Abstract
Transcriptional programs control cellular lineage commitment and differentiation during development. Understanding cell fate has been advanced by studying single-cell RNA-seq, but is limited by the assumptions of current analytic methods regarding the structure of data. We present single-cell topological data analysis (scTDA), an algorithm for topology-based computational analyses to study temporal, unbiased transcriptional regulation. Compared to other methods, scTDA is a non-linear, model-independent, unsupervised statistical framework that can characterize transient cellular states. We applied scTDA to the analysis of murine embryonic stem cell (mESC) differentiation in vitro in response to inducers of motor neuron differentiation. scTDA resolved asynchrony and continuity in cellular identity over time, and identified four transient states (pluripotent, precursor, progenitor, and fully differentiated cells) based on changes in stage-dependent combinations of transcription factors, RNA-binding proteins and long non-coding RNAs. scTDA can be applied to study asynchronous cellular responses to either developmental cues or environmental perturbations.
Introduction
The differentiation of motor neurons from neuroepithelial cells in the vertebrate embryonic spinal cordis a well characterized example of cellular lineage commitment and terminal cellular differentiation1. Neural precursor cells differentiate in response to spatiotemporally regulated morphogen gradients that are generated in the neural tube by activating a cascade of specific transcriptional programs1. A detailed understanding of this process has been hindered by the inability to isolate and purify sufficient quantities of synchronized cellular subpopulations from the developing murine spinal cord. Although in vitro approaches have been used to study both the mechanisms of motor neuron differentiation2, and motor neuron disease3, 4, alimitation of these approaches is the differential exposure of embryoid bodies (EBs) to inductive ligands and uncharacterized paracrine signaling within EBs, which lead to the generation of heterogeneous populations of differentiated cell types5. Motor neuron disease mechanisms are currently studied in a heterogeneous background of cell types whose contributions to pathogenesis are unknown. Methods to analyse the transcriptome of individual differentiating motor neurons could provide fundamental insights into the molecular basis of neurogenesis and motor neuron disease mechanisms.
Single-cell RNA-sequencing carried out over time enables the dissection of transcriptional programs during cellular differentiation of individual cells, thereby capturing heterogeneous cellular responses to developmental induction. Several algorithms for the analysis of single-cell RNA-sequencing data from developmental processes have been published, including Diffusion Pseudotime6, Wishbone7, SLICER8, Destiny9, Monocle10, and SCUBA11 (Supplementary Table 1). All of these methods can be used to order cells according to their expression profiles, and they enable the indentification of lineage branching events. However, Destiny9 lacks an unsupervised framework for determining the transcriptional events that are statistically associated with each stage of the differentiation process; and the statistical framework of Diffusion Pseudotime, Wishbone, Monocle, and SCUBA is biased, for example by assuming a differentiation process with exactly one branch event6, 7 or a tree-like structure10, 11. Although these methods can reveal the lineage structure when the biological process fits with the assumptions, an unsupervised method would be expected to have the advantage of extracting more complex relationships. For example, the presence of multiple independent lineages, convergent lineages, or the coupling of cell cycle to lineage commitment. Moreover, apart from SCUBA, these methods do not exploit the temporal information available in longitudinal single cell RNA-sequencing experiments, and they require the user to explicitly specify the least differentiated state6-10.
We present an unbiased, unsupervised, statistically robust mathematical approach to single cell RNA-sequencing data analysis that addresses these limitations. Topological data analysis (TDA) is a mathematical approach used to study the continuous structure of high-dimensional data sets. TDA has been used to study viral re-assortment12, human recombination13, 14, cancer15, and other complex genetic diseases16. scTDA is applied to study time-dependent gene expression using longitudinal single-cell RNA-seq data. Our scTDA method is a statistical framework for the detection of transient cellular populations and their transcriptional repertoires, and does not assume a tree-like structure for the expression space or a specific number of branching points. scTDA can be used to assess the significance of topological features of the expression space, such as loops or holes. In addition, it exploits temporal experimental information when available, inferring the least differentiated state from the data.
Here we apply scTDA to analyse the transcriptional programs that regulate developmental decisions as mESCs transition from pluripotency to fully differentiated motor neurons and concomitant cell types.
Results
Overview of scTDA
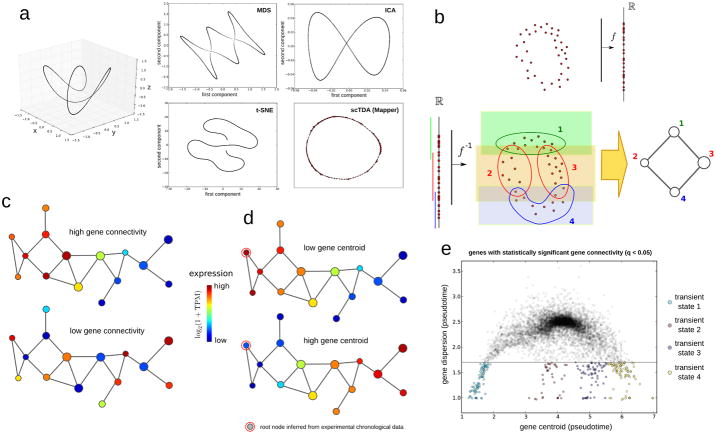
Single-cell gene expression can be represented as a sparse high-dimensional point cloud, with the number of dimensions equivalent to the number of expressed genes (∼10,000). Extracting biological information from such data requires a reduction in the dimensionality of the space. Widely-used algorithms, such as multidimensional scaling (MDS), independent component analysis (ICA), and t-distributed stochastic neighbor embedding (t-SNE), have been adapted to flow cytometry, mass spectrometry11, 17, and single-cell RNA-seq measurements10, 18. These strategies, however, are not designed to preserve continuous relationships in high dimensions. Figure 1a represents a simple example of a one-dimensional continuous manifold (a circle) twisted in three dimensions. Reduction to two dimensions using these algorithms introduces artifacts in the low-dimensional representation, including artifactual intersections (in MDS and ICA), and tearing apart the original continuous structure (in t-SNE). As cell differentiation is a continuous asynchronous process, we reasoned that longitudinal single-cell data would be best analyzed using a mathematical approach that accounts for continuous structures.
Figure 1. Topological analysis of longitudinal single-cell RNA-seq data.
a Comparison of methods for reducing dataset dimensionality. A toy example is shown, illustrating the artifacts that can emerge when standard dimensional reduction methods are used to represent differentiation trajectories. A total of 1,000 points are sampled from a twisted circle in three-dimensional space. MDS, ICA, t-SNE, and Mapper were utilized to represent the above points in two dimensions. Of these methods, only Mapper can capture the continuous circular trajectory of the three-dimensional space without introducing artificial intersections or disrupting the trajectory.
b. Mapper algorithm. Top: Mapper builds upon dimensional reduction function f mapping the high-dimensional single-cell RNA-seq point cloud data into Rk (for simplicity we take k = 1 in this figure). Bottom: under the inverse function f−1, a covering of Rk maps into a covering of the single-cell point cloud data. Clustering is performed independently in each of the induced patches in the high-dimensional space. In the low-dimensional representation, a node is assigned to each cluster of cells. If two clusters intersect, the corresponding nodes are connected by an edge. Topological features in the low dimensional representation are guaranteed to also be present in the original high-dimensional RNA-seq space.
c. Gene connectivity. Gene connectivity allows for the identification of genes that are differentially expressed by a cellular subpopulation of the differentiation process, without predefining any cellular subpopulation. Represented is a toy example of two genes with very different gene connectivity on the topological representation. Top: An example of a gene with high gene connectivity in the topological representation. This signifies that there is a set of cells with similar global expression profiles and high expression levels of the gene. Bottom: An example of a gene with low gene connectivity in the topological representation.
d. Gene centroid. The centroid of a gene, measured in pseudotime, quantifies where the expression of a gene sits in the topological representation with respect to the root node. The root node represents the least differentiated cellular state, and is determined from the experimental sampling times. A toy example of two genes with very different centroid can be used to illustrate the concept. Top: a gene with low value for the expression centroid, being mostly associated to pluripotent cells. Bottom: a gene with a high value for the centroid, being mostly associated to differentiated cells.
e. Transient cellular states. Transient cellular states can be identified in an unsupervised manner by clustering low-dispersion genes with significant gene connectivity according to their centroid in the topological representation. In the figure, an example of distribution of centroids and dispersions for genes with significant gene connectivity is shown. Four clusters of low-dispersion genes are identified, which correspond to four transient cellular states arising throughout the differentiation process.
We developed a computational approach to the analysis of longitudinal single-cell RNA-seq data based on the TDA algorithm Mapper19 (Supplementary Note 1). Mapper builds upon any given dimensional reduction algorithm, such as MDS, and produces a low-dimensional topological representation of the data that preserves locality. To that end, the projection obtained by the dimensional reduction algorithm is covered with overlapping bins, and clustering of the data within each bin is performed in the original high-dimensional space (Fig. 1b). A network is constructed by assigning a node to each cluster, and clusters that share one or more cells are connected by an edge. The result is a low-dimensional network representation of the data where nodes represent sets of cells with similar global transcriptional profiles, and edges connect nodes that have at least one cell in common (Fig. 1b). An important feature of these networks is that elements that are connected in the low-dimensional representation lie near each other in the original high-dimensional expression space, contrary to what occurs with most of the dimensional reduction algorithms currently in use, including PCA, ICA, and MDS (Fig. 1a). The scTDA approach can be applied even if there are complex structures in the high-dimensional expression space, including non-tree-like trajectories (Fig. 1a). Additionally, since nodes represent clusters of cells, the approach is highly scalable to large datasets.
We adapted topological representations so that they could be used to analyse longitudinal single-cell RNA-seq data, introducing the necessary mathematical concepts and statistics (Supplementary Fig. 1). To identify expression programs associated with different parts of the topological representation without predefining any cellular population, we developed the concept of gene connectivity. The connectivity of a gene acquires a significant value when cells that share similar global transcriptional profiles express that particular gene more than random (Fig. 1c and Online Methods). Genes with a significant connectivity are expected to be involved in a particular stage (or stages) of differentiation. To identify the least differentiated state in the topological representation, we introduced a root node, which is the node that maximizes correlation between sampling time and graph distance. Using this root node as a reference, we computed the centroid (Fig. 1d) and dispersion (the standard deviation) of genes in the topological representation (Online Methods). Genes with low centroids are upregulated in stem-like cells, whereas genes with large centroids are specific to fully differentiated cells.
To identify transient cellular subpopulations arising during differentiation, we clustered low-dispersion genes with a significant connectivity in the topological representation based on their centroid (Davies-Bouldin criterion, Fig. 1e). Finally, to assess the topological features of the representation, such as loops and holes, we used persistent homology, a framework within TDA for deriving and classifying topological features associated to data (Online Methods, Supplementary Note 1). Further details of the scTDA method and its mathematical foundations can be found in the Online Methods and Supplementary Note 1. A code implementing the scTDA method is provided as Supplementary Code.
scTDA for temporal ordering of differentiating cells
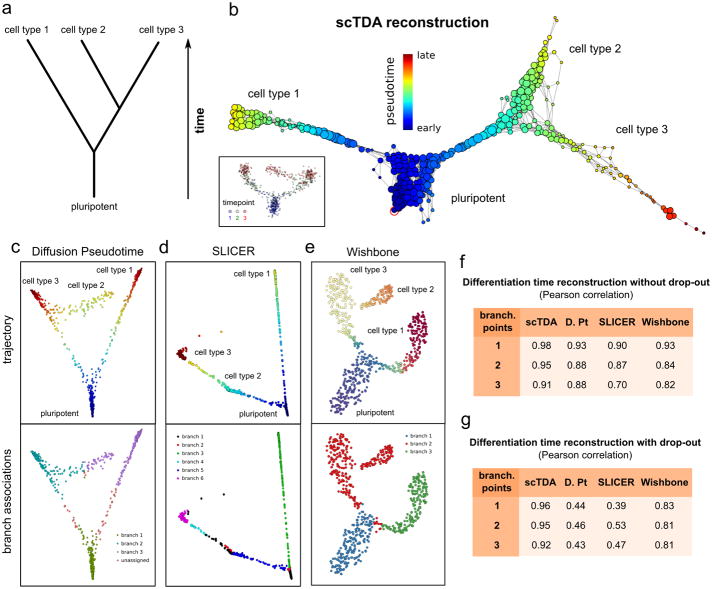
We assessed the capacity of scTDA to order asynchronously differentiating cells using a controlled setting where the truth is known. First, we simulated a noisy, branched, asynchronous cellular differentiation process with two branching points (Fig. 2a and Online Methods). 700 cells were sampled at three time points and the expression levels of 500 genes were simulated. From this data, scTDA correctly reconstructed the topological structure of the differentiation tree, and identified the most stem-like state (Fig. 2b). In contrast, the algorithms Diffusion Pseudotime6, Wishbone7 and SLICER8 failed to correctly assign some of the branches in our hands, producing artifacts in the inferred pseudo-temporal ordering of the cells (Figs. 2c to 2e), even if the most stem-like state was provided by the user of these algorithms.
Figure 2. Comparison of algorithms for ordering cellular states.
a A noisy, branched, asynchronous cellular differentiation process was simulated. The differentiation tree of the process is represented. 700 cells were sampled from this process at three time points. Using this data, we attempted to reconstruct the structure of the differentiation tree with scTDA, and compared the outcomes with reconstruction using Diffusion Pseudotime, SLICER and Wishbone, all of which rely on branch assignments for downstream statistical analysis.
b. Reconstructed differentiation trajectory using scTDA. scTDA recovered the structure of the simulated differentiation process and correctly rooted the tree using the experimental information. Nodes correspond to sets of cells sharing similar global transcriptional profiles, with the node sizes proportional to the number of cells in the node. Nodes that are connected by an edge have at least one cell in common. For reference, an inset with the latent MDS representation of the data, colored by sampling day, is also shown.
c. Reconstructed differentiation trajectory using Diffusion Pseudotime. Although the representation of the data using the first two diffusion coefficients reproduces the structure of the differentiation tree, the branches are not correctly assigned.
d. Reconstructed differentiation trajectory using SLICER. The representation constructed by SLICER using locally linear embedding was unable to capture the complete structure of the differentiation tree and branch assignments.
e. Reconstructed differentiation trajectory using Wishbone. The t-SNE representation of the data used by Wishbone reproduces the structure of the differentiation tree and correctly identifies the first branching point. However, Wishbone failed to identify the second branching point.
f, g. Pearson's correlation coefficient between the pseudo-time, inferred from the data by scTDA, Diffusion Pseudotime (D. Pt), SLICER, and Wishbone, and the actual simulated differentiation time. Cellular differentiation processes with one, two, or three branching points were considered, both in the absence (f) and the presence (g) of drop-out events.
To quantify the performance of scTDA, we extended the above simulations to processes with one, two, or three branching points (Online Methods). scTDA showed higher correlation between the inferred pseudo-time and the actual simulated differentiation time compared to the other three methods, thus reconstructing the underlying differentiation process more accurately from the data (Fig. 2f). The improvement was particularly large when drop-out events were included in the simulation (Fig. 2g), where the graphical representations produced by Diffusion Pseudotime, SLICER, and Wishbone were often unable to correctly infer the structure of the differentiation tree (Supplementary Fig. 2).
Motor neuron differentiation analysis with scTDA
We applied scTDA to longitudinal single-cell RNA-seq data from in vitro motor neuron differentiation experiments. We differentiated mESCs into motor neurons using a well-established protocol2. The mESC line expresses enhanced green fluorescent protein (eGFP) under the control of the early motor neuron-specific promoter Mnx1, allowing identification of committed motor neurons. Using a modified CEL-seq approach20 (Online Methods and Supplementary Note 2), we generated cDNA libraries from individual cells, spanning the transition from pluripotency to post-mitotic commitment. To assess reproducibility, we performed two separate differentiations. In the first differentiation (experiment 1) we sequenced 440 cells sampled throughout the entire course of motor neuron differentiation. Cells were concomitantly sampled in bulk at each time point. Using this data, we assessed library saturation and the optimal depth of sequencing coverage. In the second differentiation (experiment 2) we sought to increase statistical power and sequenced 2,304 individual with the depth of sequencing coverage near the saturation point (∼250k reads/cell). In both experiments cells were sampled each day throughout the entire course of the differentiation, from day two to six. In total we detected 15,609 and 19,009 transcripts respectively across experiments 1 and 2. Given the improved statistical power associated with an increased number of cells, we focused our attention on the results from experiment 2, and assessed the reproducibility of the results using experiment 1.
Cell populations and surface markers of differentiation
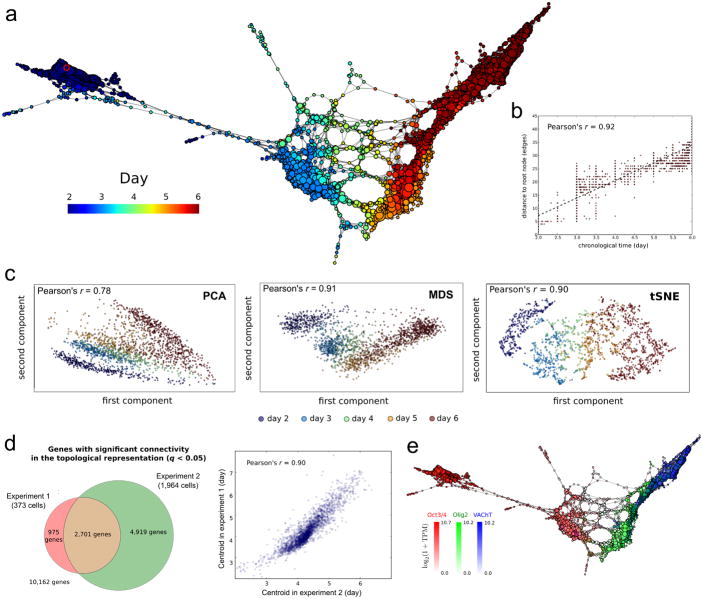
We used scTDA to analyze the two longitudinal motor neuron differentiation single-cell RNA-seq datasets. We filtered the data for cells that passed stringent quality control tests (Online Methods, Supplementary Fig. 3), retaining, respectively, 373 and 1,964 individual cells in experiments 1 and 2. An scTDA analysis recapitulated chronological order based on mRNA expression levels alone (Figs. 3a and 3b, and Supplementary Figs. 4 and 5), and simultaneously detected detailed transcriptional relationships between individual cells. We observed only a mild dependence of library complexity on timescale of our differentiation (Supplementary Fig. 6, Online Methods), unlike other experimental settings where differences in cell cycle give rise to substantial variation of the library complexity21, 22. We did not observe large batch effects (Supplementary Fig. 7). Compared with analyses of the same data sets using PCA, MDS, t-SNE, Monocle, Wishbone, Slicer and Diffusion Pseudotime, we showed that scTDA had superior ability to identify the continuous chronological structure of the differentiation process (Fig. 3c and Supplementary Figs. 8 and 9).
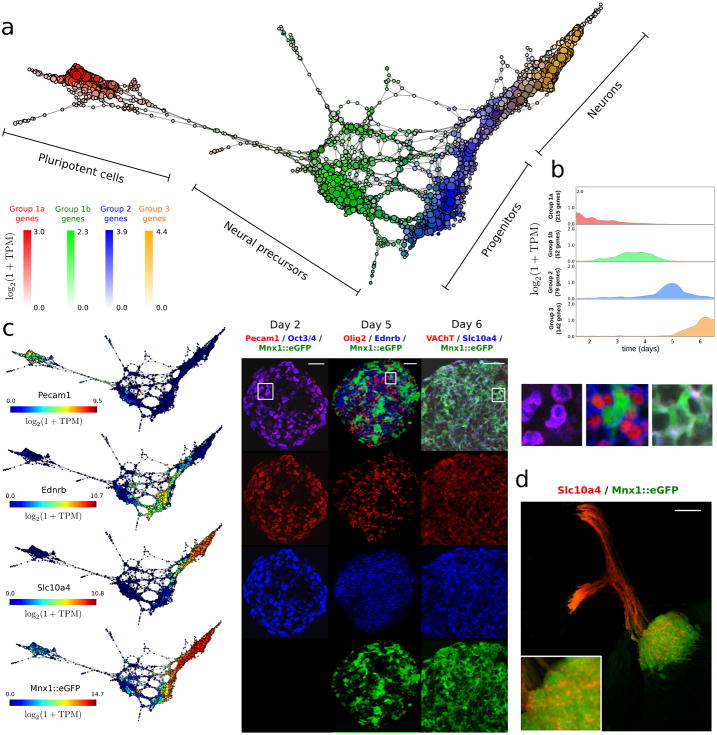
Figure 3. Differentiation of pluripotent mESCs into motor neurons (MN).
a scTDA recapitulates chronological order based on expression data alone. The topological representation of the expression data of 1,964 single cells, sampled from the differentiation of mESCs into MNs, is labeled by sampling time. The root node, inferred from the experimental chronological information, is indicated with a red circle.
b. The distance of each node to the root node, represented as a function of sampling time. The chronological time of a node is defined as the mean of the sampling times of the cells in the node.
c. Comparison with standard dimensional reduction algorithms. Dimensional reduction of the same expression data of 1,964 single cells, sampled from the differentiation of mESCs into MNs, using PCA, MDS, and t-SNE. The Pearson's correlation coefficient between the sampling time and the two-dimensional Euclidean distance to the root cell (defined as the one that maximizes this correlation) is indicated in each case.
d. Consistency between experiments 1 and 2. Left: Venn diagram of genes with significant gene connectivity (q < 0.05) in the topological representations of the two datasets. Both experiments are highly consistent in their calls (Fisher exact test p-value < 10-100). The number of significant genes is larger in experiment 2, consistent with its higher statistical power (due to the larger number of cells considered). Right: correlation between the centroid (expressed in pseudo-time) of the n = 2,701 genes with significant (q <0.05) gene connectivity in both topological representations. The distribution of gene centroids is highly consistent across the two experiments.
e. Known markers of pluripotent cells, motor neuron progenitors and post-mitotic neurons are successfully ordered. The topological representation is labeled by mRNA levels of Oct3/4 (red), Olig2 (green) and VAChT (blue).
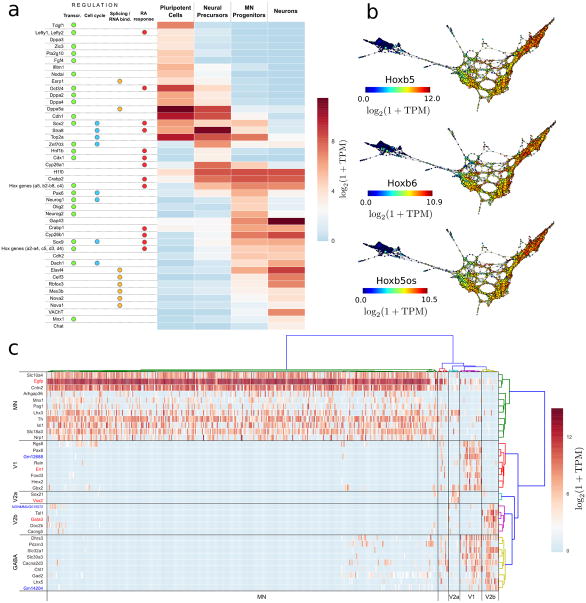
Figure 5. Differential gene expression in neurogenesis.
a Differentially expressed regulators and downstream genes in the four cell populations are shown. Genes are annotated according to their role in transcriptional, cell cycle, RNA binding protein regulation and RA response.
b. Topological representations (labeled by mRNA levels) of Hoxb5, Hoxb6 and the antisense lncRNA Hoxb5os, showing concordant expression of these transcripts during the generation of motor neurons from mESCs.
c. Post-mitotic neuronal populations. Differentially expressed genes between Vsx2+, Gata3+ and En1+ cells that were marked as post-mitotic neurons by the scTDA analysis. Hierarchical clustering of cells leads to five groups, four of which correspond to MNs, and V1, V2a and V2b interneurons. Hierarchical clustering of genes produces four groups of genes, uniquely expressed by each of the above cell types, and a fifth group associated to GABAergic neurons. lncRNAs are marked in blue.
Analysis of the data from experiment 2 identified 7,620 genes with significant gene connectivity (q-value < 0.05, Supplementary Fig. 10 and Supplementary Table 2), compromising 74% overlap with those identified from experiment 1 (p-value < 10-100, Fisher exact test, Fig. 3d). It is likely that the large number of significant genes is due to the transcriptional heterogeneity of the dataset, because the samples of cells represent multiple developmental stages. The centroids of the significant genes were consistent between the two datasets (Pearson correlation = 0.9, p-value < 10-100, Fig. 3d). We validated our findings by identifying known markers of pluripotent cells, progenitor cells and mature motor neurons in the dataset over time (Fig. 3e and Supplementary Fig. 4c). We also found that cell cycle genes were downregulated (as expected) in post-mitotic neurons (Supplementary Fig. 11). We verified that the choice of parameters for building the topological representation (number of genes, bins, and overlap between bins) did not affect the result of the analysis (Supplementary Fig. 12).
Interpretation of the distribution of the centroid and the dispersion of genes (Supplementary Fig. 13) using scTDA revealed four transcriptionally distinct cellular populations arising during differentiation from mESCs to motor neurons (Fig. 4a). 488 of 7,620 genes were identified in the four expression groups (Fig. 4b, and Supplementary Table 2). Groups 1a and 1b contain genes that are only expressed in early EBs (Oct3/4+ cells), and correspond to pluripotent and neural precursor states. Genes in groups 2 and 3 are only expressed in the progenitor (Olig2+ cells) and post-mitotic ensembles (VAChT+ cells), respectively.
Figure 4. Cellular populations during motorneuron differentiation.
a scTDA identifies four transient populations in mESC differentiation into MNs. Represented is the topological representation (colored by mRNA levels) of four groups of low-dispersion genes; pluripotent, precursor, progenitor and post-mitotic populations. In total, 488 genes were assigned to one of these four populations based on their expression profiles in the topological representation.
b. Reconstructed expression timeline for each of the four groups of low-dispersion genes.
c. Validation by detection of state-specific cell surface markers identified by scTDA. Topological representation (colored by mRNA levels) of surface proteins Pecam1, Ednrb and Slc10a4, and immunostaining of cultured EBs is shown. The scale bar in the immunostaining images denotes a length of 50 μm. Details of three regions are presented on the right. For reference, the topological representation colored by mRNA levels of the Mnx1≳eGFP reporter is also shown.
d. In vivo validation of the motor neuron surface marker Slc10a4. Spinal cord section of an E9.5 mouse immunostained for Slc10a4 (red). The pool of motor neurons is also marked by Mnx1≳eGFP expression (green). The scale bar denotes a length of 50 μm.
Ontology enrichment analyses using PANTHER23 revealed developmental genes and genes related to DNA replication in groups 1a, 1b, and 2, whereas group 3 was enriched for genes related to axonogenesis, neuron migration, and regionalization (Supplementary Table 3), consistent with the underlying cellular differentiation process.
The analysis identified 74 transcripts in the four groups that encode proteins with an extracellular domain. These are therefore putative cell surface marker candidates that could be used to purify niche populations for further study (Supplementary Table 2). Several of these surface markers have not been reported as markers of neurogenesis. We selected three of these 74 markers that had high expression level and specific antibodies available for experimental validation. The three markers were validated using immunohistochemistry in vitro in EBs (Fig. 4c, and Supplementary Fig. 14) and in vivo in the murine embryonic spinal cord (Fig. 4d).
scTDA identifies distinct proliferative states
scTDA analysis of our motor neuron differentiation data revealed numerous loops in the neural precursor population (Fig 4a) that were not present in the mESCs or the motor neurons. These loops were also observed in the scTDA analysis of experiment 1 (Supplementary Fig. 4). Neural precursor cells proliferate rapidly upon induction with retinoic acid (RA). This is in keeping with biological expectations, given that the cell cycle will give rise to periodic structures in the expression space. To evaluate whether the loops in the topological representation of neural precursors are caused by differences in the cell cycle, we built a topological representation using only cell cycle genes (Supplementary Fig. 15a, Online Methods). The cell cycle topological representation contained substantially larger loops in the same region (Supplementary Fig. 15a), separating Stra8up neural precursors and progenitors into proliferative and non-proliferative populations according to the expression of Mki67 and other markers of proliferation (Supplementary Figs. 15b and 15c). We used persistent homology to statistically assess the significance of these loops in the topological representation, asking whether loops of similar size could arise from noise effects (Online Methods). This analysis showed a strong statistical significance for some of the larger loops (q-value < 0.05, Supplementary Fig. 15d), consistent with a biological, rather than technical, origin for these features.
Characterization of developmental transitions
We next used scTDA to characterize the developmental transitions that occur during the differentiation of motor neurons. These results are available through an online database (Online Methods). We used scTDA to identify transcriptional programs that are associated with the transitions between the four transient cellular populations that we identified (Fig. 5a). These findings were consistent across both experiments.
First, the transition from a pluripotent cell to a neural precursor population is characterized by the transcriptional dynamics of pathways involved in RA signaling and downstream effector proteins24, 25 (Fig. 5a). Our analysis resolved with unprecedented resolution the timing of the transcriptional events occurring during this transition, identifying upregulation of Stra8 and downregulation of Fgf4 as some of the earliest events that mark the transition (Supplementary Fig. 16). Subsequently, there is transcriptional activation of a subset of the homeobox gene family, including Hoxa1 and Hoxb2-8, which continue to be expressed during later stages of the differentiation process, and Hoxb1, which is transiently expressed along with caudalizing transcription factors (Figs. 5a and 5b, and Supplementary Fig. 17).
A second wave of RA inducible gene activation was identified during the formation of neural progenitors. This is accompanied by transcriptional inactivation of Stra8, and activation of Crabp1 and a second set of homeobox genes, Hoxa2, Hoxa3, Hoxc5, Hoxd3 and Hoxd4 (Fig. 5a). This pattern of Hox gene activation suggests that the linear chromosomal arrangement of the Hox gene clusters does not necessitate temporal co-linearity in anterior Hox gene expression, a phenomenon that has been reported in the developing spinal cord26. Both waves of homeobox gene expression were accompanied by the upregulation of several long non-coding RNAs (lncRNAs) derived from the opposite strand (Fig. 5b, Supplementary Fig. 17, and Supplementary Table 4), consistent with previously identified lncRNA-based regulation of homeobox gene clusters27-30.
The transition to mature neurons was marked by exit from the cell cycle and post-mitotic differentiation. In keeping with expectations from previous data31, scTDA identified Neurog1/2 (modulators of neuronal specification, cyclin regulation, cell cycle exit) as well as Ascl1 and Sox9 among the known mediators of neuronal commitment, concomitant with a marked repression of topoisomerase 2A in the neuron population (Fig. 5a). We observed variability in eGFP expression levels and fluorescence intensities among differentiated neurons, which might indicate heterogeneity in later stages of differentiation. We performed differential expression analysis of En1+, Gata3+, Vsx2+ and Egfp+ cells with in this cellular population (Online Methods), and classified differentiated neurons into motor neurons (n = 343), and V1 (n = 19, En1+), V2a (n = 10, Vsx2+) and V2b interneurons (n = 15, Gata3+) (Fig. 5c), confirming the presence of cellular heterogeneity.
Interestingly, we observed the presence of several lncRNAs in the differentially expressed genes. V1 interneurons specifically express Gm12688, an intergenic lncRNA located near Foxd3, and transcribed from the opposite strand. We validated this finding using qPCR (Supplementary Fig. 18). Similarly, we identified a lncRNA in chromosome 15 (NONCODEv4 accession number NONMMUG015572) which was only expressed in V2b interneurons. Gm14204, an intergenic lncRNA located near Slc32a1 and transcribed from the opposite strand was only expressed in V1 and V2b GABAergic interneurons. These results may suggest a role for lncRNAs in neural diversification.
Among the genes identified by scTDA as being associated with developmental transitions were 20 genesencoding RNA-binding proteins (Supplementary Table 2). These include known developmental state-dependent pre-mRNA splicing factors, as well as stage-specific but uncharacterized RNA binding proteins, which may guide cellular differentiation and post-mitotic commitment. In the context of the progenitor to neuron transition, our analysis identified Nova1/2, Rbfox3, Srrm4, and the Elav-like transcripts Elavl4 and Celf3 (Fig. 5a), consistent with previous data32, 33. scTDA further revealed the upregulation of Mex3b in the progenitor and post-mitotic cell populations, and constitutive expression of Ptbp1 and Ptbp2. Previously published studies have documented Srrm4 directed inclusion of neural specific exon 10 in Ptbp134. Our results may indicate a transcriptional switch in splicing factor regulation that culminates in neural specific splicing.
scTDA of heterogeneous cellular responses
Our analyses of mESC differentiation using scTDA showed that it can chronologically order asynchronous populations of single cells, while simultaneously preserving high-dimensional relationships between their transcriptional programs. We next tested whether scTDA could be applied to other data sets to provide new insights. We analyzed three different in vivo cellular differentiation datasets35-37 (Online Methods). In each case, scTDA produced an accurate reconstruction of developmental trajectories that confirmed and extended the published observations.
First, we examined 80 single cells from the differentiating distal lung epithelium of mouse embryo35 (Supplementary Table 5). All cells were sampled from the same time point (embryonic day E18.5). As shown in Figure 6a, scTDA recapitulated the proposed relationships between differentiating cells in both the bronchiolar and alveolar lineages. Moreover, our analysis uncovered a set of alveolar type I cells in which genes associated with mitochondrial respiration are down-regulated (Supplementary Fig. 19), which might indicate cellular stress or quiescence.
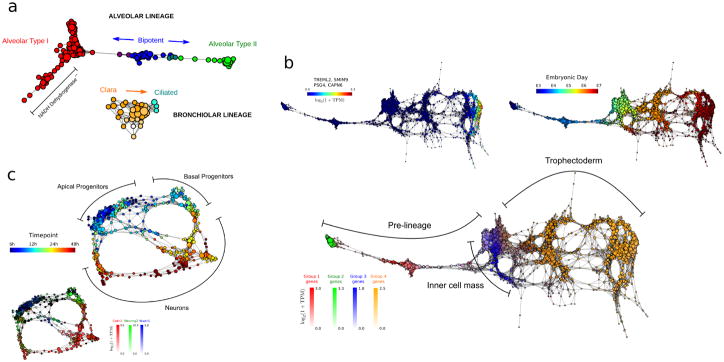
Figure 6. scTDA analysis of mouse and human developmental datasets.
a Topological representation of 80 embryonic (E18.5) mouse lung epithelial cells35 labeled according to cell type. scTDA resolves the alveolar and bronchiolar lineages that were originally identified using PCA, and identifies a putative set of cells with low NADH dehydrogenase expression that were not identified in the original analysis.
b. Topological representation of 1,529 individual cells from 88 human pre-implantation embryos36. Top-left, bottom: topological representation (labeled by expression levels of genes) associated with cellular populations identified during differentiation. scTDA resolved, without supervision, the segregation of the trophectoderm and inner cell mass from pre-lineage cells (bottom), as well as a polar trophectoderm (top-left) that were originally identified using a supervised analysis based on PCA and k-medoid clustering. Top-right: topological representation labeled by embryonic day.
c. Topological representation of 272 newborn neurons from the mouse neocortex37, labeled by sampling time after mitosis (top) and expression levels of Cntr2, Neurog2, and Wwtr1 (bottom). scTDA recapiltulated the converging developmental relations between apical and basal progenitors, and neurons that were originally identified using hierarchical clustering and additionally found lineage convergence.
Second, we applied scTDA to 1,529 cells from human pre-implantation embryos36 (Supplementary Table 6). In this case, scTDA correctly ordered cells according to embryonic developmental time, and identified the segregation of several lineages, including the inner cell mass, the early trophectoderm, and a polar trophectoderm36 (Fig. 6b). In contrast with the published analysis, the identification and characterization of these cellular populations was completely unsupervised, highlighting the value of scTDA for characterizing cell differentiation processes, a priori.
Finally, we used scTDA to analyse 272 differentiating neurons in the mouse neo-cortex37 (Supplementary Table 7). scTDA identified a continuum of cellular states with a bifurcation between apical and basal progenitors, and neurons (Fig. 6c). The topological representation more accurately reflects a basal to apical progenitor migration and a split in potency, with apical progenitors sharing transcriptional profiles that more closely reflect their neuronal counterparts. scTDA faithfully represented nonlinear and converging cellular lineages. The convergence of distinct lineages cannot be identified by tree-like based algorithms.
Discussion
In order to preserve the continuity of cellular differentiation, account for asynchronous development, and enable statistical interpretation of patterns of gene activation we used a method that exploits topological data analysis. Our method, named scTDA, enables unsupervised analyses of single-cell RNA-seq datasets over time.
We applied scTDA to study the in vitro differentiation of mESCs into neurons1, 2 and showed that it accurately recapitulated cellular differentiation, and provided the statistics needed to identify and characterize the transcriptional programs that accompany lineage commitment. We comprehensively characterized the dynamic appearance of mRNAs encoding signaling proteins, transcription factors, RNA splicing factors and long non-coding RNAs (lncRNAs). These transcripts were dynamically regulated during the transition from pluripotent cells to neural precursors, progenitors, motor neurons, and interneurons, thus providing a valuable resource for studies of stem cell differentiation and neurogenesis, and more generally to any cellular differentiation process amenable to single cell transcriptomic analysis. Inclusion of topological data enabled scTDA to identify additional processes coupled to differentiation, such as the cell cycle, which cannot be captured by tree-like structures. Furthermore, scTDA identified extensive transcriptional co-regulation of thousands of coding and noncoding genes in precursor, progenitor and neuronal populations. Some of these, such as the surface markers Pecam1, Ednrb, and Slc10a4, respectively for the pluripotent, neural progenitor, and mature motor neuron populations, as well as the unique expression of the lncRNA Gm12688 by V1 interneurons were validated.
In principle, scTDA can be applied to any biological system responding to inductive cues or environmental perturbations (Fig. 6). For example, scTDA could be used to study cellular differentiation processes such as hematopoiesis, the evolution of cancer cells, neurodegeneration, or developmental disorders, all of which arise from extracellular signals and mutations that culminate in heterogeneous transcriptional responses and cellular behavior.
Accession Codes
We deposited the single-cell RNA-seq data and read counts from the two motor neuron differentiation experiments in the NCBI Gene Expression Omnibus (GEO) database, with accession number GSE94883. The single-cell RNA-seq data from the differentiating distal lung epithelium of mouse embryo35 can be accessed through the GEO database, accession number GSE52583. The single-cell RNA-seq data from human pre-implantation embryos36 can be accessed through the Array Express database, with accession number E-MTAB-3929. The single-cell RNA-seq data from differentiating neurons in the mouse neo-cortex37 can be accessed at https://vpn.ias.edu/science2016/,DanaInfo=genebrowser.unige.ch+telley_govindan_science2016.tgz.
Online Methods
Cell culture and single cell isolation
Murine embryonic stem cell (mESC) based differentiations were performed using the method of Wichterle et al.2. In brief, stem cell colonies are expanded on an adherent substrate, after which they are dissociated and monodispersed in a serum-free suspension (day 0). Individual stem cells aggregate into embryoid bodies (EBs), which maintain exclusive expression of pluripotency markers until they are induced down the neuron lineage by addition of RA and Smoothened Agonist (SAG) (day 2). Metabolized culture medium is replenished on day 5, coincident with the appearance of early eGFP positive cells within the EBs. EBs were dissociated into single cells using the Worthington Biochemical Papain Dissociation System (LK003178). Single cell deposition was accomplished using a Beckman Coulter MoFlo Astrios EQ cell sorter into 96 well plates. Cells were then snap-frozen and subsequently lysed.
Single cell library generation
To obtain single cell expression profiles, modified CEL-Seq20 was carried out as described in the Supplementary Note 2. Briefly, we reverse transcribed the mRNA in each cell lysate with barcoded primers, pooled the single cell samples, synthesized second strand cDNA, and then linearly amplified by in vitro transcription with T7 RNA polymerase. The amplified RNA (aRNA) was chemically fragmented and T4 RNA ligase was used to ligate Illumina 3′-RNA adapters. The aRNA was then run on an Agilent Bioanalyzer to assess proper fragmentation and then reverse transcribed to generate cDNA and subjected to PCR enrichment. The subsequent multiplexed samples were paired end sequenced (2×125 bps) on an Illumina HiSeq 2500. Bulk RNA samples were purified from 2×106 cells in 1 mL of Trizol and standard Qiaquick RNA extraction protocols with a RIN 9.8 or higher. Stranded RNA-seq libraries were generated at the New York Genome Center using a TruSeq Stranded Total RNA Library Prep Kit. Stranded cDNA libraries were paired end sequenced (2×125 bps) on a HiSeq 2500, operating in high output mode, yielding 30M reads per indexed library.
Immunofluorescence
EBs were washed three times in ice cold PBS and fixed for two hours at room temperature in 4% PFA. They were then washed with ice cold PBS and either embedded into OCT and stored at -80°C for sectioning, or stored in PBS at 4°C for whole EB staining. 20um sections were cut using a Leica CM 1950 cryotome and mounted onto a glass slide. Whole and sectioned EBs were incubated in blocking solution (0.1% tween-20, 5% donkey serum, 1% BSA) for two hours at room temperature followed by primary antibodies diluted in blocking solution overnight. Primary antibodies used were: goat-αSlc9a1 (Santa Cruz sc-16097, 1:100), rabbit-αOlig2 (Millipore AB9610, 1:100), mouse-αOlig2: clone 21F1.1 (Millipore MABN50, 1:100), rabbit-αEdnrb (Novus NLS54, 1:200), guinea pig-αVAChT (gift from Dr. Neil Schneider's lab, 1:400), rabbit-αSlc10a4 (Novus Biologicals NBP1-81134, 1:100), mouse-αPou5f1 (BD Biosciences 611202, 1:100), goat-αPECAM1 (Santa Cruz sc-1506, 1:100), sheep-αCD44 (R&D Systems AF6127, 1:250), goat-αFoxc1 (Santa Cruz sc-21396, 1:50), and goat-αSstr2 (Santa Cruz sc-11606, 1:50). Alexa Fluor-conjugated secondary antibodies from Life Technologies were used at a 1:1000 dilution for two hours at room temperature. CD44, Foxc1 and Sstr2 were respectively conjugated to Cy-5, Alexa-405, and Cy-3 fluorophores using the DyLight system from Abcam (ab201798, ab188287, ab188288). Coverslips were mounted with Vectashield and EBs were imaged on an Olympus Fluoview FV1000 microscope using Olympus Fluoview v4.1.
Processing of single cell RNA-seq data
Paired-end 125bp reads were de-multiplexed, trimmed, and mapped to the UCSC mouse reference (mm10) using Tophat38. Gene expression was quantified using transcript read counts as derived from HTSeq39. Read counts were normalized as,
where ri denotes the unambiguous read count for transcript i, and r denotes the total number of reads that are mapped to transcripts in the cell. A strategy based on spike-in read counts, as described by Stegle et al.40, was implemented to filter out cells with a low content of mapped RNA and/or low sequencing depth. Specifically, the ratio ∊1 between average spike-in reads in the cell and average spike-in reads in the library (for spike-ins with an average of more than five reads in the library) was used to discard cells with low sequencing coverage. Similarly, the ratio ∊2 between the total number of spike-in reads in the cell and the total number of mapped reads was used to discard cells with a low number of mapped reads, relative to the sequencing coverage. Those cells showed very low expression of house-keeping genes, possibly representing cells under stressed conditions and/or with large amounts of degraded RNA. Based on the distribution of ∊1 and ∊2 in each of the two experiments (Supplementary Fig. 2a), cells with ∊1 > 5 · 10−4 and 0.7 > ∊2 > 0.01 in the experiment 1, and cells with 4.0 > ∊1 > 0.05 and 1.0 > ∈2 in experiment 2, were kept for subsequent analysis. The distribution of filtered out cells across different libraries was uniform in both experiments (Supplementary Fig. 2b). To reduce the noise observed near detection threshold in experiment 1, read counts with ri < 5 were set to zero. Additionally, one of the libraries (RPI36) was discarded from subsequent analysis, because it presented a large batch effect. To assess the dependence of the library complexity on the differentiation time (Supplementary Fig. 3), we computed at each time point the distribution of the geometric library size, defined as the sum of log expression values over all genes in a cell22.
Topological representation
The algorithm Mapper19 (Supplementary Note 1) was used to build topological representations of the RNA-seq data, through the implementation provided by Ayasdi Inc. Several open-source implementations of Mapper are available (https://github.com/MLWave/kepler-mapper, http://danifold.net/mapper/, https://github.com/RabadanLab/sakmapper, https://github.com/paultpearson/TDAmapper). In brief, the processed RNA-seq data was endowed with a dissimilarity matrix by taking pairwise correlation distance (1 - Pearson correlation). To minimize the effect of drop-out events present in single-cell data, we only considered the 5,000 genes (for experiment 1) and the 4,600 genes (for experiment 2) with highest variance across each dataset. These are highly expressed transcripts for which the probability of not being captured by the RNA amplification (drop-out events) is small41-44. The space was reduced to R2 using MDS, as displayed in Fig. 3c. A covering of R2 consisting of 26×26 (62×62) rectangular patches was considered for experiment 1 (2), respectively. The size of the patches was chosen such that the number of cells in each row or column of patches was the same, hence avoiding sampling density biases. The overlap between patches was 66% (on average). Single linkage clustering was performed in each of the pre-images of the patches using the algorithm described in Singh et al.19. A network was constructed in which each vertex corresponds to a cluster, and edges correspond to non-vanishing intersections between clusters. We checked for the absence of batch effects in the topological representation by verifying that batches from the same timepoint had a substantial overlap in the representation (Supplementary Figs. 3d and 7) and the stability against different choices for the threshold for the number of genes used to compute the distance matrix and for the covering of R2 (Supplementary Fig. 12).
We verified that our analysis was reproducible using a freely-available open-source implementation of Mapper. To that end, we repeated the analysis of experiment 1 using a topological representation built with Sakmapper (https://github.com/RabadanLab/sakmapper) (Supplementary Fig. 20). Comparison with the results of the scTDA analysis presented in the main text shows a clear consistency between the two analyses. In particular, 78% of the genes with significant gene connectivity (q-value < 0.05) in the representation generated with Sakmapper also displayed significant gene connectivity in the Ayasdi representation (Fisher exact test p-value < 10-100). The centroid and dispersion of those genes were also consistent between the two topological representations, with Pearson's correlation coefficient r = 0.98 (p-value < 10-100) for the centroid (Supplementary Fig. 20b), and r = 0.94 (p-value < 10-100) for the dispersion (Supplementary Fig. 20c) of significant genes.
Gene connectivity, centroid and dispersion within the topological representation
A notion of gene connectivity in the topological representation was introduced, defined as
where ei,α represents the average expression of gene i in node α of the topological representation, normalized as described in the paragraph “Processing of single cell RNA-seq data”; Γ denotes the set of nodes of the topological representation; Aαβ is its adjacency matrix; and N the total number of nodes. With this normalization, si takes values between 0 and 1.
The gene connectivity score depends on the distribution of expression values of the specific gene (Supplementary Fig. 10), and therefore genes cannot be ranked accordingly to their gene connectivity score in a meaningful way. To assess the magnitude of the connectivity score relative to genes with the same expression profile, and rank genes accordingly, we introduced a non-parametric statistical test. We tested for the null hypothesis of a randomly expressed gene with the same distribution of expression values having a higher gene connectivity score. To that end, a null-distribution was built for each gene i using a permutation test. Cell labels were randomly permuted 5,000 times for each gene, computing si after each permutation. A p-value was estimated by counting the fraction of permutations that led to a larger value of si than the original one. Gene connectivity and its statistical significance were computed for each gene expressed in at least three cells. The resulting p-values were adjusted for multiple testing by using Benjamini-Hochberg procedure for controlling the false discovery rate.
To establish a pseudo-temporal ordering within the topological representation, a notion of root node was introduced. The latter was defined as the node that maximizes the function,
where corr(x,y) denotes Pearson's correlation coefficient between x and y; dα is the graph distance function to node α, that assigns to each node of the topological representation a value corresponding to the number of edges that are crossed in the shortest path from that node to node α; and t is the chronological sampling time function, that assigns to each node of the topological representation the average sampling time (expressed in days) of the cells contained in the node.
Least-squares linear regression was performed to determine the best fit for the coefficients a0 and a1 in the relation
where droot is the graph distance function to the root node, determined in the previous paragraph. These coefficients were used to define the centroid and dispersion of each gene in the topological representation, expressed in days, and given respectively by
and
Such normalization, i.e. using coefficients a0 and a1 to express the centroid and dispersion in units of pseudo-time (days), allows comparing the connectivity and dispersion of a gene across different topological representations or studies.
Significance of topological features
We computed the first persistent homology group45, 46 using the graph distance of the topological representation. Given the pairwise distances of a set of points sampled from a space, persistent homology enables the quantification of topological features (connected components, loops, cavities, etc., preserved under continuous deformations of the space) compatible with the data at each scale. The first homology group, in particular, classifies loops of the space (Supplementary Note 1). We used persistent homology death times as a proxy of the size of the loops, and evaluated their statistical significance using a permutation test. To that end, we randomly permuted the labels of the genes 500 times, for each cell independently. For each permutation we built a topological representation using the same parameters as in the original representation and computed the first persistent homology group. A p-value for each of the loops was estimated from the distribution of the number of loops as a function of their death time. The resulting p-values were adjusted for multiple testing by using Benjamini-Hochberg procedure for controlling the false discovery rate.
Comparison with other methods for analyzing longitudinal single-cell RNA-seq data
We dimensionally reduced the processed single cell RNA-seq data of experiment 2 using MDS, ICA and tSNE. For MDS and ICA we used the same set of highly expressed, highly variant genes that we used for building the topological representation. For tSNE, we used the top 10 principal components of the gene space. In each representation we determined the cell that maximized the Pearson correlation coefficient between the two-dimensional Euclidean distance to the cell and chronological sampling time, corresponding to the least differentiated cellular state.
Additionally, we compared scTDA to the single cell software Monocle10, based on ICA and minimum-spanning trees, Diffusion Pseudotime6 and Wishbone7, based on diffusion coefficients, and SLICER8, based on locally linear embedding. We followed all recommendations in the documentation of these algorithms. In our tests, Monocle failed in running over the complete dataset from experiment 2, consisting of 2,304 cells, and only a partial set of 834 cells, sampled from all time points was analyzed.
Simulated data
Noisy, branched asynchronous cellular differentiation processes were simulated, from which 700 cell were sampled at three time points. To that end, we used the following strategy:
- We simulated a noisy branched tree-like structure with three parameters (t, u, and v), and performed a non-linear transformation into four-dimensional embedding space (spanned by the variables g(1), g(2), g(3), and g(4)). This space provides the structure for four different groups of correlated genes.
- We randomly sampled 700 points from this space, corresponding to 700 cells, and assigned a sampling day based on a multinomial distribution with probabilities given by a logistic function of t, to simulate asynchrony.
- We simulated the expression of 300 genes driven by the variables g(1), g(2), g(3), and g(4), in addition to 200 genes with non-correlated expression, sampled from normal distributions.
- We simulated the effect of drop-out events using the standard logistic dependence of the drop-out probability as a function of expression.
In what follows, we provide a detailed description of each of these steps.
First, we simulated four groups of correlated genes. These were defined by the equations,
where , are normally-distributed random variables with mean μ = 150 and standard deviation σ = 8. The index i = 1, …, 700 runs across the sampled cells, and ui, vi, and ti are randomly sampled from
where only the first 2 + r equations are considered in simulated differentiation processes with r lineage branching points. The variable ti represents the differentiation pseudo-time of cell i at the time of sampling. To simulate asynchrony, each sampled cell was assigned a sampling day accordingly to
where pi ∈ [0,1] is a random number uniformly distributed, and τ is the logistic function
We simulated 75 genes in each of the four groups of genes, with expression values given by
where and are normally-distributed random variables with mean μ = 1 and standard deviation σ = 0.2. In addition, we simulated 200 extra genes with non-correlated expression
with and normally-distributed random variables with mean μ = 200 and 1, and standard deviation σ = 50 and 0.2, respectively. Hence, a total of 500 genes were simulated in each of the 700 cells. To model the effect of drop-out events, we randomly set to zero the expression of some of the genes in some of the cells, with probability
where m is the original expression value of the gene in the cell.
Apart from scTDA, we ran the algorithms Diffusion Pseudotime6, Wishbone7, and SLICER8 in the simulated datasets. We followed all the recommendations in the documentation of these algorithms. We assessed the performance of each algorithm by computing the Pearson's correlation coefficient between the cell pseudotime inferred by each of these algorithms and the simulated variable t.
Gene ontology annotation
Gene ontologies were obtained from EMBL-EBI QuickGO47. Specifically, categories GO:0006355 “Regulation of transcription, DNA-templated”, GO:0008380 “RNA splicing”, GO:0044822 “Poly(A) RNA binding”, GO:0051726 “Regulation of cell cycle”, and GO:0007049 “Cell cycle” were used to annotate genes. Expression of genes associated to DNA replication was analyzed by considering the 99 genes in the category GO:0006260 “DNA replication” expressed in less than 1,400 cells in experiment 2. Genes coding for proteins in the cellular surface were identified by looking in UniProtKB database48 for proteins annotated with an extracellular topological domain. Gene ontology enrichment analysis was performed using PANTHER classification system23.
Transient cellular populations
Low-dispersion genes (ki < 1.7 days and ki < 2.25 days respectively in experiments 2 and 1) with significant gene connectivity (q < 0.05) in the topological representation were clustered according to their centroid using k-means clustering (Supplementary Fig. 13b and 13c). The optimal number of clusters according to Davies-Bouldin index was four in experiment 2 (three in experiment 1) (Supplementary Fig. 13a), as it was also evidenced from visual inspection of the centroid distribution for low-dispersion genes. A state r = 1, …, 4 was assigned to each node of the topological representation based on the average expression of each cluster of low-dispersion genes in the node (Supplementary Fig. 13d). Genes with significant gene connectivity according to the permutation test described in the paragraph “Gene connectivity, centroid, and dispersion within the topological representation” were assigned to each of the four populations based on the number of cells expressing the gene in each state r. Only genes expressed in at least 80 cells and at most 1,500 cells were considered.
Analysis of long non-coding RNAs
The coordinates of intergenic and antisense lncRNAs were downloaded from NONCODEv449, and read counts were obtained using HTSeq (http://www-huber.embl.de/users/anders/HTSeq). The connectivity and statistical significance of each long non-coding gene in the topological representation was computed using scTDA. Only lncRNAs that were significant (q < 0.05) in both experiments 1 and 2, and that were supported by at least 50 reads in the longitudinal stranded RNA-seq data were kept. Curation was performed to remove lncRNAs whose 3′-end overlapped the 3′-end of another gene, gene assignment therefore being ambiguous, or that corresponded to possible miss-annotations of the 3′ UTR of a nearby gene.
Characterization of interneuron populations
Differential expression analysis between En1+, Gata3+, Vsx2+ and Egfp+ cells in nodes characterized as post-mitotic (Supplementary Fig. 13d, state 3) was performed using the software SCDE42 with default parameters.
Analysis of mouse and human developmental datasets
We downloaded the read count data of the single cell RNA-seq experiments performed in refs. 35 (mouse distal lung epithelium), 36 (human preimplantation embryos), and 37 (mouse neocortex), normalized it as described in the paragraph “Processing of single cell RNA-seq data”, and anlysed it following the same steps described in the paragraphs “Topological representation” and “Gene connectivity, centroid and dispersion within the topological representation”. The topological representations were built using the Ayasdi implementation of Mapper, using correlation distance as metric and two dimensional MDS as dimensional reduction algorithm. We used a covering of R2 consisting of 33×33, 52×52, and 30×30 rectangular patches with an average overlap of 73%, 70%, and 71%, respectively for the data from mouse distal lung epithelium, human preimplantation embryos, and mouse neocortex. In the case of the data from human preimplantation embryos, we also performed an analysis of transient cellular populations, following the same steps described in paragraph “Transient cellular populations” and setting a threshold of 2.95 in gene dispersion.
scTDA software
The algorithms described in this work were implemented and documented in an object oriented python library for topological data analysis of high-throughput longitudinal single-cell RNA-seq data, called scTDA. scTDA is provided as Supplementary Code and is publicly available at https://github.com/RabadanLab/scTDA.
Online database
We established a database for the topological representations and statistics of the two motor neuron differentiation datasets. The database is publicly available at https://rabadan.c2b2.columbia.edu/motor_neurons_tda.
Supplementary Material
Acknowledgments
We thank Tom Jessell, Nicole Francis, and Hemali Phatnani for critical reading of the manuscript. A.H.R. and T.M. thank the New York Genome Center and D. Goldstein for sequencing support, Susan Morton for providing Engrailed antibody and P. Sims for experimental discussions. P.G.C. and R.R. thank A. J. Levine, F. Abate, I. Filip, S. Zairis, U. Rubin, and P. van Nieuwenhuizen for useful comments and discussions, and O. T. Elliott for technical support with the online database. The work of P.G.C and R.R. is supported by the NIH grants U54-CA193313-01 and R01GM117591. The work of A.H.R., E.K.K., T.J.R. and T.M. is supported by ALS Therapy Alliance grant ATA-2013-F-056 and NIH grant NS088992.
Footnotes
Author Contributions: P.G.C. and R.R. developed the topology-based computational approach (scTDA) and applied it to single cell RNA sequencing data. A.H.R., E.K.K, and T.M. designed all experiments. A.H.R., E.K.K., and T.J.R. conducted experiments. I.S. conducted all flow cytometry. A.H.R., P.G.C., E.K.K., T.M., and R.R. analyzed the data and wrote the manuscript.
Competing Financial Interests Statement: The authors declare no competing financial interests.
References
- 1.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 2.Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 3.Sances S, et al. Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nature neuroscience. 2016;16:542–553. doi: 10.1038/nn.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phatnani HP, et al. Intricate interplay between astrocytes and motor neurons in ALS. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E756–765. doi: 10.1073/pnas.1222361110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bratt-Leal AM, Carpenedo RL, McDevitt TC. Engineering the embryoid body microenvironment to direct embryonic stem cell differentiation. Biotechnology progress. 2009;25:43–51. doi: 10.1002/btpr.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haghverdi L, Buttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nature methods. 2016;13:845–848. doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- 7.Setty M, et al. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat Biotechnol. 2016;34:637–645. doi: 10.1038/nbt.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welch JD, Hartemink AJ, Prins JF. SLICER: inferring branched, nonlinear cellular trajectories from single cell RNA-seq data. Genome biology. 2016;17:106. doi: 10.1186/s13059-016-0975-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angerer P, et al. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics. 2016;32:1241–1243. doi: 10.1093/bioinformatics/btv715. [DOI] [PubMed] [Google Scholar]
- 10.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marco E, et al. Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proc Natl Acad Sci U S A. 2014;111:E5643–5650. doi: 10.1073/pnas.1408993111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan JM, Carlsson G, Rabadan R. Topology of viral evolution. Proc Natl Acad Sci U S A. 2013;110:18566–18571. doi: 10.1073/pnas.1313480110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camara PG, Levine AJ, Rabadan R. Inference of Ancestral Recombination Graphs through Topological Data Analysis. PLoS computational biology. 2016;12:e1005071. doi: 10.1371/journal.pcbi.1005071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camara PG, Rosenbloom DI, Emmett KJ, Levine AJ, Rabadan R. Topological Data Analysis Generates High-Resolution, Genome-wide Maps of Human Recombination. Cell Syst. 2016;3:83–94. doi: 10.1016/j.cels.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicolau M, Levine AJ, Carlsson G. Topology based data analysis identifies a subgroup of breast cancers with a unique mutational profile and excellent survival. Proc Natl Acad Sci U S A. 2011;108:7265–7270. doi: 10.1073/pnas.1102826108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, et al. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci Transl Med. 2015;7:311ra174. doi: 10.1126/scitranslmed.aaa9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bendall SC, et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157:714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh G, Mémoli F, Carlsson GE. SPBG 91-100. Citeseer; 2007. [Google Scholar]
- 20.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: Single-Cell RNA-Seq by Multiplexed Linear Amplification. Cell Reports. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Finak G, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome biology. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDavid A, Finak G, Gottardo R. The contribution of cell cycle to heterogeneity in single-cell RNA-seq data. Nat Biotechnol. 2016;34:591–593. doi: 10.1038/nbt.3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balmer JE. Gene expression regulation by retinoic acid. The Journal of Lipid Research. 2002;43:1773–1808. doi: 10.1194/jlr.r100015-jlr200. [DOI] [PubMed] [Google Scholar]
- 25.Rhinn M, Dolle P. Retinoic acid signalling during development. Development. 2012;139:843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- 26.Gaunt SJ, Strachan L. Temporal colinearity in expression of anterior Hox genes in developing chick embryos. Developmental dynamics : an official publication of the American Association of Anatomists. 1996;207:270–280. doi: 10.1002/(SICI)1097-0177(199611)207:3<270::AID-AJA4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Weissman SM, Newburger PE. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biology. 2014;11:777–787. doi: 10.4161/rna.28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin M, et al. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PloS one. 2011;6:e23356. doi: 10.1371/journal.pone.0023356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mallo M, Alonso CR. The regulation of Hox gene expression during animal development. Development. 2013;140:3951–3963. doi: 10.1242/dev.068346. [DOI] [PubMed] [Google Scholar]
- 30.Dinger ME, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukas Sommer QM, David J. Anderson neurogenins, a Novel Family of atonal-Related bHLH Transcription Factors, Are Putative Mammalian Neuronal Determination Genes That Reveal Progenitor Cell Heterogeneity in the Developing CNS and PNS. Molecular and Cellular Neuroscience. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- 32.Darnell RB. RNA protein interaction in neurons. Annu Rev Neurosci. 2013;36:243–270. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quesnel-Vallieres M, Irimia M, Cordes SP, Blencowe BJ. Essential roles for the splicing regulator nSR100/SRRM4 during nervous system development. Genes & development. 2015;29:746–759. doi: 10.1101/gad.256115.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calarco JA, et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 35.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petropoulos S, et al. Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell. 2016;165:1012–1026. doi: 10.1016/j.cell.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Telley L, et al. Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science. 2016;351:1443–1446. doi: 10.1126/science.aad8361. [DOI] [PubMed] [Google Scholar]
Rk f−1 Rk Methods-only References
- 38.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- 41.Grun D, Kester L, van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nature methods. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- 42.Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nature methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shalek AK, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brennecke P, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nature methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- 45.Edelsbrunner H, Letscher D, Zomorodian A. Topological persistence and simplification. Discrete and Computational Geometry. 2002;28:511–533. [Google Scholar]
- 46.Zomorodian A, Carlsson G. Computing persistent homology. Discrete & Computational Geometry. 2005;33:249–274. [Google Scholar]
- 47.Binns D, et al. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics. 2009;25:3045–3046. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Y, et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.