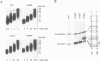Abstract
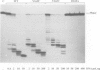
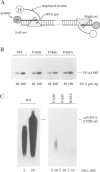
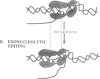
In this report we present the alignment of one of the most conserved segments (Exo III) of the 3'-5' exonuclease domain in 39 DNA polymerase sequences, including prokaryotic and eukaryotic enzymes. Site-directed substitutions of the two most conserved residues, which form the Exo III motif Tyr-(X)3-Asp of phi 29 DNA polymerase, did not affect single-stranded DNA binding, DNA polymerization, processivity or protein-primed initiation. In contrast, substitution of the highly conserved Tyr residue by Phe or Cys decreased the 3'-5' exonuclease activity to 7.5 and 4.1%, respectively, of the wild-type activity. Change of the highly conserved Asp residue into Ala resulted in almost complete inactivation (0.1%) of the 3'-5' exonuclease. In accordance with the contribution of the 3'-5' exonuclease to the fidelity of DNA replication, the three mutations in the Exo III motif (Y165F, Y165C and D169A) produced enzymes with an increased frequency of misinsertion and extension of DNA polymerization errors. Surprisingly, the three mutations in the Exo III motif strongly decreased (80- to 220-fold) the ability to replicate phi 29 DNA, this behaviour being due to a defect in the strand displacement activity, an intrinsic property of phi 29 DNA polymerase required for this process. Taking these results into account, we propose that the strand displacement activity of phi 29 DNA polymerase resides in the N-terminal domain, probably overlapping with the 3'-5' exonuclease active site.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argos P. Analysis of sequence-similar pentapeptides in unrelated protein tertiary structures. Strategies for protein folding and a guide for site-directed mutagenesis. J Mol Biol. 1987 Sep 20;197(2):331–348. doi: 10.1016/0022-2836(87)90127-6. [DOI] [PubMed] [Google Scholar]
- Beese L. S., Steitz T. A. Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991 Jan;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad A., Blanco L., Lázaro J. M., Martín G., Salas M. A conserved 3'----5' exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989 Oct 6;59(1):219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- Bernad A., Blanco L., Salas M. Site-directed mutagenesis of the YCDTDS amino acid motif of the phi 29 DNA polymerase. Gene. 1990 Sep 28;94(1):45–51. doi: 10.1016/0378-1119(90)90466-5. [DOI] [PubMed] [Google Scholar]
- Bernad A., Lázaro J. M., Salas M., Blanco L. The highly conserved amino acid sequence motif Tyr-Gly-Asp-Thr-Asp-Ser in alpha-like DNA polymerases is required by phage phi 29 DNA polymerase for protein-primed initiation and polymerization. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4610–4614. doi: 10.1073/pnas.87.12.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad A., Zaballos A., Salas M., Blanco L. Structural and functional relationships between prokaryotic and eukaryotic DNA polymerases. EMBO J. 1987 Dec 20;6(13):4219–4225. doi: 10.1002/j.1460-2075.1987.tb02770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco L., Bernad A., Blasco M. A., Salas M. A general structure for DNA-dependent DNA polymerases. Gene. 1991 Apr;100:27–38. doi: 10.1016/0378-1119(91)90346-d. [DOI] [PubMed] [Google Scholar]
- Blanco L., Bernad A., Lázaro J. M., Martín G., Garmendia C., Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. 1989 May 25;264(15):8935–8940. [PubMed] [Google Scholar]
- Blanco L., Bernad A., Salas M. Evidence favouring the hypothesis of a conserved 3'-5' exonuclease active site in DNA-dependent DNA polymerases. Gene. 1992 Mar 1;112(1):139–144. doi: 10.1016/0378-1119(92)90316-h. [DOI] [PubMed] [Google Scholar]
- Blanco L., Bernad A., Salas M. Transition from initiation to elongation in protein-primed phi 29 DNA replication: salt-dependent stimulation by the viral protein p6. J Virol. 1988 Nov;62(11):4167–4172. doi: 10.1128/jvi.62.11.4167-4172.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco L., Salas M. Characterization and purification of a phage phi 29-encoded DNA polymerase required for the initiation of replication. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5325–5329. doi: 10.1073/pnas.81.17.5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco L., Salas M. Replication of phage phi 29 DNA with purified terminal protein and DNA polymerase: synthesis of full-length phi 29 DNA. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6404–6408. doi: 10.1073/pnas.82.19.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco M. A., Bernad A., Blanco L., Salas M. Characterization and mapping of the pyrophosphorolytic activity of the phage phi 29 DNA polymerase. Involvement of amino acid motifs highly conserved in alpha-like DNA polymerases. J Biol Chem. 1991 Apr 25;266(12):7904–7909. [PubMed] [Google Scholar]
- Burgers P. M., Bambara R. A., Campbell J. L., Chang L. M., Downey K. M., Hübscher U., Lee M. Y., Linn S. M., So A. G., Spadari S. Revised nomenclature for eukaryotic DNA polymerases. Eur J Biochem. 1990 Aug 17;191(3):617–618. doi: 10.1111/j.1432-1033.1990.tb19165.x. [DOI] [PubMed] [Google Scholar]
- Carthew R. W., Chodosh L. A., Sharp P. A. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell. 1985 Dec;43(2 Pt 1):439–448. doi: 10.1016/0092-8674(85)90174-6. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Chung D. W., Zhang J. A., Tan C. K., Davie E. W., So A. G., Downey K. M. Primary structure of the catalytic subunit of human DNA polymerase delta and chromosomal location of the gene. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11197–11201. doi: 10.1073/pnas.88.24.11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton L. K., Goodman M. F., Branscomb E. W., Galas D. J. Error induction and correction by mutant and wild type T4 DNA polymerases. Kinetic error discrimination mechanisms. J Biol Chem. 1979 Mar 25;254(6):1902–1912. [PubMed] [Google Scholar]
- Derbyshire V., Freemont P. S., Sanderson M. R., Beese L., Friedman J. M., Joyce C. M., Steitz T. A. Genetic and crystallographic studies of the 3',5'-exonucleolytic site of DNA polymerase I. Science. 1988 Apr 8;240(4849):199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- Derbyshire V., Grindley N. D., Joyce C. M. The 3'-5' exonuclease of DNA polymerase I of Escherichia coli: contribution of each amino acid at the active site to the reaction. EMBO J. 1991 Jan;10(1):17–24. doi: 10.1002/j.1460-2075.1991.tb07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher M. P., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 28. The pyrophosphate exchange and pyrophosphorolysis reactions of deoxyribonucleic acid polymerase. J Biol Chem. 1969 Jun 10;244(11):3019–3028. [PubMed] [Google Scholar]
- Freemont P. S., Friedman J. M., Beese L. S., Sanderson M. R., Steitz T. A. Cocrystal structure of an editing complex of Klenow fragment with DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmendia C., Bernad A., Esteban J. A., Blanco L., Salas M. The bacteriophage phi 29 DNA polymerase, a proofreading enzyme. J Biol Chem. 1992 Feb 5;267(4):2594–2599. [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Gibbs J. S., Weisshart K., Digard P., deBruynKops A., Knipe D. M., Coen D. M. Polymerization activity of an alpha-like DNA polymerase requires a conserved 3'-5' exonuclease active site. Mol Cell Biol. 1991 Sep;11(9):4786–4795. doi: 10.1128/mcb.11.9.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose F., Yamaguchi M., Nishida Y., Masutani M., Miyazawa H., Hanaoka F., Matsukage A. Structure and expression during development of Drosophila melanogaster gene for DNA polymerase alpha. Nucleic Acids Res. 1991 Sep 25;19(18):4991–4998. doi: 10.1093/nar/19.18.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inciarte M. R., Viñuela E., Salas M. Transcription in vitro of phi29 DNA and EcoRI fragments by Bacillus subtilis RNA polymerase. Eur J Biochem. 1976 Dec;71(1):77–83. doi: 10.1111/j.1432-1033.1976.tb11091.x. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Leegwater P. A., Strating M., Murphy N. B., Kooy R. F., van der Vliet P. C., Overdulve J. P. The Trypanosoma brucei DNA polymerase alpha core subunit gene is developmentally regulated and linked to a constitutively expressed open reading frame. Nucleic Acids Res. 1991 Dec 11;19(23):6441–6447. doi: 10.1093/nar/19.23.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Morrison A., Bell J. B., Kunkel T. A., Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3'----5' exonuclease activity. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Patel S. S., Wong I., Johnson K. A. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991 Jan 15;30(2):511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- Peñalva M. A., Salas M. Initiation of phage phi 29 DNA replication in vitro: formation of a covalent complex between the terminal protein, p3, and 5'-dAMP. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5522–5526. doi: 10.1073/pnas.79.18.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignède G., Bouvier D., de Recondo A. M., Baldacci G. Characterization of the POL3 gene product from Schizosaccharomyces pombe indicates inter-species conservation of the catalytic subunit of DNA polymerase delta. J Mol Biol. 1991 Nov 20;222(2):209–218. doi: 10.1016/0022-2836(91)90207-m. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz L. J. Amino acid changes coded by bacteriophage T4 DNA polymerase mutator mutants. Relating structure to function. J Mol Biol. 1988 Aug 20;202(4):711–724. doi: 10.1016/0022-2836(88)90552-9. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz L. J. Locations of amino acid substitutions in bacteriophage T4 tsL56 DNA polymerase predict an N-terminal exonuclease domain. J Virol. 1989 Nov;63(11):4762–4766. doi: 10.1128/jvi.63.11.4762-4766.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reha-Krantz L. J., Stocki S., Nonay R. L., Dimayuga E., Goodrich L. D., Konigsberg W. H., Spicer E. K. DNA polymerization in the absence of exonucleolytic proofreading: in vivo and in vitro studies. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2417–2421. doi: 10.1073/pnas.88.6.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley R. G., White J. H., McAleese S. M., Goman M., Alano P., de Vries E., Kilbey B. J. DNA polymerase delta: gene sequences from Plasmodium falciparum indicate that this enzyme is more highly conserved than DNA polymerase alpha. Nucleic Acids Res. 1991 Dec 25;19(24):6731–6736. doi: 10.1093/nar/19.24.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- Simon M., Giot L., Faye G. The 3' to 5' exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991 Aug;10(8):2165–2170. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss E. C., Kobori J. A., Siu G., Hood L. E. Specific-primer-directed DNA sequencing. Anal Biochem. 1986 Apr;154(1):353–360. doi: 10.1016/0003-2697(86)90536-1. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Studwell P. S., O'Donnell M. Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J Biol Chem. 1990 Jan 15;265(2):1171–1178. [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. Selective inactivation of the exonuclease activity of bacteriophage T7 DNA polymerase by in vitro mutagenesis. J Biol Chem. 1989 Apr 15;264(11):6447–6458. [PubMed] [Google Scholar]
- Zaballos A., Lázaro J. M., Méndez E., Mellado R. P., Salas M. Effects of internal deletions on the priming activity of the phage phi 29 terminal protein. Gene. 1989 Nov 30;83(2):187–195. doi: 10.1016/0378-1119(89)90104-2. [DOI] [PubMed] [Google Scholar]
- Zhang J., Chung D. W., Tan C. K., Downey K. M., Davie E. W., So A. G. Primary structure of the catalytic subunit of calf thymus DNA polymerase delta: sequence similarities with other DNA polymerases. Biochemistry. 1991 Dec 24;30(51):11742–11750. doi: 10.1021/bi00115a002. [DOI] [PubMed] [Google Scholar]