crocodilla), G. gallus (Galliformes) and D. rerio (Cypriniformes). Finally, the nematode species selected for this analysis were C. elegans (Rhabditidae), A. suum (Ascaridia), B. malayi (Spiurida) and N. americanus (Strongylida). The number of data points used for pairwise and tree-based comparison of argonaute sequences within the Drosophila genus are n = 10 and n = 8, respectively (Fig. 2A and B). For the tree-based comparison of argonaute sequences of different insect orders n = 6 data points were used in the dataset. The ago4 sequence of G. gorilla was not included in the dataset, due to the presence of a single insertion that disturbed correct alignments. The number of data points used for the estimation of Ka/Ks values of primate argonautes was n = 16 for pairwise and n = 12 for tree-based comparison of ago4 sequence, and n = 21 and n = 14 for pairwise and tree-based comparison of the other primate argonaute sequences. Finally, the Ka/Ks values that were estimated by means of tree-based comparison generated 6 different data points (n = 6) for the Caenorhabditis and nematode data sets. Significant differences between Ka/Ks values of the argonaute sequences and the housekeeping genes glyceraldehyde 3-phosphate dehydrogenase and apha-tubulin1a was determined with the MWU-test.
Site-specific selection analyses
Codeml provides several models that allow the Ka/Ks ratio to vary among sites, which were used to test for codon-specific directional selection. For this purpose, control codon based phylogenetic ML-tree reconstructions were made with MEGA7 in combination with the codon-based nucleic acid sequence alignments used for global Ka/Ks estimations. Comparisons of model 7 (Neutral, beta) and model 8 (Positive Selection, beta and Ka/Ks > 1) were used to test for signs of positive selection. Significant difference between these models was determined in R using the Chi-Square Distribution test (df = 2) for the Likelihood ratio test (LRT) values.
Data Availability
All sequence information is publicly available on Genbank, flybase and wormbase. The accession numbers of all Argonaute sequences used for the construction of the phylogenetic tree our summarized in Supplementary Table S1.
Electronic supplementary material
Acknowledgements
The authors gratefully acknowledge the K.U. Leuven Research Foundation (3E150454), the Research Foundation of Flanders (FWO-Flanders; G028013N) for financial support.
Author Contributions
Conceptualization, Investigation and writing original draft: N.W; Review & Editing: D.S and J.V.B.; Funding acquisition: N.W. and J.V.B.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-08043-5
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kashi K, Henderson L, Bonetti A, Carninci P. Discovery and functional analysis of lncRNAs: Methodologies to investigate an uncharacterized transcriptome. Biochimica et biophysica acta. 2016;1859:3–15. doi: 10.1016/j.bbagrm.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 4.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wynant N, Santos D, Vanden Broeck J. Biological mechanisms determining the success of RNA interference in insects. International review of cell and molecular biology. 2014;312:139–167. doi: 10.1016/B978-0-12-800178-3.00005-1. [DOI] [PubMed] [Google Scholar]
- 6.Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol. 2008;23:578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck AH, Blaxter M. Functional diversification of Argonautes in nematodes: an expanding universe. Biochem Soc Trans. 2013;41:881–886. doi: 10.1042/BST20130086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 9.Yigit E, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 10.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Gene Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilkins C, et al. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436:1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 13.van Rij RP, et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Gene Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szittya G, Burgyan J. RNA interference-mediated intrinsic antiviral immunity in plants. Curr Top Microbiol Immunol. 2013;371:153–181. doi: 10.1007/978-3-642-37765-5_6. [DOI] [PubMed] [Google Scholar]
- 15.Cullen BR, Cherry S. & tenOever, B. R. Is RNA interference a physiologically relevant innate antiviral immune response in mammals? Cell Host Microbe. 2013;14:374–378. doi: 10.1016/j.chom.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 16.Backes S, et al. The Mammalian response to virus infection is independent of small RNA silencing. Cell Rep. 2014;8:114–125. doi: 10.1016/j.celrep.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogerd HP, et al. Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs. Journal of virology. 2014;88:8065–8076. doi: 10.1128/JVI.00985-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. The Journal of general virology. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 19.Parameswaran P, et al. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. Plos Pathog. 2010;6:e1000764. doi: 10.1371/journal.ppat.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Lu J, Han Y, Fan X, Ding SW. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui L, et al. The Nucleocapsid Protein of Coronaviruses Acts as a Viral Suppressor of RNA Silencing in Mammalian Cells. Journal of virology. 2015;89:9029–9043. doi: 10.1128/JVI.01331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conine CC, et al. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. P Natl Acad Sci USA. 2010;107:3588–3593. doi: 10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su H, Trombly MI, Chen J, Wang XZ. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Gene Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song J, Guo W, Jiang F, Kang L, Zhou S. Argonaute-1 is indispensable for juvenile hormone mediated oogenesis in the migratory locust, Locusta migratoria. Insect Biochem Molec. 2013;43:879–887. doi: 10.1016/j.ibmb.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 25.Dueck A, Ziegler C, Eichner A, Berezikov E, Meister G. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 2012;40:9850–9862. doi: 10.1093/nar/gks705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnettler E, et al. Induction and suppression of tick cell antiviral RNAi responses by tick-borne flaviviruses. Nucleic Acids Res. 2014;42:9436–9446. doi: 10.1093/nar/gku657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang C, Wang J, Long M, Fan C. gKaKs: the pipeline for genome-level Ka/Ks calculation. Bioinformatics. 2013;29:645–646. doi: 10.1093/bioinformatics/btt009. [DOI] [PubMed] [Google Scholar]
- 28.Karlikow M, Goic B, Saleh MC. RNAi and antiviral defense in Drosophila: setting up a systemic immune response. Dev Comp Immunol. 2014;42:85–92. doi: 10.1016/j.dci.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 29.de Vries W, Berkhout B. RNAi suppressors encoded by pathogenic human viruses. Int J Biochem Cell Biol. 2008;40:2007–2012. doi: 10.1016/j.biocel.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, et al. Hepatitis C virus core protein is a potent inhibitor of RNA silencing-based antiviral response. Gastroenterology. 2006;130:883–892. doi: 10.1053/j.gastro.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 31.Haasnoot J, Berkhout B. RNAi and cellular miRNAs in infections by mammalian viruses. Methods Mol Biol. 2011;721:23–41. doi: 10.1007/978-1-61779-037-9_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cardenas WB, et al. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. Journal of virology. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang HW, Watson JC, Jacobs BL. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. P Natl Acad Sci USA. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartman AL, et al. Inhibition of IRF-3 activation by VP35 is critical for the high level of virulence of ebola virus. Journal of Virology. 2008;82:2699–2704. doi: 10.1128/JVI.02344-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katze MG, He YP, Gale M. Viruses and interferon: A fight for supremacy. Nat Rev Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 36.Carter WA, De Clercq E. Viral infection and host defense. Science. 1974;186:1172–1178. doi: 10.1126/science.186.4170.1172. [DOI] [PubMed] [Google Scholar]
- 37.Miesen P, Joosten J, van Rij RP. PIWIs Go Viral: Arbovirus-Derived piRNAs in Vector Mosquitoes. Plos Pathog. 2016;12:e1006017. doi: 10.1371/journal.ppat.1006017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petit M, et al. piRNA pathway is not required for antiviral defense in Drosophila melanogaster. P Natl Acad Sci USA. 2016;113:E4218–4227. doi: 10.1073/pnas.1607952113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 40.Sarkies P, et al. Ancient and novel small RNA pathways compensate for the loss of piRNAs in multiple independent nematode lineages. PLoS biology. 2015;13:e1002061. doi: 10.1371/journal.pbio.1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baum JA, et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol. 2007;25:1322–1326. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
- 42.Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS biology. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cullen BR. Viruses and RNA interference: issues and controversies. Journal of virology. 2014;88:12934–12936. doi: 10.1128/JVI.01179-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong MT, Chen SS. Emerging roles of interferon-stimulated genes in the innate immune response to hepatitis C virus infection. Cell Mol Immunol. 2016;13:11–35. doi: 10.1038/cmi.2014.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- 47.Wynant N, Verlinden H, Breugelmans B, Simonet G, Vanden Broeck J. Tissue-dependence and sensitivity of the systemic RNA interference response in the desert locust. Schistocerca gregaria. Insect Biochem Mol Biol. 2012;42:911–917. doi: 10.1016/j.ibmb.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 48.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence information is publicly available on Genbank, flybase and wormbase. The accession numbers of all Argonaute sequences used for the construction of the phylogenetic tree our summarized in Supplementary Table S1.


