Abstract
The zebrafish has become an increasingly popular and valuable cancer model over the past few decades. While most zebrafish cancer models are generated by expressing mammalian oncogenes under tissue-specific promoters, here we describe a method that allows for the precise optical control of oncogene expression in live zebrafish. We utilize this technique to transiently or constitutively activate a typical human oncogene, kRASG12V, in zebrafish embryos and investigate the developmental and tumorigenic phenotypes. We demonstrate the spatiotemporal control of oncogene expression in live zebrafish, and characterize the different tumorigenic probabilities when kRASG12V is expressed transiently or constitutively at different developmental stages. Moreover, we show that light can be used to activate oncogene expression in selected tissues and single cells without tissue-specific promoters. Our work presents a novel approach to initiate and study cancer in zebrafish, and the high spatiotemporal resolution of this method makes it a valuable tool for studying cancer initiation from single cells.
Introduction
For the past 60 years, zebrafish have been widely used as an important vertebrate model organism. Approximately 70% of human genes have at least one zebrafish ortholog, including 84% of the genes associated with human diseases1. In addition, many other advantages such as high fecundity, rapid extra-uterine development, larval transparency, inexpensive husbandry, and scalability make zebrafish a very attractive animal model2. Zebrafish models have provided valuable insights into pathways that underlie human disease and contributed to therapeutic discovery2–4. In cancer research, zebrafish have also become a popular animal model5, 6, with multiple transgenic lines generated that faithfully recapitulate human cancers including leukemia7, melanoma8, rhabodomyosarcoma9, liver cancer10, 11, pancreatic cancer12 and others13–16. Most of these models utilize tissue-specific promoters to drive the expression of mammalian oncogenes, and they therefore have limited spatial and/or temporal resolution, even when paired with chemical inducers10, 17. Moreover, specific promoters are not yet available for certain organs and tissues, hindering investigations of tumorigenesis in those tissues.
The initiation of cancer is a rare event, taking place at the level of individual transformed cells18–20, and only a subset of these transformed cells eventually lead to tumorigenesis21. In a recent study of a melanoma zebrafish model, it was reported that among all the transformed cells expressing human B-RAFV600E, only those expressing crestin, a gene usually expressed in neural crest progenitors, were able to give rise to melanoma22. However, why and how the crestin expression was triggered in specific transformed cells has yet to be determined. Most likely, various molecular pathways are responsible for the oncogenic transformation of different cell types, and understanding these early steps will require technologies that can target these initiating transformed cells in live organisms. An ability to control cancer induction in a spatiotemporally precise manner (i.e., in single cells of different tissues at different times) would thus significantly advance our understanding of tumorigenesis. To address these challenges, we developed a novel approach that allows us to optically control genetic expression with high spatio-temporal resolution. This approach does not require tissue-specific promoters and long exposures to possibly detrimental illumination. It relies on:
-
(i)
the ERT2 modified ligand-binding domain of the estrogen receptor23 allowing nuclear translocation of fused protein such as a Cre-recombinase or a Gal4 transcriptional activator upon tamoxifen addition, and
-
(ii)
caged cyclofen, a photo-stable analog of tamoxifen which can be photoactivated by short (few seconds) UV or two-photon illumination24, 25 as shown in Fig. 1A & B.
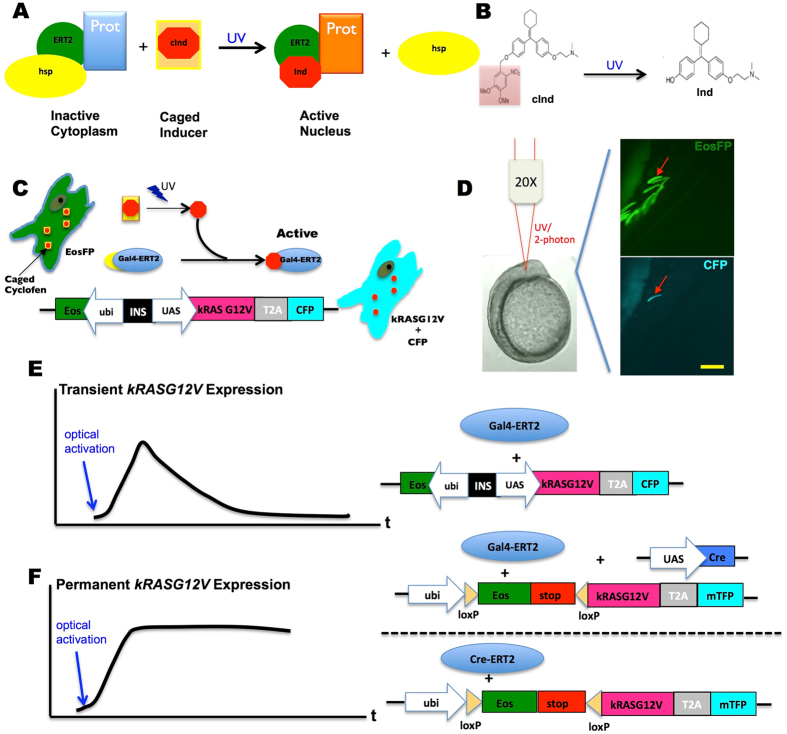
Figure 1.
Schematic principle and system design of photo-control over specific oncogene expression. (A) A protein (Prot) of interest genetically fused with the estrogen receptor (ERT2) is inactivated by the complex it forms with cytoplasmic chaperones (hsp). This complex can be dissociated by binding of a specific inducer (e.g. cyclofen) to ERT2. (B) A caged inducer (caged cyclofen, cInd) is chemically synthesized and can be uncaged by illumination at ~370 nm (~740 nm with a two-photon source). (C) The scheme describes how the expression of an oncogene can be photo-activated in a live cell transfected with dual fluorescent markers (EosFP and CFP). (D) This example shows that expression of the oncogene was induced selectively in one muscle fiber cell through localized uncaging of cInd in a developing zebrafish embryo. The injected embryo mosaically expressed EosFP, but only the light-activated cell expressed CFP (arrow denoted). (E,F) Two systems were used to control the transient or constitutive expression of kRASG12V. Scale bar: 200 μm.
We have used this approach to induce tumors in zebrafish embryos by photo-inducing the expression of human kRASG12V in vivo, either transiently (with a Gal4-ERT2/UAS system) or constitutively (with a Cre-ERT2/loxP system). We demonstrate that this optogenetic technique enables rapid kRASG12V activation in specific regions and single cells without restriction to a specific cell type, while simultaneously fluorescently labelling these oncogene-expressing cells. We emphasize that the method developed here is suitable for spatially controlled light activation of any gene of interest.
Using this approach, we find that while transient kRASG12V expression during early embryogenesis (first few hours post fertilization, hpf) disrupts tissue patterning and promotes tumorigenesis, transient kRASG12V expression at later stages (1–2 days post fertilization, dpf) does not give rise to tumors. Periodic kRASG12V expression in juvenile zebrafish can lead to tumor, albeit at a very low frequency. On the other hand, we find that constitutive kRASG12V expression increases the probability of tumorigenesis during larval development, and may result in tumor formation in the adult fish. By analysing the correlation of tumorigenesis probability and the number of kRASG12V expressing cells, we deduced that the probability of tumorigenesis due to a single transformed cell at 3dpf is about 0.5% in this specific experimental context. Our approach therefore provides experimental access to the very early stages of cancer initiation and could facilitate new strategies for cancer prevention, early diagnosis and treatment.
Results
Design of optically controlled expression of a kRASG12V oncogene in live zebrafish embryos
The main motivation of this work is to develop an optical method that allows for cancer induction in a live animal, at a high spatiotemporal resolution. Our approach involves fusing a specific protein to the modified ligand binding domain of human estrogen receptor (ERT2)23. As shown in Fig. 1A, such a fusion protein is sequestered in the cytoplasm by the complex it forms with chaperones. The fused protein can be released from this complex upon binding of a ligand (e.g. tamoxifen) to the ERT2 domain. We have shown that cyclofen24, 25, a photo-stable analog of tamoxifen, has similar affinity to the ERT2 domain, and can be chemically caged and uncaged (i.e. photo-activated) upon UV (375 nm) or two-photon (750 nm) illumination (Fig. 1B). Binding of (uncaged) cyclofen to the ERT2 domain subsequently releases the fused protein from its chaperone complex, thereby effectively turning on its activity. Here, we use this system to optically turn on or off the expression of the human oncogene, kRASG12V. We engineered a Gal4-ERT2/UAS system to photo-activate the expression of kRASG12V and a co-expressed cyan fluorescent protein marker, CFP or mTFP (Fig. 1C). Figure 1D demonstrated the transient optical activation of kRASG12V in a single cell of a transgenic mosaic embryo upon 1 minute local UV illumination at 10 hpf. To test whether transient or constitutive expression of kRASG12V could induce tumorigenesis, we created two systems to optically control its expression.
To transiently activate kRASG12V expression (Fig. 1E), we injected a ubi:Eos; UAS:kRASG12V-T2A-CFP plasmid together with Tol2 transposase mRNA into Tg(ubi:Gal4-ERT2) embryos at the one-cell stage. Introducing cyclofen or photo-activating caged-cyclofen releases the Gal4-ERT2 protein from its chaperone complex and turns on the expression of kRASG12V. Once cyclofen is removed (or diffuses out following photo-activation) the expression of the oncogene is turned off and its products (mRNAs and proteins) are slowly degraded, resulting in a transient expression of kRASG12V (Fig. S1A,C,E).
To constitutively activate the oncogene, we designed a photo-activable Cre/loxP recombination system (Fig. 1F). Either UAS:Cre and ubi:loxP-Eos-stop-loxP-kRASG12V-T2A-mTFP plasmids were co-injected into Tg(ubi:Gal4-ERT2) embryos, or ubi:loxP-Eos-stop-loxP-kRASG12V-T2A-mTFP plasmid was injected into Tg(ubi:Cre-ERT2) embryos at the one-cell stage. Adding (or photo-activating) cyclofen activates the recombinase that remodels the genome so that expression of kRASG12V is turned on constitutively (Fig. S1B,D,F).
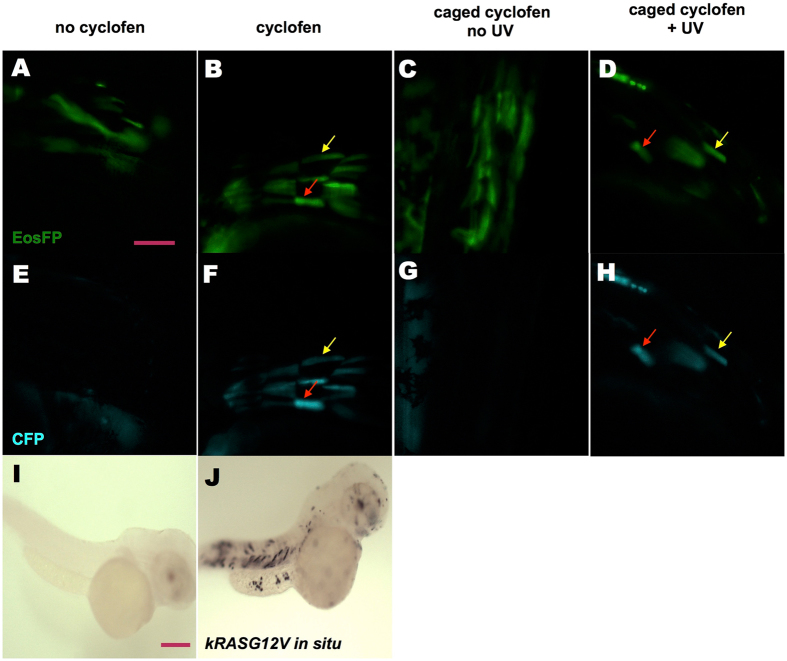
First, we wanted to confirm that the approach described above allowed for (photo-) activable expression of kRASG12V. To test this, we injected a ubi:Eos; UAS:kRASG12V-T2A-CFP plasmid into Tg(ubi:Gal4-ERT2) embryos at the one-cell stage. Healthy, normal embryos were selected at 24 hpf (hours post fertilization) and were transferred to fresh E3 medium. At 32 hpf, some injected embryos were incubated with 2 μM cyclofen, while others were left in the E3 medium as control. Twelve hours later, despite auto-fluorescence from the yolk, substantial CFP expression was only detected in the cells of embryos treated with cyclofen (Fig. 2B,F), but not in the control embryos (Fig. 2A,E). The fluorescence of CFP co-localized with the fluorescence of EosFP (Eos fluorescent protein), indicating that, as designed, they were co-expressed in the same cell (denoted by arrows). Beyond fluorescent markers, we also looked for more direct evidence of kRASG12V expression. To that purpose we used in-situ hybridization to probe for the presence of kRASG12V mRNA in embryos activated or not with cyclofen. Indeed, the oncogene mRNA could only be detected in the injected embryos treated with cyclofen (Fig. 2J), but not in the embryos without cyclofen treatment (Fig. 2I). After verifying that the expression of the oncogene could be induced by cyclofen, we tested the photo-control of transient oncogenic expression. The same injected embryos were incubated at 32 hpf in 4 μM caged cyclofen for 4 hours. They were then rinsed and transferred into fresh E3 medium. To photo-activate the caged cyclofen, the embryos were illuminated for 2 minutes with a benchtop UV lamp (~365 nm). Twelve hours later, injected embryos treated with caged cyclofen without UV illumination expressed only EosFP, but no CFP (Fig. 2C,G). Embryos that were UV illuminated displayed a clear expression of CFP that co-localized with EosFP expression (Fig. 2D,H), demonstrating the successful photo-induction of kRASG12V expression. We also validated both the transient and constitutive expression systems by analyzing the expression over-time of kRASG12V and fluorescent markers (Supplementary Information and Figs S1, S2).
Figure 2.
KRASG12V induction by incubation in cyclofen or photo-activation of caged cyclofen. (A,E) Injecting UAS:kRASG12V-T2A-CFP; ubi:Eos plasmid into Tg(ubi:Gal4-ERT2) embryos resulted in a mosaic expression of EosFP protein, but (in absence of free cyclofen) no expression of CFP. (B,F) Embryos treated with 2 μM cyclofen at 32hpf display expression of CFP co-localized with EosFP expression (denoted by arrows). (C,G) Incubation of caged cyclofen (with no UV) did not activate CFP expression while (D,H) 2 min UV uncaging effectively activated the expression of CFP 12 hours later (denoted by arrows). The induction of kRASG12V by cyclofen was confirmed by kRASG12V mRNA in situ hybridization. Human kRASG12V mRNA was transcribed only in the embryo incubated in cyclofen (J), but not in the control embryo (I). Scale bar: 200 μm.
Tumor induction by transient and constitutive expression of kRASG12V
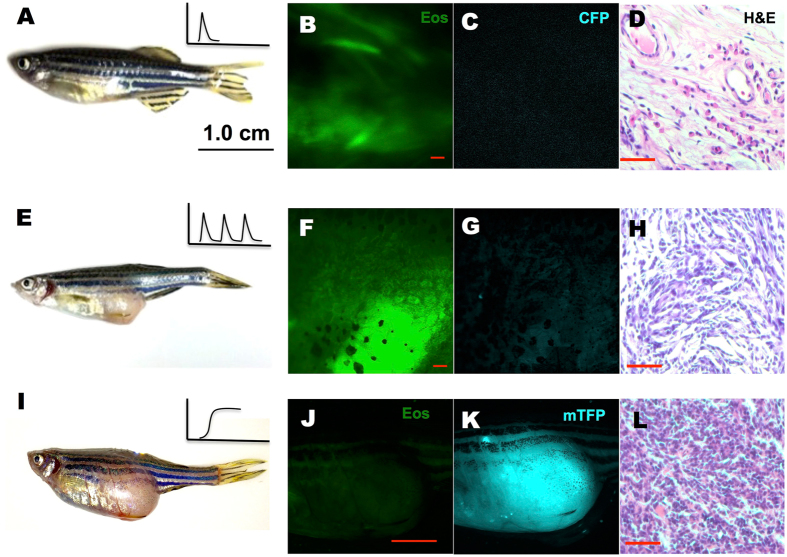
Having validated our photo-activation approaches, we tested the possibility of inducing a tumor by expressing the oncogene in zebrafish. First, a transient activation of kRASG12V was performed in 1 dpf Tg(ubi:Gal4-ERT2) embryos injected with a ubi:Eos; UAS:kRASG12V-T2A-CFP plasmid at the one-cell stage and incubated for 4 hours in cyclofen or caged cyclofen with photo-activation. Since injection procedures sometimes result in developmental defects, we only selected healthily developing embryos at 3 dpf for further observations. We observed no obvious tumorigenesis up to 10 dpf. Fish were then housed in regular fish system and monitored biweekly for gross tumor morphology. Even after one year, no tumors were detected in any of the 82 cyclofen treated fish or the 68 photo-induced fish that we monitored. Representative fish were taken out for imaging and pathological analysis. We found normally developing fish (Fig. 3A) with visible expression of EosFP (Fig. 3B), but no more CFP (Fig. 3C). Hematoxylin-Eosin (H&E) staining also showed no noticeable tumor histology (Fig. 3D). Since a pulse of kRASG12V induction at 1 dpf failed to induce a tumor, we attempted to activate kRASG12V expression at an earlier stage. Thus 2 μM cyclofen was added immediately after injection at the one-cell stage for 24 hours. Again, only healthy embryos were selected at 3 dpf, and embryos were carefully monitored for 2 weeks. We found severe developmental defects among many (23/124) cyclofen treated fish through 5–8 dpf (Fig. S3A & B). In addition, a small portion (9/124) of the fish displayed obvious tumorigenic phenotypes, with hyperplasic tissues strongly expressing CFP (and thus the oncogene) even at 6 dpf (Fig. S3C–E). All the embryos that displayed severe developmental or tumorigenic defects did not survive their first two weeks. However, all the fish that survived grew normally with no apparent tumors detected over a year. These results imply that a short pulse of kRASG12V expression before 1 dpf is not able to induce a tumor as the fish grows into adulthood. We then wondered whether prolonged activation of kRASG12V expression could make a difference. Thus we periodically activated kRASG12V by treating the fish in 2 μM cyclofen for 24 hours every five days for two months. By the end of these two months, while the fish did display CFP fluorescence (and thus co-expression of kRASG12V), we did not observe any evident tumors. The fish were subsequently left growing under normal aquatic condition. Within a period of 12 months, a few of these fish (2/64) did develop tumors. A representative tumor (Fig. 3E) near the urinary bladder was excised for H&E staining (Fig. 3H). We observed a pack of homogeneous cells with condensed nuclei and increased nuclei-cytoplasm ratio. These cells are disorganized and do not display the texture of normal tissue in the same region (Fig. 3D). These histological characteristics reproduce the typical morphology and growth patterns of tumors seen in zebrafish26. Notice however that although the histo-pathological examination revealed typical tumor morphology, the expression of CFP (and the concomitant oncogene) was no longer detected (Fig. 3F,G).
Figure 3.
Tumor induced by periodic or constitutive, but not transient activation of kRASG12V expression. In the latter system, one-time activation of kRASG12V expression was induced by transient cyclofen incubation or photo-activation of caged cyclofen at 1dpf (A). Periodic kRASG12V expression was achieved by 1 day incubation in 2 μM cyclofen every 5 days for a course of two months (E). (B–D) Transient induction of kRASG12V did not affect normal development (as shown by the histopathological staining) with no CFP expression in one-year old fish. (E–H) Tumors were observed (albeit rarely) in fish periodically treated with cyclofen. A representative one-year old fish displayed a clear tumor (E) revealed by histopathological staining (H). Interestingly, the cells in the tumor still expressed EosFP (F), but lost CFP expression (G). (I–L) Constitutive kRASG12V expression following light-activation of caged cyclofen induced tumor (I) in 5 month-old fish as revealed by both fluorescent markers (J,K) and H&E staining (L). Scale bar: B, D, F, H, L 200 μm; J, 1 cm.
In parallel to transient activation of kRASG12V, we also investigated tumor development caused by constitutive expression of the oncogene. To induce its constitutive activation, ubi:loxP-Eos-stop-loxP-kRASG12V-T2A-mTFP plasmid together with Tol2 mRNA were injected into Tg(ubi:Cre-ERT2) embryos at the one-cell stage. Healthy embryos at 1 dpf expressing EosFP were treated with 2 μM cyclofen or with 4 μM caged cyclofen for 4 hours. Those embryos were then transferred to E3 medium and photo-activation was conducted with 2-minute illumination by a UV lamp. Tumorigenic phenotypes in some embryos could be readily observed at 5 dpf (Fig. S4). Moreover, tumors were observed in adult fish (8/76) through 2 months to 1 year. High expression of mTFP was also detected in those tumors (Fig. 3I–L and Fig. S5).
Characterizing the correlation between the probability of tumorigenesis and the number of kRASG12V expressing cells
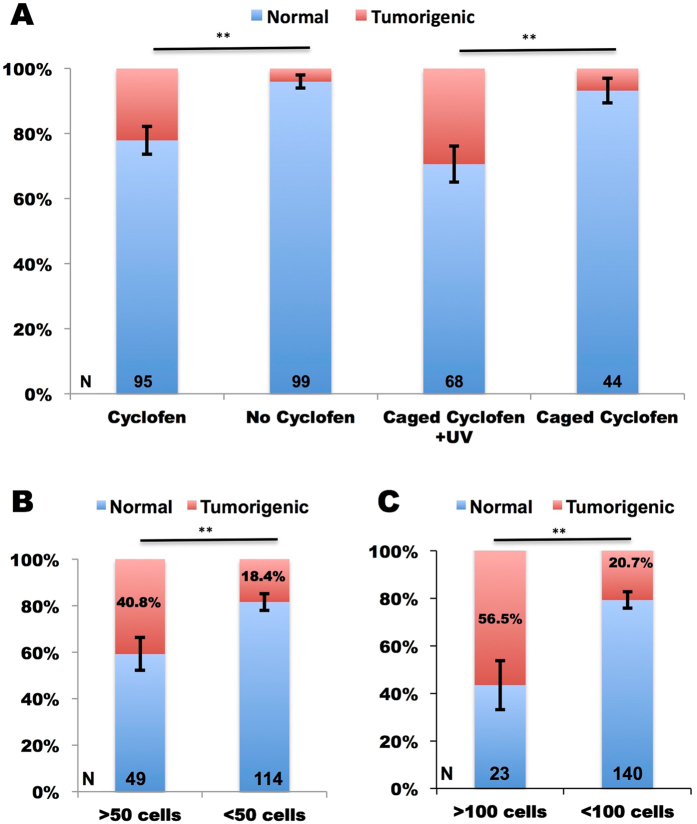
Thus far, we have seen that constitutive induction of kRASG12V expression in early developing embryos, sometimes resulted in tumor growth. We wondered why certain fish, but not others developed a tumor, even while the oncogene was turned on constitutively in all the embryos. Our initial hypothesis was to link tumorigenic probability to the number of cells in which kRASG12V expression was activated. To test the hypothesis, we only focused on constitutive activation of kRASG12V since transient activation was much less efficient in causing tumorigenesis. In this experiment, we injected ubi:loxP-Eos-stop-loxP-kRASG12V-T2A-mTFP plasmid together with the tol2 mRNA into Tg(ubi:Cre-ERT2) embryos at the one-cell stage. This resulted in mosaic embryos expressing the Eos protein in some of their cells. Constitutive expression of kRASG12V (and the concomitant expression of mTFP) was activated at 1 dpf by either cyclofen or photo-activation of caged cyclofen. Only healthily developing embryos at 3 dpf were selected for continuing analysis, and tumorigenesis probability was measured till 10 dpf. Here, tumorigenesis was defined when obvious hyperplasic tissues were observed. First we noticed that activation of kRASG12V, either by native cyclofen or by photo-activation of caged cyclofen, resulted in significant increase of tumorigenesis (Fig. 4A). Then following cyclofen activation we split the embryos into four subgroups based on the number of mTFP-expressing cells they had at 3 dpf: two sub-groups with more (or less) than 50 cells and two sub-groups with more (or less) than 100 cells. We found that the groups with more mTFP-expressing cells at 3 dpf exhibited a much higher tumorigenic rate than the groups with less mTFP-expressing cells (Fig. 4B,C). Besides the observation of a high correlation between the number of cells expressing kRASG12V and early tumorigenesis probability, we used these data to estimate the probability of tumorigenesis for a single transformed cell, assuming every cell could equally contribute to tumorigenesis. Thus for the 50-cells groups, the upper bound on the probability can be estimated as (see Fig. 4B): = 40.8/50 × 100% = 0.82%, and the lower bound as = 18.4/50 × 100% = 0.37%. Similarly, based on the 100-cell groups, we estimated the upper and lower bound probability as 0.56% and 0.21%. Thus, we deduce that the probability of tumorigenesis from a single cell is about 0.5%.
Figure 4.
Statistics of early tumorigenesis in embryos with constitutive activation of kRasG12V. Tg(ubi:Cre-ERT2) embryos injected with ubi:loxP-Eos-stop-loxP-kRASG12V-T2A-mTFP plasmid were split into cyclofen, no cyclofen, caged cyclofen + UV and caged cyclofen groups. Tumorigenic probabilities were measured for each group from 3 dpf to 10 dpf (A). For the cyclofen group, embryos were separated into four sub-groups based on the number of cells expressing mTFP at 3dpf (more or less than 50 (B) and more or less than 100 cells (C)), and tumorigenic probabilities were assessed for each sub-group (B,C). Statistical significance: **p < 0.01.
Precise optical activation of kRASG12V expression in live zebrafish embryos
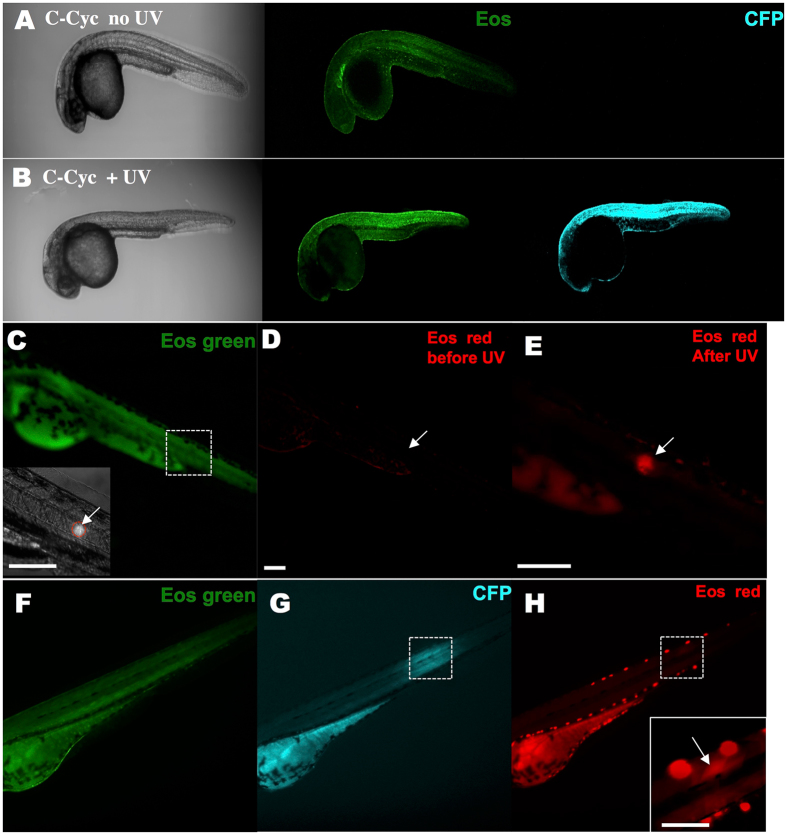
The fundamental challenge in understanding cancer initiation and evolution lies in the difficulties identifying and visualizing the very early tumorigenic event in single cells. We therefore investigated whether our method could precisely enable spatio-temporal targeting of kRASG12V activation. In this case, a stable Tg(ubi:Eos;UAS:kRASG12V-T2A-CFP) fish line was generated and crossed with the Tg(ubi:Gal4-ERT2) line. Again, we showed that caged cyclofen only was not able to activate CFP expression at 1 dpf (Fig. 5A), but that 2 minutes global UV illumination could effectively turned on CFP expression (Fig. 5B). Then we tested local light activation of transient kRASG12V expression by photoactivation of caged cyclofen. We generated a focalized beam of UV light (a circular region about 40 μm in diameter; peak around 396 nm) within the 1dpf embryo (Fig. 5C). The photo-switchable EosFP protein27 was efficiently converted from green to red after 1 min UV illumination (Fig. 5D,E indicated by arrows). Eighteen hours later, we observed local expression of CFP in the illuminated region implying local activation of kRASG12V expression (Fig. 5F,G). While the expression of the photo-activated EosFP red decreased over time, it remained correlated with the expression of CFP (Fig. 5H), indicating a successful photo-activation of kRASG12V expression in the illuminated region only. Furthermore, we also tested the activation of kRASG12V expression at different developmental stages by a UV laser illumination. We illuminated a small square region (40 × 40 μm) for 5-second with the 405-nm laser line. Embryos were thus photo-activated at 10 hpf (Fig. S6A) or 32 hpf (Fig. S6B,C) within different tissues. Clear local expression of CFP indicated precise light activation of oncogene expression. In agreement with our previous experiments with transient expression of kRASG12V, we found that local transient activation of the oncogene led to normal development, and all embryos developed into adults without any tumorigenic phenotypes. In contrast, global photo-activation of kRASG12V expression at 10 hpf resulted in developmental defects at 5dpf (Fig. S6D) in many embryos (24/87), and hence their death within 10 days.
Figure 5.
KRASG12V activation and tracking in a small group of cells within a live zebrafish embryo. A stable Tg(ubi:Eos;UAS:kRASG12V-T2A-CFP) line was generated and crossed with the Tg(ubi:Gal4-ERT2) line. Caged cyclofen alone at 1 dpf was not able to induce CFP expression (A), while 2-minute UV illumination (B) effectively turned on CFP after 18 hours. (C) Local activation of kRASG12V expression was achieved by introducing a focalized UV beam onto embryos pre-incubated with caged cyclofen. (D,E) 1 minute UV effectively converted EosFP from green to red. (F,G) After 18 hours, CFP expressed in a localized region of the illuminated embryo and weak remaining EosFP red signal was detected in the same region (H). Scale bar: 200 μm.
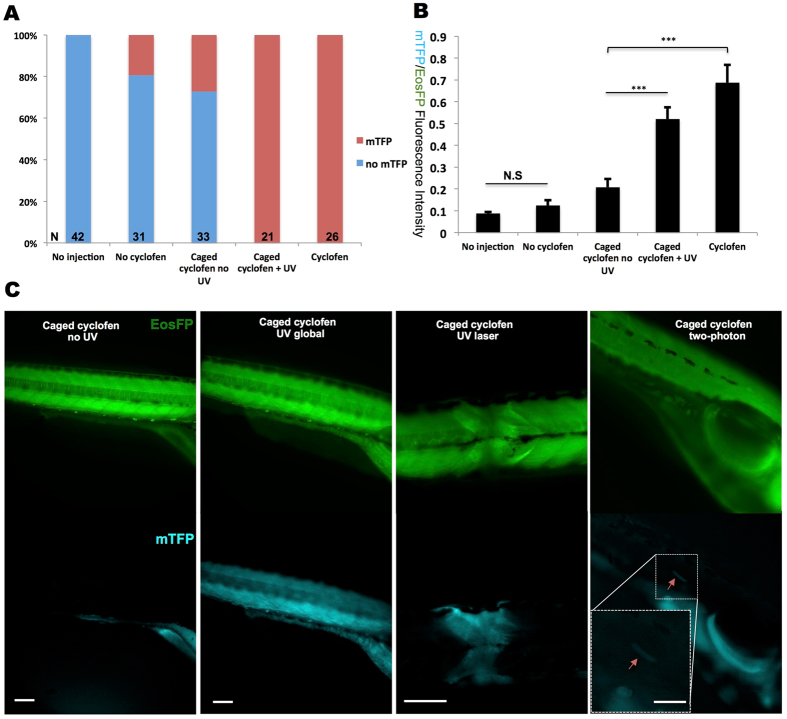
To allow precise constitutive activation of kRASG12V expression by light, we also generated a stable Tg(ubi:loxP-Eos-stop-loxP-kRASG12V-T2A-mTFP) line. We first crossed this line with the Tg(ubi:Cre-ERT2) line, but found that mTFP expression could be hardly detected after cyclofen activation (Fig. S7A). Since we also observed a low EosFP expression, we suspected that crossing these two lines-resulting in a double heterozygous line- would decrease the expression of both Cre-ERT2 and fluorescent proteins. To circumvent this issue, we directly injected about 20 pg Cre-ERT2 mRNA into the Tg(ubi:loxP-Eos-stop-loxP-kRASG12V-T2A-mTFP) embryos at the one-cell stage. Although we found about 20–30% of injected embryos showed leakage of mTFP expression without photo-activation (probably due to too large a dose of injected Cre-ERT2 mRNA), most of the injected embryos expressed mTFP only after cyclofen activation or caged cyclofen photo-activation (Fig. 6A). Distinct fluorescent patterns of non-activated, leaky and globally-activated embryos are shown in Fig. S7B. We also measured the mTFP/Eos fluorescent ratio to quantify the signals. The non-injected group set the auto-fluorescence baseline. Despite minor leakage, the mTFP/Eos ratio could be significantly increased by both cyclofen and photo-activation of caged cyclofen (Fig. 6B). Local activation was performed with 405 nm laser at 10 hpf using the Leica SP5-Blue microscope as before. We found that a 5-second laser illumination locally activated mTFP expression with a notable decrease of EosFP fluorescence in the same region, confirming recombination by Cre and activation of the oncogene (Fig. 6C).
Figure 6.
Constitutive photo-activation of kRASG12V expression in localized tissue and single cells. To induce constitutive activation of kRASG12V, Cre-ERT2 mRNA was injected into Tg(ubi:loxP-Eos-stop-loxP-kRASG12V-T2A-mTFP) embryos at the one-cell stage. (A) Despite some minor light independent activation of Cre, mTFP expression could be fully induced by both cyclofen and caged cyclofen photo-activation. (B) Quantification of kRASG12V activation signal via the ratio of the fluorescence of the co-expressed mTFP protein and the (partially) floxed Eos protein. (C) UV (lamp or laser) and two-photon activation were performed at 10hpf to achieve global (with the UV lamp), local (with the UV laser) and single cell (with 2-photon laser) activation. Fish were imaged at 2dpf. Scale bar: 200 μm. Data are presented as mean ± SEM. ***p < 0.001; N.S, not significant.
To achieve local (possibly single cell) control of gene expression with this approach, a possible concern is the diffusion of the cyclofen molecule upon photo-activation for a prolonged period. Indeed the diffusion constant of cyclofen is about 400 μm2/sec.28. Illumination with a weak UV source for 5 sec or 1 min (as mentioned earlier) results in cyclofen uncaging over a region of typical radius ~50 μm or ~160 μm respectively. Such diffusion can make the light activation of kRASG12V expression in a single cell challenging. One solution is the use of a stronger localized UV illumination (e.g. a focused UV laser beam) over a shorter time (<1 sec). Another possibility is to use two-photon uncaging, which has been shown to result in the release of cyclofen in a much more confined region, down to a single cell25. To test that later option, we performed two-photon activation to reach single cell resolution. We injected Cre-ERT2 mRNA into Tg(ubi:loxP-Eos-stop-loxP-kRASG12V-T2A-mTFP) embryos at the one-cell stage. The embryos were subsequently photo-activated at 10 hpf for 10 seconds with the two-photon (750 nm) beam of the microscope. At 2 dpf, weak mTFP expression in single cells could be detected (Fig. 6C noted by arrow). For the 19 embryos tested for two-photon activation, we found that 6 displayed single cell mTFP expression, 3 showed leaky expression (i.e. mTFP expression in non-activated cells) and 10 had no any mTFP expression (Fig. S7C). While two-photon activation led to higher mTFP/Eos fluorescence ratio than for the non-activated group, the mTFP/EOS ratio was still significantly lower than by direct cyclofen activation (Fig. S7D).
To test whether global and local activation would induce tumor development, those activated fish that developed normally were kept growing to adulthood. However, although mTFP was still expressed in those fish, no tumor was observed within one year. We found that the expression of kRASG12V in these fish was significantly lower at 5dpf and 5mpf than in those that were injected with the equivalent plasmid (Fig. S8A), as estimated from both the mTFP raw intensity (Fig. S8B) and the mTFP/Eos fluorescence ratio (Fig. S8C).
Discussion
A variety of approaches to control protein activity by light have been developed for over a decade29. These methods have greatly accelerated our understanding of biological systems. However, as far as in vivo applications in live organisms are concerned, these tools have been used mainly in neuroscience30. Most optogenetic approaches adopt light-sensitive proteins that usually have some activity even without light29 and may require long (hours) illumination times31–33. This critical drawback makes it rather difficult to precisely photo-manipulate biological processes such as cancer induction. In this work, we demonstrate a powerful new optical tool to manipulate tumor induction in live zebrafish. Our approach utilizes caged-cyclofen, a synthetically modified estrogen receptor inducer, which allows for stringent light-dependent activation of proteins fused to the binding domain of the modified estrogen receptor (ERT2). In absence of ligand these protein constructs are sequestered by cytoplasmic chaperones. Uncaging of the ligand (cyclofen) releases the fused proteins from their chaperone complex, allowing them to diffuse into the nucleus and control the expression of the desired genes.
Our experiments highlight two inducible systems for the transient or constitutive expression of kRASG12V. We validated these two systems by micro-injection of the appropriate gene construct at the one-cell stage, obtaining mosaic embryos expressing the oncogene in some of their cells. We find that a short pulse (less than 24 hours) of kRASG12V expression at 1 dpf in those embryos fails to induce tumor formation. This is not so surprising since cancer results from a prolonged process of cumulative mutations34, 35. The decrease of CFP fluorescence within a few days confirms the loss of kRASG12V expression and activity. Therefore a time-limited activation of kRASG12V might not be enough to drive further oncogenic mutations, hence no tumor. Notice however that an early (<1dpf) short pulse of kRASG12V results in severe developmental defects and embryonic lethality. These phenotypes are probably the result of the teratogenic effects of over-expression of kRASG12V, which is known to play a critical role during early embryogenesis. Notice that embryonic defects caused by kRASG12V over-expression have been previously reported in both mouse36 and zebrafish37.
In contrast, constitutive activation of kRASG12V at >1dpf, does not result in noticeable developmental defects but increases early tumorigenic rates and causes tumor in adult fish. Surprisingly, we find that periodic activation of kRASG12V for 2 months (24 hours every 5 days) does not lead to tumor formation by the end of the treatment. However, a few fish (2/64) do develop tumors (albeit without CFP and hence kRas expression) after one year. This result suggests that periodic, but not necessarily constitutive activation of the oncogene could also result in tumorigenesis. The precise mechanism that underlies this phenomenon needs further elucidation.
On the other hand, permanent activation of kRASG12V from 1dpf in mosaic embryos causes early tumorigenesis with a probability that correlates with the number of cells expressing the oncogene. This is expected because the more numerous the cells with oncogenic activation the greater the probability of passenger mutations and tumor growth. This result is also in agreement with previous finding that reported significant liver hyperplasia and tumor after continuous induction of kRASG12V in the zebrafish liver11. By examining the number of cells expressing mTFP at 3dpf and analyzing the probability of tumorigenesis at 10dpf, we estimated that the probability of tumorigenesis due to a single transformed cell is as low as 0.5%. The analysis assumes that in this specific case every transformed cell can equally contribute to early tumorigenesis. However since the transgenic line did not develop tumors following activation of the oncogene, we deduce that the aforementioned probability may depend on the strength of oncogene expression, context, cell type, etc. This important issue deserves further study.
In the transgenic lines we engineered, our approach has shown precise control of kRASG12V induction in different regions and single cells of developing zebrafish embryos. This level of control has not been achieved by any other inducible cancer models reported so far. However, we find that the constitutive photo-activation of kRASG12V in this transgenic line is not efficient to induce a tumor. The lower expression level of mTFP in the photo-activated transgenic fish as compared to the injected fish (with mosaic expression of the oncogene) points to a lower expression of the oncogene. This fact may explain why these fish, even with active oncogenic expression, fail to develop tumors within one year. Our observations suggest that not only the number of oncogene expressing cells, but also the level of expression of the oncogene may be important in contributing to tumor induction. Engineering a similar line albeit with a stronger promotor should allow us to investigate that important issue.
In summary, our study presents the first light-inducible tumor model in a vertebrate. We have demonstrated the success of light activation of the human kRASG12V oncogene in the zebrafish and have observed the multiple tumors formed over a one-year period. Our technique should be helpful in evaluating how different organs and tissues respond to the same oncogenic induction. More importantly, successful demonstration of activating and labeling kRASG12V expression in a small number of cells by light makes this technique powerful in the study of the early initiation and evolution of cancer. This novel light-inducible model should help investigate and understand cancer initiation, and ultimately lead to the development of novel and effective barriers to, and treatments for cancer.
Materials and Methods
Plasmid construction and in vitro transcription
A 564 bp DNA sequence of human kRASG12V was slightly modified to optimize its expression in zebrafish (see below for a full sequence). Cre, kRASG12V and loxP-Eos-stop-loxP-kRASG12V-T2A-mTFP sequences were ordered from Eurogentec and were inserted in pUC57 vector. The ubi:Eos; UAS:kRASG12V-T2A-CFP plasmid was cloned by inserting the kRASG12V sequence into a UAS:fgf8a-T2A-CFP; ubi:Eos plasmid (unpublished; construct sequence is available upon request), using the KpnI and FspAI restriction sites to replace the fgf8a. Similarly, we made the UAS:Cre;ubi:Eos plasmid by replacing fgf8a-T2A-CFP with Cre. Based on the UAS:fgf8a-T2A-CFP; ubi:Eos plasmid, ubi:loxP-Eos-stop-loxP-kRASG12V-T2A-mTFP plasmid was cloned by first replacing Eos with loxP-Eos-stop-loxP-kRASG12V-T2A-mTFP sequence, using BamHI and EcoRV sites, and then by deleting the fgf8a-T2A-CFP sequence, using KpnI and FspA1 sites, followed by quick blunting and ligation. A ubi:Gal4-ERT2 plasmid was derived from the ubi:Cre-ERT2 plasmid24 by substituting Cre sequence with Gal4-FF between NcoI and SmaI. All nucleotide sequences and details of the cloning steps are available on request. To make Tol2 transposase mRNA, a pCS-TP Tol2 plasmid was first linearized with NotI and was used to synthesize mRNA using a mMESSAGE mMACHINE® SP6 Transcription Kit (Thermo Fisher Scientific). Similar to Tol2 mRNA, Cre-ERT2 mRNA was made with another pCSCre-ERT2 plasmid.
Fish lines and maintenance
With the plasmids we constructed, we generated three stable fish lines via the Tol2 transposon system38: Tg(ubi:Gal4-ERT2), Tg(ubi:Eos;UAS:kRASG12V-T2A-CFP) and Tg(ubi:loxP-Eos-stop-loxP-kRASG12V-T2A-mTFP). The transgenic line Tg(ubi:Cre-ERT2) 39 was kindly provided by Dr. D. Traver. All the fish lines were raised at UCLA Zebrafish Core Facility. The facility also provided regular maintenance of all zebrafish lines we generated. The overall health of the animals in the fish-facility is under the supervision of UCLA veterinary advice. Animal care and all experimental procedures were performed in accordance with the protocol approved by UCLA Institutional Animal Care and Use Committee (ARC# 2001–074–33).
Drug treatments and UV uncaging
The Jullien lab synthesized native cyclofen and caged cyclofen in powder form. For long-term storage, both native and caged cyclofen were kept at −80 °C. Stock of 2 mM cyclofen and 4 mM caged cyclofen in DMSO were prepared and also kept at −80 °C in darkness. To induce kRASG12V expression, embryos were either directly incubated in 1, 2, 10 μM native cyclofen for 2–24 hours, or incubated in 4, 10 μM caged cyclofen for 2–24 hours and photo-activated by UV before being transferred back to E3 medium. Periodic induction of kRASG12V expression was carried out by treating Tg(ubi:Gal4-ERT2) embryos injected with UAS:kRASG12V-T2A-CFP;ubi:Eos plasmid with 2 μm cyclofen for 24 hours every five days until two-month post fertilization. For whole embryo uncaging, we used a benchtop UV lamp (Fisher VL-6-L) which emits a peak wavelength at 365 nm with a FWHM (full width at half maximum) of 40 nm, and delivers on the illuminated sample a typical photon flux of ~4.3 Einstein/(s·m2)40. Injected embryos pre-treated with caged cyclofen were briefly rinsed in E3 medium before they were illuminated by the benchtop UV lamp for 2 minutes in a 60 mm × 15 mm petri dish. Illuminated fish were transferred back to E3 medium, covered with aluminum foil and were incubated at 28 °C. Localized uncaging was achieved on a Nikon microscope equipped with an adjustable iris and a lumencor LED light source with a UV light peaking at 396 nm. The maximum relative spectral power of the UV light was 27 mW/nm. The size of the uncaging region was controlled by an iris that defines a circular illumination area ranging from 50 μm to 400 μm in diameter. Uncaging was performed for 20 seconds to 2 minutes at the maximum power of the UV LED light on each embryo. After uncaging, embryos were transferred to a 12-well plate with E3 medium and incubated in total darkness. Laser uncaging was conducted with the 405 nm laser equipped on a Leica SP5-BLUE confocal microscope (UCLA CNSI core facility). Two-photon activation was performed using a Ti-Sapphire laser25 with a 40×, 0.8 NA water immersion objective. The laser beam (200 fs, 76 MHz, 20 mW, 750 nm) was used to photo-activate caged-cyclofen for 10 s in 10hpf embryos.
RT-qPCR and in-situ hybridization
Expression of kRASG12V mRNA was profiled on embryos from one-day to two-weeks post fertilization. At each time point, five to seven injected embryos were collected for mRNA extraction. Total extracted mRNA was purified as previously described41, and stored at −80 °C. SuperScript® III First-Strand Synthesis System (Invitrogen) was used to synthesize and purify cDNA from the frozen samples of mRNA. Fast SYBR Green Master Mix (Thermo Fisher Scientific) was used for qPCR. Primer sets used for Eos and kRASG12V cDNA detection were: Eos forward (GCAACAAAGCCATGTGAATATGA), Eos reverse (CAAACTTTCCCG CCAATGGTCCA), kRASG12V forward (AACCAATGTATAGAAGGCATCAT), kRASG12V reverse (TAAATGTGATTTGCCTTCAAGAA). To account for priming differences, titrations of the ubi:Eos; UAS:kRASG12V-T2A-CFP and ubi:loxP-Eos-stop-loxP-kRASG12V-T2A-mTFP plasmids were used as reference templates. The difference (ΔCt) in the number of amplification cycles between kRASG12V and Eos (used as a reference gene) was measured and normalized to reference templates in triplicate PCR assays. The ratio of the expression of kRASG12V to Eos was quantified as: 2−ΔCt. Whole-mount in situ hybridization with digoxigenin-labeled riboprobes was performed at 2 dpf as described previously42. Riboprobes for kRASG12V mRNA were synthesized from the UAS:kRASG12V-T2A-CFP plasmid.
Microinjection and microscopic imaging
Exogenous genes were incorporated into embryo genomes at the one-cell stage using the Tol2 transposon system43. DNA constructs (final concentration 25 ng/μl) were mixed with Tol2 mRNA (final concentration 100 ng/μl), and about 200 picoliters mixed solutions were micro-injected into embryos at the one-cell stage, following protocols described earlier44. Alternatively, about 200 picoliters Cre-ERT2 mRNA (final concentration 50 ng/μl) was injected into the Tg(ubi:loxP-Eos-stop-loxP-kRASG12V-T2A-mTFP) embryos at the one-cell stage. After injection, embryos were incubated in E3 medium at 28 °C. Healthy, normal embryos were selected at 24 hpf and 3 dpf for further observation and analysis. Fluorescent imaging was performed on a Nikon Eclipse Ti microscope equipped with a C-HGFI Intensilight Fiber illuminator, and confocal imaging was acquired on a Leica SP5-STED microscope. Embryos and adult zebrafish were anesthetized first in tricaine (Sigma-Aldrich), and aligned in a small liquid drop on a cover slide for imaging. The fluorescence intensity of individual images was quantified with ImageJ64 after background subtraction. In situ hybridization and H&E samples were imaged on a Leica LMD (Laser Micro Dissection) 7000 Microscope.
Histological analysis
Ten days post fertilization embryos and adult zebrafish were first anesthetized with tricaine. Selected tissues bearing tumor or normal tissues from control were dissected. The embryos and dissected tissues were fixed in 4% paraformaldehyde overnight at 4 °C prior to rinsing in 70% ethanol. Fixation and decalcification of adult fish were performed as previously reported45. Fish samples were then sent to UCLA Translational Pathology Core Laboratory (TPCL) for sectioning and H&E staining. Stained slides were imaged on a Leica LMD (Laser Micro Dissection) 7000 Microscope.
Statistical analyses
RT-qPCR data were collected from three independent experiments and presented as mean ± standard error of mean (SEM). Statistical significance was analyzed using Fisher’s exact test in Fig. 4, and Student’s two-tailed t-test or one-way ANOVA in other figures as appropriate. P < 0.05 was indicated as *; P < 0.01 was indicated as **; P < 0.001 was indicated as *** and N.S. means “not significant”.
Electronic supplementary material
Acknowledgements
We thank Dr. David Traver and Dr. Christian Mosiman for the Tg(ubi:Cre-ERT2) line; Dr. Thomas Le Saux, Dr. Xavier Michalet for providing technical assistance; Dr. Rui Li for tumor histological analysis; the California NanoSystems Institute (CNSI) Advanced Light Microscopy/Spectroscopy Shared Resource Facility at UCLA for microscopy support and Dr. James K. Chen for suggestions and comments on the manuscript. This work was supported by CNRS under a LIA between CNRS and UCLA and by the Partner University Fund a program of the French American Culture Exchange. D.B. and L.J. acknowledge the financial support of the IReSP within the Plan Cancer (grant PhotoCancer) and the Structuration de la Recherche PSL 2015 (grant Microgut). D.B., S.V. and M.V. acknowledge support under the program “Investissements d’Avenir” launched by the French Government and implemented by the ANR with the references: ANR-10-LABX-54 MEMO LIFE and ANR-11-IDEX-0001–02 PSL* Research University. L.J. acknowledges ANR FranceBioImaging for financial support. Z.F. was supported by Dean Willard Chair fund, CSC scholarship and UCLA Dissertation Year fellowship.
Author Contributions
Z.F., D.B., S.W. designed the experiments. Z.F., S.N., F.H. and B.D. conducted the experiments. I.A. synthesized cyclofen and caged cyclofen. Z.F. and D.B. analyzed and interpreted the data. S.V., M.V., L.J. and S.L. advised on experiments and data interpretation. Z.F. and D.B. wrote the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-09697-x
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhiping Feng, Email: zpfeng@stanford.edu.
Shimon Weiss, Email: sweiss@chem.ucla.edu.
David Bensimon, Email: david@lps.ens.fr.
References
- 1.Howe K, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santoriello C, Zon LI. Hooked! Modeling human disease in zebrafish. J Clin Invest. 2012;122:2337–2343. doi: 10.1172/JCI60434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dooley K, Zon LI. Zebrafish: a model system for the study of human disease. Curr Opin Genet Dev. 2000;10:252–256. doi: 10.1016/S0959-437X(00)00074-5. [DOI] [PubMed] [Google Scholar]
- 4.Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 5.Langenau, D. M. Cancer and Zebrafish. Mechanisms, Techniques, and Models. Vol. 36 (Springer, 2016). [PubMed]
- 6.Stanton MF. Diethylnitrosamine-Induced Hepatic Degeneration and Neoplasia in the Aquarium Fish, Brachydanio Rerio. J Natl Cancer Inst. 1965;34:117–130. doi: 10.1093/jnci/34.1.117. [DOI] [PubMed] [Google Scholar]
- 7.Langenau DM, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299:887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- 8.Patton EE, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15:249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 9.Langenau DM, et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007;21:1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen AT, et al. An inducible kras(V12) transgenic zebrafish model for liver tumorigenesis and chemical drug screening. Disease models & mechanisms. 2012;5:63–72. doi: 10.1242/dmm.008367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, et al. Inducible and repressable oncogene-addicted hepatocellular carcinoma in Tet-on xmrk transgenic zebrafish. Journal of hepatology. 2012;56:419–425. doi: 10.1016/j.jhep.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 12.Liu S, Leach SD. Screening pancreatic oncogenes in zebrafish using the Gal4/UAS system. Methods in cell biology. 2011;105:367–381. doi: 10.1016/B978-0-12-381320-6.00015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu S, et al. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell. 2012;21:362–373. doi: 10.1016/j.ccr.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berghmans S, et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci USA. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leacock SW, et al. A zebrafish transgenic model of Ewing’s sarcoma reveals conserved mediators of EWS-FLI1 tumorigenesis. Disease models & mechanisms. 2012;5:95–106. doi: 10.1242/dmm.007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill JA, et al. Enforced expression of Simian virus 40 large T-antigen leads to testicular germ cell tumors in zebrafish. Zebrafish. 2010;7:333–341. doi: 10.1089/zeb.2010.0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le, X. et al. Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish. Proceedings of the National Academy of Sciences of the United States of America104, 9410-9415, doi:0611302104 [pii]10.1073/pnas.0611302104 (2007). [DOI] [PMC free article] [PubMed]
- 18.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 19.Crespi B, Summers K. Evolutionary biology of cancer. Trends in ecology & evolution. 2005;20:545–552. doi: 10.1016/j.tree.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 21.Bissell MJ, Hines WC. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufman CK, et al. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science. 2016;351 doi: 10.1126/science.aad2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feil R, et al. Ligand-activated site-specific recombination in mice. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinha DK, et al. Photoactivation of the CreER T2 recombinase for conditional site-specific recombination with high spatiotemporal resolution. Zebrafish. 2010;7:199–204. doi: 10.1089/zeb.2009.0632. [DOI] [PubMed] [Google Scholar]
- 25.Sinha DK, et al. Photocontrol of protein activity in cultured cells and zebrafish with one- and two-photon illumination. Chembiochem: a European journal of chemical biology. 2010;11:653–663. doi: 10.1002/cbic.201000008. [DOI] [PubMed] [Google Scholar]
- 26.Yan C, et al. Chemical inhibition reveals differential requirements of signaling pathways in krasV12- and Myc-induced liver tumors in transgenic zebrafish. Sci Rep. 2017;7 doi: 10.1038/srep45796. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Culbertson CT, Jacobson SC, Michael Ramsey J. Diffusion coefficient measurements in microfluidic devices. Talanta. 2002;56:365–373. doi: 10.1016/S0039-9140(01)00602-6. [DOI] [PubMed] [Google Scholar]
- 29.Gautier A, et al. How to control proteins with light in living systems. Nature chemical biology. 2014;10:533–541. doi: 10.1038/nchembio.1534. [DOI] [PubMed] [Google Scholar]
- 30.Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu Rev Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 32.Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nature chemical biology. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomasetti C, Marchionni L, Nowak MA, Parmigiani G, Vogelstein B. Only three driver gene mutations are required for the development of lung and colorectal cancers. P Natl Acad Sci USA. 2015;112:118–123. doi: 10.1073/pnas.1421839112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hornsby C, Page KM, Tomlinson IP. What can we learn from the population incidence of cancer? Armitage and Doll revisited. The Lancet. Oncology. 2007;8:1030–1038. doi: 10.1016/S1470-2045(07)70343-1. [DOI] [PubMed] [Google Scholar]
- 36.Tuveson DA, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/S1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 37.Ju B, et al. Oncogenic KRAS promotes malignant brain tumors in zebrafish. Molecular cancer. 2015;14 doi: 10.1186/s12943-015-0288-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosimann C, et al. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development. 2011;138:169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu L, et al. Spatiotemporal manipulation of retinoic acid activity in zebrafish hindbrain development via photo-isomerization. Development. 2012;139:3355–3362. doi: 10.1242/dev.077776. [DOI] [PubMed] [Google Scholar]
- 41.Peterson SM, Freeman JL. RNA isolation from embryonic zebrafish and cDNA synthesis for gene expression analysis. Journal of visualized experiments: JoVE. 2009 doi: 10.3791/1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prince VE, Moens CB, Kimmel CB, Ho RK. Zebrafish hox genes: expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development. 1998;125:393–406. doi: 10.1242/dev.125.3.393. [DOI] [PubMed] [Google Scholar]
- 43.Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome biology. 2007;8(Suppl 1) doi: 10.1186/gb-2007-8-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosen JN, Sweeney MF, Mably JD. Microinjection of zebrafish embryos to analyze gene function. Journal of visualized experiments: JoVE. 2009 doi: 10.3791/1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore JL, Aros M, Steudel KG, Cheng KC. Fixation and decalcification of adult zebrafish for histological, immunocytochemical, and genotypic analysis. BioTechniques. 2002;32:296–298. doi: 10.2144/02322st03. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.