Abstract
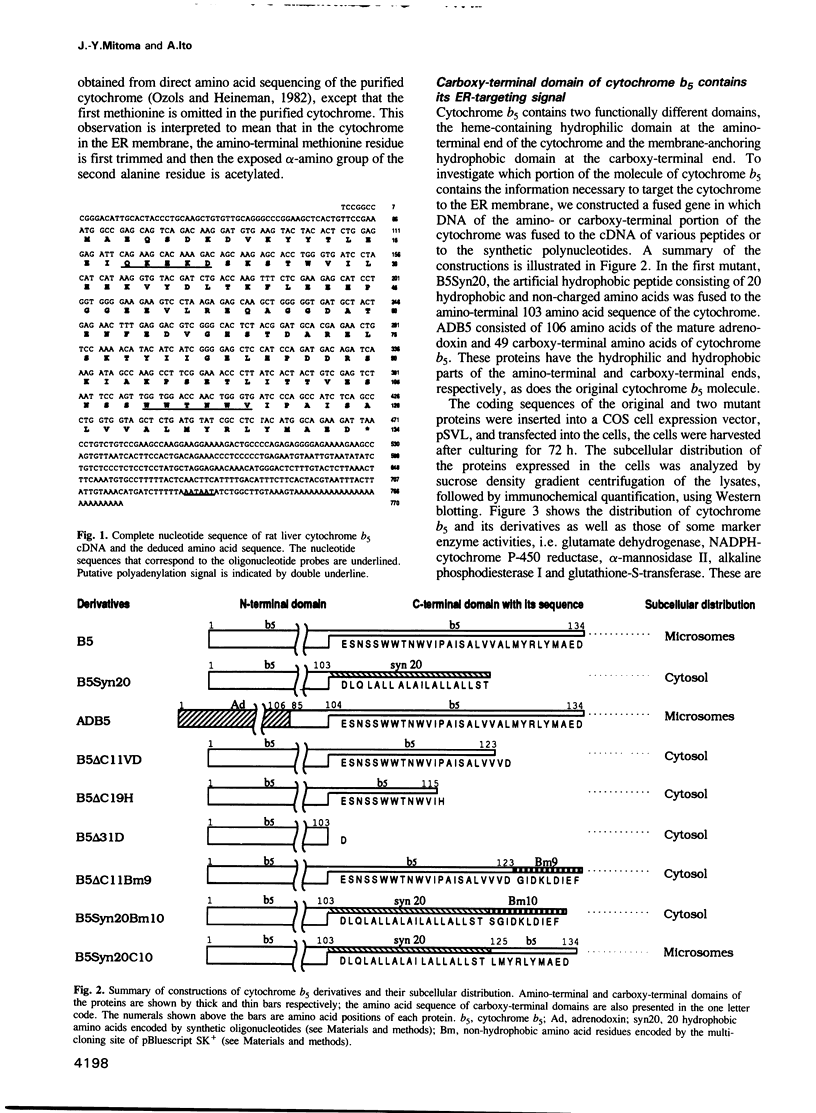
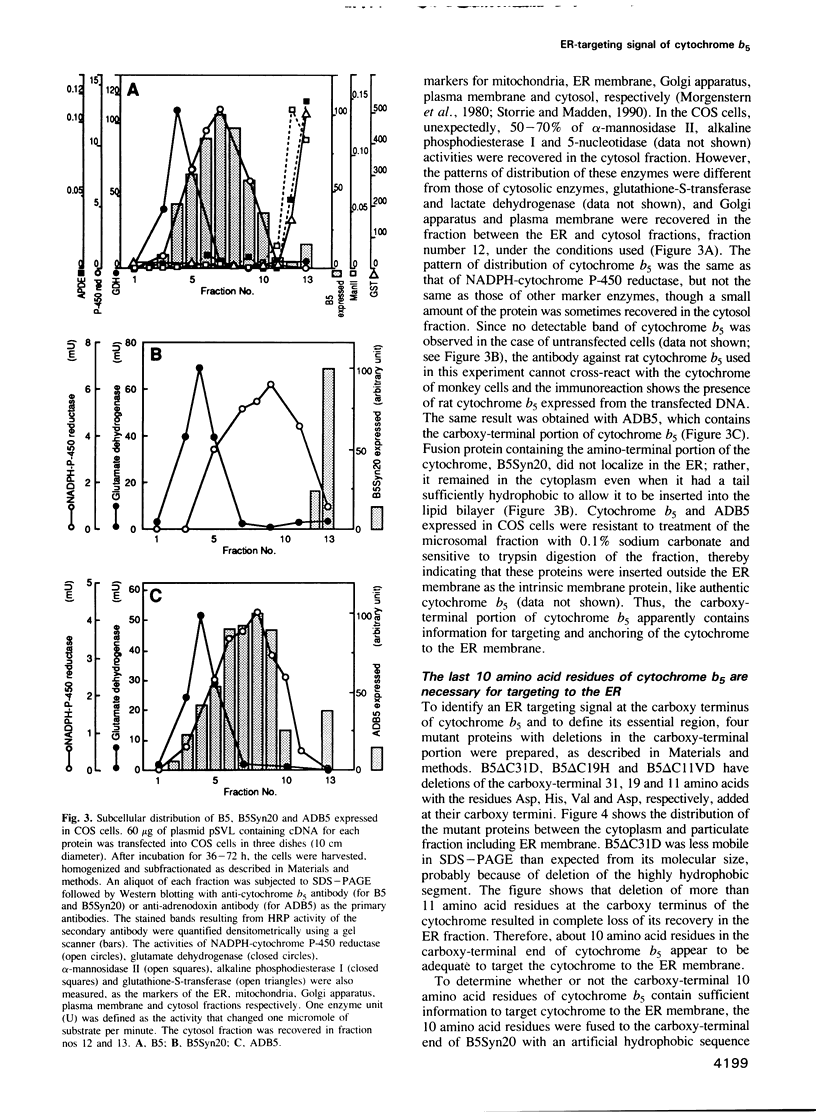
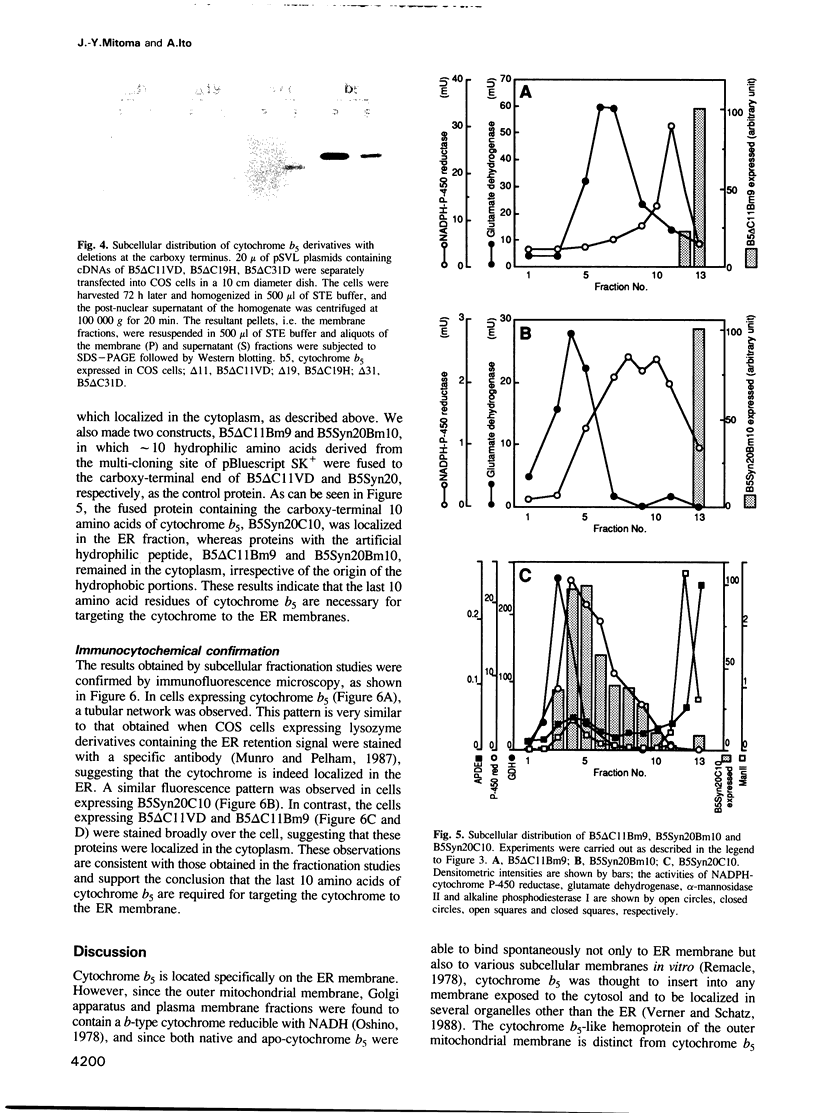
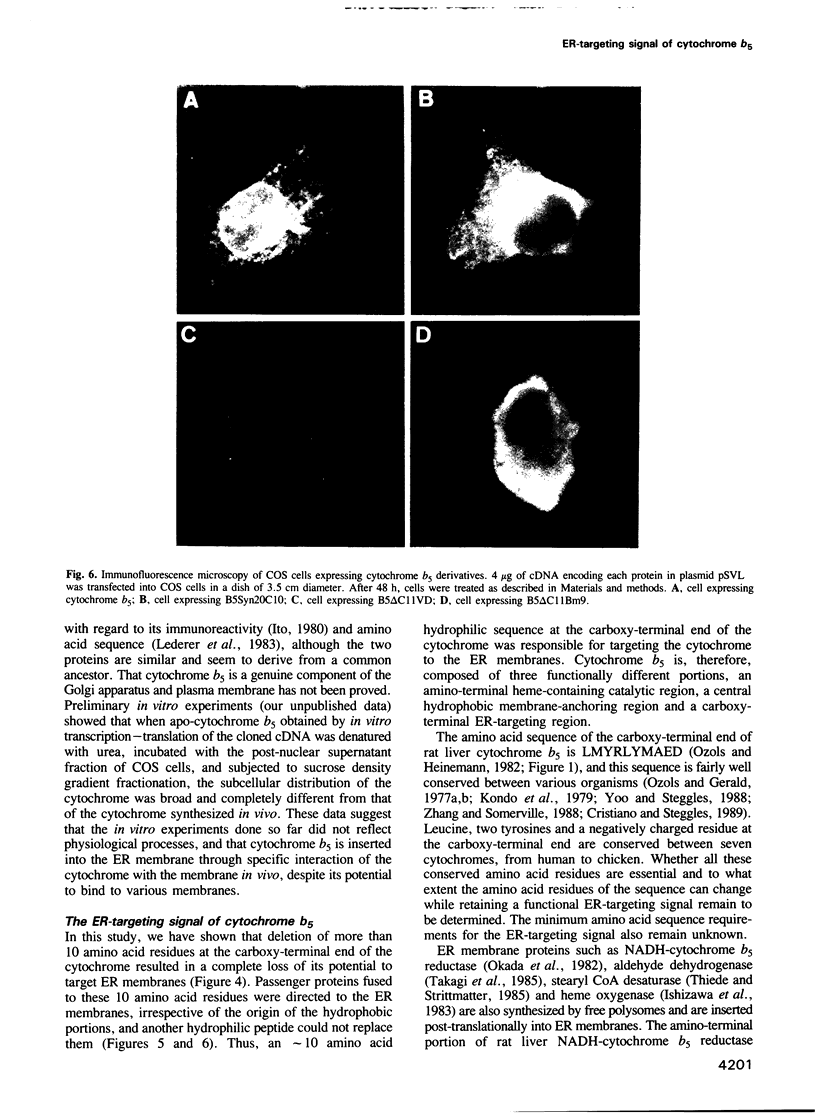
Cytochrome b5 is an integral membrane protein located on the outer surface of the endoplasmic reticulum (ER). This cytochrome is considered to be synthesized on free ribosomes and to be inserted post-translationally into the ER membrane, without participation of a signal recognition particle. To elucidate the signal responsible for targeting of cytochrome b5 to the ER membrane in vivo, DNAs encoding various derivatives of the cytochrome were constructed and introduced into cultured mammalian COS cells, and the subcellular distributions of the derivatives expressed in the cells were then analyzed. The deletion of more than 11 amino acid residues at the carboxy-terminal end of cytochrome b5 abolished the targeting and anchoring of the cytochrome to the ER membrane. Fusion proteins consisting of the carboxy-terminal 10 amino acid residues of cytochrome b5 and passenger proteins with the hydrophobic portion could be localized in the ER membrane. Thus, the last 10 amino acid residues of cytochrome b5 carry information necessary for the cytochrome to be targeted to the ER membrane.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. J., Mostov K. E., Blobel G. Mechanisms of integration of de novo-synthesized polypeptides into membranes: signal-recognition particle is required for integration into microsomal membranes of calcium ATPase and of lens MP26 but not of cytochrome b5. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7249–7253. doi: 10.1073/pnas.80.23.7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews D. W., Lauffer L., Walter P., Lingappa V. R. Evidence for a two-step mechanism involved in assembly of functional signal recognition particle receptor. J Cell Biol. 1989 Mar;108(3):797–810. doi: 10.1083/jcb.108.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold H., Maier K. P. Crystallization and some properties of glutamate dehydrogenase from rat liver. Biochim Biophys Acta. 1971 Nov 19;251(2):133–140. doi: 10.1016/0005-2795(71)90095-x. [DOI] [PubMed] [Google Scholar]
- Bendzko P., Prehn S., Pfeil W., Rapoport T. A. Different modes of membrane interactions of the signal sequence of carp preproinsulin and of the insertion sequence of rabbit cytochrome b5. Eur J Biochem. 1982 Mar;123(1):121–126. doi: 10.1111/j.1432-1033.1982.tb06507.x. [DOI] [PubMed] [Google Scholar]
- Blobel G. Intracellular protein topogenesis. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristiano R. J., Steggles A. W. The complete nucleotide sequence of bovine liver cytochrome b5 mRNA. Nucleic Acids Res. 1989 Jan 25;17(2):799–799. doi: 10.1093/nar/17.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch H. G., Fleming P. J., Strittmatter P. The binding of cytochrome b5 to phospholipid vesicles and biological membranes. Effect of orientation on intermembrane transfer and digestion by carboxypeptidase Y. J Biol Chem. 1979 Jul 25;254(14):6483–6488. [PubMed] [Google Scholar]
- Enomoto K., Sato R. Incorporation in vitro of purified cytochrome b 5 into liver microsomal membranes. Biochem Biophys Res Commun. 1973 Mar 5;51(1):1–7. doi: 10.1016/0006-291x(73)90498-1. [DOI] [PubMed] [Google Scholar]
- Felgner P. L., Gadek T. R., Holm M., Roman R., Chan H. W., Wenz M., Northrop J. P., Ringold G. M., Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habig W. H., Pabst M. J., Jakoby W. B. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J Biol Chem. 1974 Nov 25;249(22):7130–7139. [PubMed] [Google Scholar]
- Ishizawa S., Yoshida T., Kikuchi G. Induction of heme oxygenase in rat liver. Increase of the specific mRNA by treatment with various chemicals and immunological identity of the enzymes in various tissues as well as the induced enzymes. J Biol Chem. 1983 Apr 10;258(7):4220–4225. [PubMed] [Google Scholar]
- Ito A. Cytochrome b5-like hemoprotein of outer mitochondrial membrane; OM cytochrome b. I. Purification of OM cytochrome b from rat liver mitochondria and comparison of its molecular properties with those of cytochrome b5. J Biochem. 1980 Jan;87(1):63–71. doi: 10.1093/oxfordjournals.jbchem.a132753. [DOI] [PubMed] [Google Scholar]
- Ito A., Miyazoe R., Mitoma J., Akao T., Osaki T., Kunitake T. Synthetic cationic amphiphiles for liposome-mediated DNA transfection. Biochem Int. 1990 Oct;22(2):235–241. [PubMed] [Google Scholar]
- Ito A., Sato R. Proteolytic microdissection of smooth-surfaced vesicles of liver microsomes. J Cell Biol. 1969 Jan;40(1):179–189. doi: 10.1083/jcb.40.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A., Sato R. Purification by means of detergents and properties of cytochrome b5 from liver microsomes. J Biol Chem. 1968 Sep 25;243(18):4922–4923. [PubMed] [Google Scholar]
- Kondo K., Tajima S., Sato R., Narita K. Primary structure of the membrane-binding segment of rabbit cytochrome b5. J Biochem. 1979 Oct;86(4):1119–1128. doi: 10.1093/oxfordjournals.jbchem.a132606. [DOI] [PubMed] [Google Scholar]
- Kumamoto T., Ito A., Omura T. Critical region in the extension peptide for the import of cytochrome P-450(SCC) precursor into mitochondria. J Biochem. 1989 Jan;105(1):72–78. doi: 10.1093/oxfordjournals.jbchem.a122622. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lederer F., Ghrir R., Guiard B., Cortial S., Ito A. Two homologous cytochromes b5 in a single cell. Eur J Biochem. 1983 Apr 15;132(1):95–102. doi: 10.1111/j.1432-1033.1983.tb07330.x. [DOI] [PubMed] [Google Scholar]
- Leto T. L., Roseman M. A., Holloway P. W. Mechanism of exchange of cytochrome b5 between phosphatidylcholine vesicles. Biochemistry. 1980 Apr 29;19(9):1911–1916. doi: 10.1021/bi00550a028. [DOI] [PubMed] [Google Scholar]
- Morgenstern R., Meijer J., Depierre J. W., Ernster L. Characterization of rat-liver microsomal glutathione S-transferase activity. Eur J Biochem. 1980 Feb;104(1):167–174. doi: 10.1111/j.1432-1033.1980.tb04412.x. [DOI] [PubMed] [Google Scholar]
- Munro S., Pelham H. R. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987 Mar 13;48(5):899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- Müller G., Zimmermann R. Import of honeybee prepromelittin into the endoplasmic reticulum: structural basis for independence of SRP and docking protein. EMBO J. 1987 Jul;6(7):2099–2107. doi: 10.1002/j.1460-2075.1987.tb02476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Frey A. B., Guenthner T. M., Oesch F., Sabatini D. D., Kreibich G. Studies on the biosynthesis of microsomal membrane proteins. Site of synthesis and mode of insertion of cytochrome b5, cytochrome b5 reductase, cytochrome P-450 reductase and epoxide hydrolase. Eur J Biochem. 1982 Feb;122(2):393–402. doi: 10.1111/j.1432-1033.1982.tb05894.x. [DOI] [PubMed] [Google Scholar]
- Omura T., Takesue S. A new method for simultaneous purification of cytochrome b5 and NADPH-cytochrome c reductase from rat liver microsomes. J Biochem. 1970 Feb;67(2):249–257. doi: 10.1093/oxfordjournals.jbchem.a129248. [DOI] [PubMed] [Google Scholar]
- Ozols J., Carr S. A., Strittmatter P. Identification of the NH2-terminal blocking group of NADH-cytochrome b5 reductase as myristic acid and the complete amino acid sequence of the membrane-binding domain. J Biol Chem. 1984 Nov 10;259(21):13349–13354. [PubMed] [Google Scholar]
- Ozols J., Gerard C. Covalent structure of the membranous segment of horse cytochrome b5. Chemical cleavage of the native hemoprotein. J Biol Chem. 1977 Dec 10;252(23):8549–8553. [PubMed] [Google Scholar]
- Ozols J., Heinemann F. S. Chemical structure of rat liver cytochrome b5. Isolation of peptides by high-pressure liquid chromatography. Biochim Biophys Acta. 1982 May 21;704(1):163–173. doi: 10.1016/0167-4838(82)90143-1. [DOI] [PubMed] [Google Scholar]
- Remacle J. Binding of cytochrome b5 to membranes of isolated subcellular organelles from rat liver. J Cell Biol. 1978 Nov;79(2 Pt 1):291–313. doi: 10.1083/jcb.79.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini D. D., Kreibich G., Morimoto T., Adesnik M. Mechanisms for the incorporation of proteins in membranes and organelles. J Cell Biol. 1982 Jan;92(1):1–22. doi: 10.1083/jcb.92.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M., Mihara K., Sato R. Signal recognition particle is required for co-translational insertion of cytochrome P-450 into microsomal membranes. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3361–3364. doi: 10.1073/pnas.81.11.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara S., Müller R., Taguchi H., Yoshida T. Cloning and expression of cDNA for rat heme oxygenase. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7865–7869. doi: 10.1073/pnas.82.23.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatz L., Strittmatter P. A form of cytochrome b5 that contains an additional hydrophobic sequence of 40 amino acid residues. Proc Natl Acad Sci U S A. 1971 May;68(5):1042–1046. doi: 10.1073/pnas.68.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storrie B., Madden E. A. Isolation of subcellular organelles. Methods Enzymol. 1990;182:203–225. doi: 10.1016/0076-6879(90)82018-w. [DOI] [PubMed] [Google Scholar]
- Strittmatter P., Rogers M. J., Spatz L. The binding of cytochrome b 5 to liver microsomes. J Biol Chem. 1972 Nov 25;247(22):7188–7194. [PubMed] [Google Scholar]
- Takagi Y., Ito A., Omura T. Biogenesis of microsomal aldehyde dehydrogenase in rat liver. J Biochem. 1985 Dec;98(6):1647–1652. doi: 10.1093/oxfordjournals.jbchem.a135435. [DOI] [PubMed] [Google Scholar]
- Thiede M. A., Ozols J., Strittmatter P. Construction and sequence of cDNA for rat liver stearyl coenzyme A desaturase. J Biol Chem. 1986 Oct 5;261(28):13230–13235. [PubMed] [Google Scholar]
- Thiede M. A., Strittmatter P. The induction and characterization of rat liver stearyl-CoA desaturase mRNA. J Biol Chem. 1985 Nov 25;260(27):14459–14463. [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verner K., Schatz G. Protein translocation across membranes. Science. 1988 Sep 9;241(4871):1307–1313. doi: 10.1126/science.2842866. [DOI] [PubMed] [Google Scholar]
- Yoo M., Steggles A. W. The complete nucleotide sequence of human liver cytochrome b5 mRNA. Biochem Biophys Res Commun. 1988 Oct 14;156(1):576–580. doi: 10.1016/s0006-291x(88)80881-7. [DOI] [PubMed] [Google Scholar]
- Zhang H., Somerville C. The primary structure of chicken liver cytochrome b5 deduced from the DNA sequence of a cDNA clone. Arch Biochem Biophys. 1988 Jul;264(1):343–347. doi: 10.1016/0003-9861(88)90603-0. [DOI] [PubMed] [Google Scholar]





