Abstract
An important question for bacterial cell division is how the invaginating septum can overcome the turgor force generated by the high osmolarity of the cytoplasm. I suggest that it may not need to. Several studies in Gram-negative bacteria have shown that the periplasm is isoosmolar with the cytoplasm. Indirect evidence suggests that this is also true for Gram-positive bacteria. In this case the invagination of the septum takes place within the uniformly high osmotic pressure environment, and does not have to fight turgor pressure. A related question is how the V-shaped constriction of Gram-negative bacteria relates to the plate-like septum of Gram-positive bacteria. I collect evidence that Gram-negative bacteria have a latent capability of forming plate-like septa, and present a model in which septal division is the basic mechanism in both Gram-positive and Gram-negative bacteria.
Keywords: bacteria, division, cytokinesis, FtsZ, tubulin, peptidoglycan, turgor
Introduction
In bacterial cytokinesis FtsZ provides the cytoskeletal framework for the Z ring, which serves as the docking site for up to two dozen other division proteins. The Z ring assembles early in the cell cycle and remains in place without any obvious constriction for most of the cycle. Near the end of the cell cycle the Z ring, including all the downstream proteins, constricts over a period of minutes to pinch the cell in two. The mechanisms of FtsZ have been investigated intensively since the discovery 25 years ago that FtsZ is a homolog of eukaryotic tubulin, and that it is localized at the site of constriction [1-5].
Bacteria are surrounded by a cell envelope. In Gram-positive bacteria the cell envelope consists of an inner lipid membrane (IM) and a rigid cell wall composed primarily of peptidoglycan (the abbreviation PG will refer to the cell wall, implicitly including all other components). Gram-negative bacteria have an additional outer lipid membrane (OM), which is closely attached to the PG. The PG layer is separated from the IM by a variable space termed the periplasm, which is a major focus of the present analysis. To achieve division all layers of the cell envelope must invaginate into the division site, although not at the same time, and eventually fully cover the daughter cells.
In 2008 Osawa et al. [6] demonstrated that FtsZ-mts could assemble Z rings when reconstituted in tubular liposomes. The mts (membrane-targeting sequence) is an amphipathic helix that can tether FtsZ directly to membrane. These reconstituted Z rings could constrict the thick walls of multilamellar liposomes. Both Z-ring assembly and constriction were achieved by FtsZ-mts alone – no other division protein was needed. This discovery elevated FtsZ as the prime candidate for generating the constriction force in bacterial cell division.
How could FtsZ constrict membranes? Previous work had demonstrated that FtsZ protofilaments can switch conformation from straight to curved, and it was suggested that the curved protofilaments could generate a bending force on the membrane and pull it inward [7, 8]. Additional experiments with “inside-out Z rings” supported this mechanism [9-12].
Recently this Z-centric hypothesis has been questioned. Coltharp and Xiao [13, 14] suggested that the force from bending FtsZ protofilaments would be insufficient to constrict the IM against the turgor pressure generated by the high osmolarity of the bacterial cytoplasm. They suggested that the major force for constriction is likely generated by inward growth of the PG wall, pushing the IM from the outside. In this mechanism the role of FtsZ is primarily to serve as a docking site for the PG synthetic enzymes, and to regulate assembly of the PG.
Here I am not arguing for one constriction mechanism or the other. I present the hypothesis that the essential steps of cell division, constricting the IM and building new PG to form a septum, likely take place completely within the high osmolarity environment, and never have to resist the turgor pressure. This would mean that both potential mechanisms, FtsZ bending and cell wall ingrowth, would need only modest forces to achieve division.
The cell envelope of Gram-positive and Gram-negative bacteria visualized by cryoEM
The structural relationships of the membranes and PG wall have been determined by thin section electron microscopy (EM). In the early days of EM, bacteria were chemically fixed, dehydrated and infiltrated with plastic, and thin sections were cut. Aldehydes and OsO4 were the most popular fixation agents. There was, however, always concern about changes in the structure that might occur during the time for the fixation to work and during subsequent processing.
Rapid freezing offered the promise of instantaneous fixation, but ice crystal formation disrupted tissue structures more than a few μm from the freezing surface. Cryoprotectants could reduce ice crystals, but might alter the cellular structures. A major advance was the development of high pressure freezing; see McDonald for a review of the history and applications [15]. Briefly, the sample is subjected to ∼2,000 bar pressure simultaneously with rapid freezing, both being achieved in ∼30 msec. The high pressure prevents formation of ice crystals, and the rapid freezing preserves the organic structures. The frozen specimen can then be sectioned with a cryo microtome. This generally involves no chemical stain and relies on the natural contrast of the membranes and protein structures with surrounding water. Most of the results discussed below (Figs. 1-3) were obtained from high pressure freezing followed by cryosectioning. An alternative to cryosectioning is cryoEM tomography, where the whole bacterium is imaged at multiple tilts and a 3-D image is reconstructed (Fig. 4). The thin samples for tomography are typically prepared by plunge freezing without the need for high pressure.
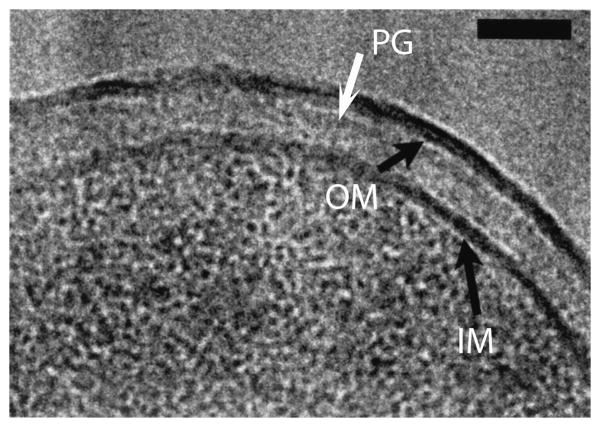
Figure 1.
CryoEM (high pressure freezing and cryosection) of the cell envelope of E. coli K-12. The black arrows point to the IM and OM. The PG is the lightly stained narrow band indicated by the white arrow. The periplasm is the lighter stained zone between the PG and IM. Bar is 50 nm. Reprinted from [16].
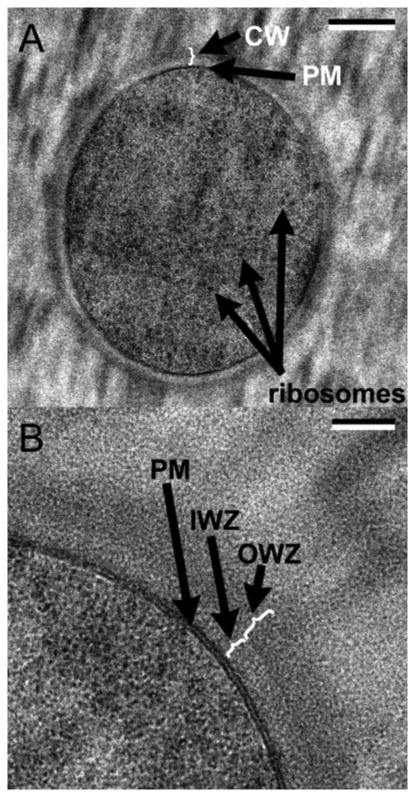
Figure 2.
CryoEM of the cell envelope of B. subtilis. A: cross section through the middle of a whole cell. B: higher magnification view. IM is designated PM (plasma membrane), periplasm is designated IWZ and the PG layer is OWZ. Bars are 200 nm (A) and 50 nm (B). Reprinted from [18].
Figure 3.
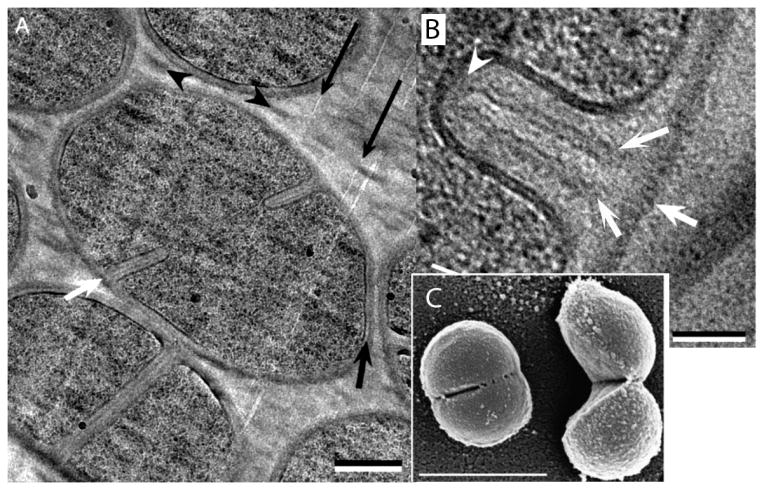
Septal division of S. aureus. A: The bacterium at lower left has a completed septum but has not separated. The bacterium in the center has a septum partially invaginated. B: A newly forming septum in cross-section. White arrows indicate the PG wall of the mother cell and the two forming daughter PG layers. (C) A scanning EM of a cell just beginning to pop, and one that has just popped. (A and B) are from [33], (C) is from [34]. Bars are (A) 250 nm, (B) 50 nm, (C) 1,000 nm.
Figure 4.
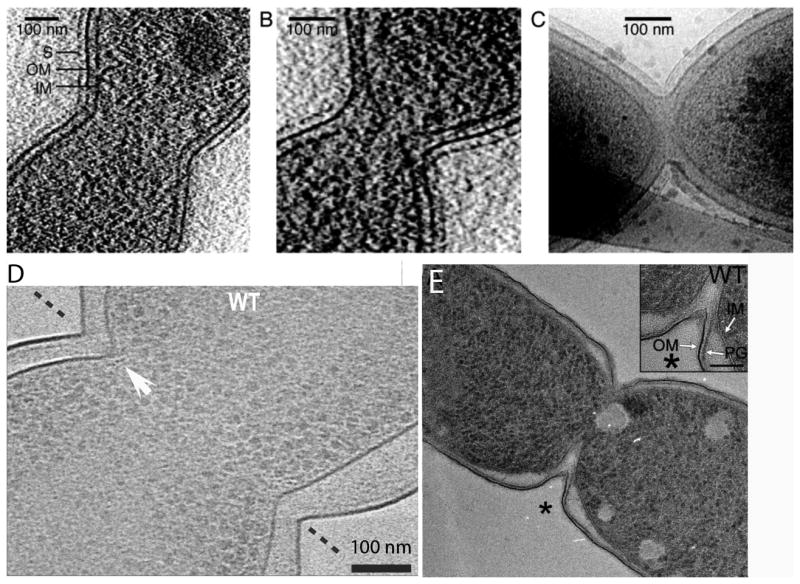
A-C: Division in C. crescentus imaged by cryoEM tomography (reprinted from [37]). The shallow V shape was interpreted to indicate a constrictive mode of division. A and B are slices through a 3D tomogram, while (C) is a projection through the whole bacterium. D: Division of E. coli imaged by cryoEM tomography, showing a slice through the 3D tomogram (reprinted from [39]. The white arrow points to small dots identified as FtsZ filaments in cross-section. This cell has a sharp, V-shaped constriction, typical of constrictive division. The periplasm of this cell is considerably expanded, especially at lower right. E: A dividing E. coli cell prepared by high pressure freezing, freeze substitution (reprinted from [40]. The division shows a V-shaped constriction and a clearly extended invagination of the IM, forming a septum.
Matias, Beveridge and colleagues applied the technique of high pressure freezing and cryosectioning to bacteria in a series of seminal papers from 2003-2008. Fig. 1 shows the cell envelope of E. coli K12 [16]. The IM and OM are each seen as a single, thin, dark staining layer. The PG appears as a line of slightly enhanced density close to the OM. The periplasm is the lighter staining space between the IM and the PG. Matias et al. [16] noted two important features of the periplasmic space: its width varied by ∼10% over non-compressed sections of the cell envelope; and it could be significantly compressed by the sectioning knife. They suggested that the periplasmic space has inherent compressibility or flexibility that allows the IM to move closer or farther away from the PG. It is not a rigid gel.
The term “periplasm” has had two different meanings in the literature. A number of authors use the term to refer to the entire space between the IM and OM, with the PG layer being contained within this periplasm. However, we will use the terms periplasm or periplasmic space to refer specifically to the space between the PG and IM. This designation was used in the original study of osmolarity of the compartments [17], and it is also consistent with the definition of periplasm in Gram-positive bacteria.
Early studies had suggested that in Gram-positive bacteria the IM was pressed in tight apposition to the relatively thick PG cell wall [18]. In 2005 Matias and Beveridge [18] overturned this model when they applied cryosectioning to image the envelope of Bacillus subtilis. Fig. 2 reproduces their definitive images. The cell envelope comprises the IM, resolved here as bilayer; an “intermediate wall zone” identified here as the periplasm; and a medium density “outer wall zone” that can be identified as the PG layer.
Several other Gram-positive bacteria have been imaged by the same technique, and dimensions of their cell envelopes are given in Table 1. For most of these the periplasm is ∼20 nm thick vs ∼10 nm for E. coli, and the PG is much thicker, ∼20-30 nm vs 6.4 nm for E. coli. An exception is Mycobacterium bovis, whose periplasm and PG are closer to the dimensions of E. coli. Interestingly, M. bovis and related Corynebacterineae were shown to have an outer lipid bilayer, chemically different but structurally similar to the OM of E. coli [19].
Table 1.
Dimensions of the cell envelope of various bacteria determined from cryoEM of cryosections following high pressure freezing.
| Bacterial strain | Thickness (nm) | Ref. | ||
|---|---|---|---|---|
| IM | periplasm | PG | ||
| E. coli K12 | 5.8 | 9.3 | 6.4 | [16] |
| E. coli K12 | 6.3 | 11.5 | 6.4 | [19] |
| P. aeruginosa PAO1 | 6.0 | 15 | 2.4 | [16] |
| B. subtilis 168 | 6.6 | 22 | 33 | [18] |
| S. aureus D2C | 5.4 | 16 | 19 | [49] |
| B. subtilis 168 | 20 | 30 | [35] | |
| B. subtilis W23 | 20 | 33 | [35] | |
| S. gordonii | 16 | 26 | [35] | |
| M. bovis | 6.3 | 14 | 6.3 | [19] |
The periplasm of Gram-negative bacteria is isoosmotic with the cytoplasm
An important concern for bacterial division is how it deals with the turgor force of the bacterial cytoplasm. To understand this we need to know which elements of the cell envelope generate and contain the turgor force.
In Gram-negative bacteria the OM is tightly coupled to the PG by the very abundant lipoprotein Lpp (other attachments exist but Lpp is the most abundant). Lpp is a triple helical rod, 8.3 nm long with three fatty acid chains on its N-terminus that insert into the OM [20]. About 1/3 of the chains are covalently attached to PG at the C terminus, so most of the trimers should form a tight bridge from the OM to PG. There are 2.4 × 105 Lpp trimers per cell [20], so they should be closely spaced on the OM, with ∼30 nm2 per Lpp trimer. In view of this tight coupling, the OM-PG will be considered to be a unit that creates the turgor pressure and provides the mechanical support to contain it. Note that this is the first filtration barrier an external osmolyte would encounter. If it passes this barrier it will enter the periplasm, where it will then encounter the more selective barrier of the IM.
The filtration barrier of the OM-PG layer is based primarily on the size of the osmolyte. Decad and Nikaido [21] assayed its permeability to sugars and concluded that “the cell wall acts as a molecular sieve, with an exclusion limit near 550 to 650 Daltons for saccharides.” The limit of 550-650 Da is probably imposed by the porins in the OM. Sucrose, at 342 Da, easily passes through the OM-PG wall into the periplasm. Importantly, the IM is impermeable to sucrose. This means that when sucrose is added to the outside medium it will diffuse through the OM-PG layer into the periplasm, but will not move across the IM into the cytoplasm.
The cytoplasm has a high concentration of charged macromolecules and salts to neutralize them, creating a high osmolarity and a turgor pressure. Over the years there have been two models for how this turgor pressure is supported. In one model the turgor pressure is generated by the IM. The IM is not structurally rigid, so it must be supported by the rigid PG layer. Since EM shows that the IM is separated from the PG wall by a 10 nm periplasmic space, this would require that the periplasm be a rigid, inelastic gel that can support the turgor pressure on the IM. In the second model the turgor pressure is generated by the OM-PG layer, and the cell maintains the periplasm in a high osmolar state to match that of the cytoplasm. The IM then experiences no turgor pressure, and floats freely between the cytoplasm and periplasm. This second model is now widely accepted, based on work discussed next.
Stock et al. [17] did the pioneering study of the periplasm of E. coli and Salmonella typhimurium using radiolabeled probes to determine the fraction of water in the cytoplasm and periplasm. They found that the periplasmic volume increased rapidly in response to an abrupt increase in sucrose in the external medium. As noted above, sucrose crosses the OM-PG barrier and rapidly increases the osmolarity of the periplasm, but cannot cross the IM. This causes water to flow from the cytoplasm, increasing its osmotic pressure until it matches the newly increased level of the periplasm. The flow of water from the cytoplasm reduces cytoplasmic volume and pulls the IM inward, substantially increasing the periplasmic volume. The IM does not retract uniformly, but forms a small number of “plasmolysis spaces,” often at the cell poles and division sites. Schwarz and Koch [22] imaged these expanded pockets of periplasm by EM, and Sochacki et al. [23] used periplasmic GFP to image them in living cells. Stock et al. concluded that the “cytoplasmic membrane is flexible and unable to support a pressure gradient…Under all conditions the periplasm and cytoplasm remained isoosmotic” [17].
The osmolarity of the Gram-negative periplasm is thought to be achieved by membrane-derived oligosaccharides (MDOs). MDOs are anionic glucose oligomers, ∼2,300 Da with an average –3 charge, that are too large to pass through the small pores of the OM-PG layer [24-26]. They create a Donnan equilibrium with an osmolarity that equals that of the cytoplasm. Upon plasmolysis the expanded spaces persist for 30 min or more, but if cells are maintained in a high osmolar medium they down-regulate synthesis of MDOs [24-26], the cytoplasm swells and the periplasm shrinks.
Cayley et al. [26] expanded the study to measure effects of osmotic shock on cells growing in low to high osmolar medium. This is now the definitive study of osmolarity of the periplasm. They concluded that “the periplasm and cytoplasm are isoosmotic, and that E. coli maintains turgor pressure across the cell wall and not across the cytoplasmic membrane” [26].
The periplasm of Gram-positive bacteria is probably isoosmotic with the cytoplasm
Since Gram-positive bacteria have no OM, the outer semipermeable barrier is the PG wall itself. The PG layer in Gram-positive bacteria is much thicker than in Gram-negative, and probably operates as a molecular sieve. The question is, what size molecules can pass through this sieve and what size are blocked?
Scherrer and Gerhardt [27] measured the ability of dextrans and polyglycols to permeate into living Bacillus megaterium. That paper is sometimes misunderstood as demonstrating that the “number average molecular weight” exclusion limit of the Bacillus cell wall is 70,000 to 120,000 Da. That was indeed their initial finding based on tests of unfractionated polymers. But the important point of their study was to recognize that the dextrans and polyglycols are polydisperse, and that the apparent passage is highly tilted toward the smaller molecules in the sample. Based on fractionated polyglycols they concluded that “…monodisperse molecules can penetrate the cell wall only if equivalently smaller than a glycol of Mn = 1,200 Da” [27]. This is about twice the size that can penetrate the OM-PG of Gram-negative bacteria.
Demchick and Koch [28] reported a much larger permeability barrier of 30-40 kDa for isolated B. subtilis PG sacculi. They used dextrans that were size fractionated, so their measures should not be distorted by polydispersity. However, the sacculi they assayed were prepared by treatment with boiling SDS and then with trypsin, which would remove all wall proteins. This may have increased the pore size.
Another indication of larger pore size is that Gram-positive bacteria are susceptible to antimicrobial peptides and proteins. The antimicrobial peptides are 10-50 amino acids (1,100-5,500 Da), which must traverse the peptidoglycan cell wall to reach the plasma membrane target [29]. Antimicrobial proteins, such as the 14 kDa phospholipase A2 are even larger [30]. However, in this case antimicrobial activity depended on cell growth and autolysins, suggesting that the large protein traversed the PG wall by active and perhaps transient openings during PG remodeling, not passive transport through fixed pores [30].
Perhaps the best evidence for an isoosmolar periplasm are the cryoEM images showing that the IM of Gram-positive bacteria is not plastered against the PG cell wall, but is separated by a ∼20 nm periplasmic space (Fig. 2) [18]. This means that, to support the cytoplasmic turgor pressure, the periplasm must either be a rigid gel or be isoosmotic with the cytoplasm. Evidence against a rigid gel was obtained by plasmolysis experiments [18]. Sudden treatment with 20% glycerol plus 5% NaCl caused the cytoplasm to shrink and the periplasmic space to expand. Some expanded periplasmic spaces contained small vesicles that could easily distort the space. Matias and Beveridge concluded that “Whatever is in the IWZ [periplasm], it is quite compactable…and can be deformed by the action of the vesicles” [18]. They suggested that there must be a substance in the periplasm that can balance the turgor pressure of the cytoplasm, even if it is not sufficiently concentrated to generate visible density in the EM. These structural observations suggest that the periplasm is isoosmotic with the cytoplasm in Gram-positive bacteria, as it is in Gram-negative bacteria.
It is remarkable that 40 years after the definitive demonstration of isoosmolarity for Gram-negative bacteria [17], there is no comparable measurement for Gram-positive bacteria. We really need a definitive measure of the pore size of the PG wall in living Gram-positive bacteria, and a measure of the osmolarity of the periplasm. Another important question is the nature of the osmolytes in the Gram-positive periplasm. MDOs have not been reported for Gram-positive bacteria, but teichoic acids are a candidate [31,32].
Gram-positive bacteria divide by septation
Several species of Gram-positive bacteria show clear evidence for division by a septation mechanism, in which the invaginating IM forms a thin plate. The PG wall of the mother cell remains in place around the outside, and new PG wall for the daughter cells grows into the septum. Matias and Beveridge [33] obtained superb images of septa in Staphylococcus aureus using high pressure freezing and cryosectioning. Fig. 3A shows bacteria with forming and completed septa and Fig. 3B shows a high magnification cross section of a newly forming septum. The IM is resolved as a bilayer, defining the edges of the septal plate, which has a uniform width of 75 nm. Cell wall material is identified as two “high-density zones”, which are the future outer wall PG of the daughter cells. They are apparently fused to the outer wall PG, although the EM images do not inform on the molecular linkage. The septum was held together primarily by its attachment to the outer cell wall PG layer. The PG layer was 19 nm thick over most of the bacterium, but increased to 40 nm at the septum, the “outer wall bridge.” This bridge presumably holds the septum together until it has completed invagination, and it is then disrupted to permit the daughter cells to separate.
Recent work has shown that in S. aureus this bridge is not digested gradually. It fails abruptly in a dramatic “popping” event lasting ∼ 1 msec [34]. The failure is mechanical, beginning as a crack at one point on the ring and propagating around, leaving the daughter cells connected at a hinge (Fig. 3C). Note that the new cell wall retains the flat shape it had in the septum, and is only gradually remodeled to the ovoid shape. Overall the picture seems very clear for S. aureus. Zuber et al. [35] reported a very similar septal structure in Enterococcus gallinarum, also using high pressure freezing and cryosectioning. Division by invagination of a thin, plate-like septum seems to be the general mechanism for Gram-positive bacteria.
Constrictive division in Gram-negative bacteria may be a modified septation mechanism
Based on early EM studies, Steed and Murray in 1966 summarized the consensus opinion that “Gram-negative bacteria in general,…seem to show a process of constriction involving all layers at once as if a noose were being tightened around the equator of the cell” [36]. This results in a V-shaped constriction of all layers (Figs. 4A, D; 5A), very different from the septal plate established for Gram-positive bacteria.
Figure 5.
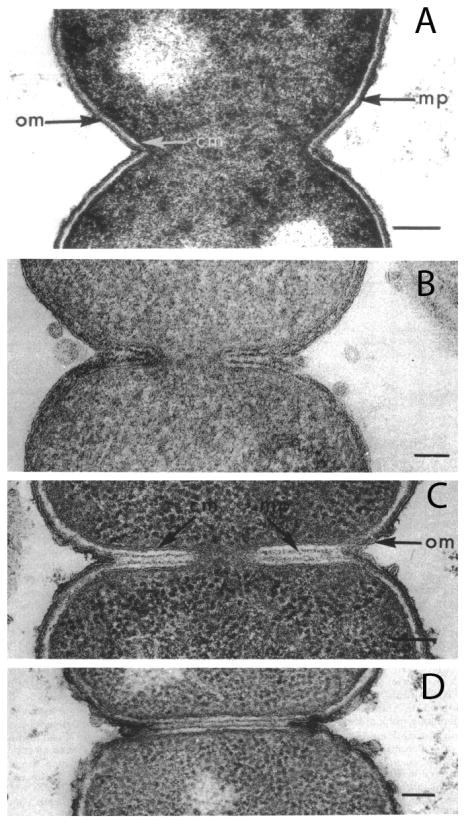
E. coli division imaged by conventional embedding and sectioning following different fixation methods. A: Fixation with OsO4, the standard method in the 1970's, gives the conventional picture of constrictive division. B: Acrolein plus glutaraldehyde, a novel fixative introduced by Burdett and Murray, preserved clear septa. The two dark lines within the septa are the newly forming PG layers. C, D: E. coli CRT 97 is a chain-forming strain apparently deficient in murein hydrolases. Septa in these strains can be preserved by any fixation, OsO4 being shown here. Reprinted from [38].
Recent cryoEM provides support for constrictive division. In 2005 Judd et al. [37] undertook a study of Caulobacter crescentus division using cryoEM tomography. Their images showed broad constrictions that initially maintained the 30 nm spacing of the IM and OM. This was consistent with the classical constriction mechanism. As constriction progressed, however, the IM was seen to move ahead of the OM (Fig. 4B). Their images did not show a distinct density for the PG layer, but if C. crescentus is similar to E. coli, the PG layer is tightly attached to the OM. In this case, as the IM invaginates ahead of the OM-PG it expands the periplasmic space. Eventually the IM fuses to form around each daughter cell, well before the OM-PG has finished constricting (Fig. 4C). It should be noted that Judd et al. did not consider their images representative of Gram-negative bacteria in general. They were aware of the work of Burdett and Murray [38] showing a septation mechanism for E. coli (discussed below) and they concluded that the constrictive division they found was specific for C. crescentus [37].
However, a recent cryoEM study provided evidence for a constrictive division in E. coli (Fig. 4D) [39]. The main focus of that study was to image FtsZ filaments, and this was its major success; note the rows of dots under the IM in Fig. 4D, which are identified as ribbons of FtsZ protofilaments in cross-section. The authors did not address the question of constrictive vs. septal division, but Fig. 4D shows a classic V-shaped constriction. The PG layer is a thin, lightly contrasted line close to the OM, as seen in Fig. 1. In this image the periplasmic space seems to have been greatly expanded, suggesting possible plasmolysis. Also, in the upper left constriction the IM at the tip of the V has advanced ahead of the OM, similar to Fig. 4B.
In contrast, one recent image of dividing E. coli prepared by high pressure freezing that shows an apparent septum (Fig. 4E) [40]. This is a clear example of what we will term “septation-constriction.” The OM-PG wall has formed a V-shaped constriction, but at the bottom of the V the IM has invaginated much further, forming a thin septum. We will next present additional evidence that this structure may be the underlying mechanism of division in E. coli.
In 1966 Steed and Murray presented evidence that Gram-negative bacteria actually did have septa at the division site, at least under certain conditions [36]. Specifically, they found septa in 50% of E. coli cells grown at 45° C, using conventional fixation. Burdett and Murray [38] made a major advance when they explored a wide range of fixatives. Fixation with OsO4 or glutaraldehyde alone, the favored fixatives at that time, showed dividing cells with the characteristic V shape of constrictive division (Fig. 5A). However, a mixture of 5% acrolein plus 0.25 % glutaraldehyde fixative preserved a large fraction of dividing cells with a clear septum (Fig. 5B). Importantly, the cells still had the V shape characteristic of constrictive division, but at the bottom of the V a septum continued toward the interior of the cell. The septum was characterized as an annular plate, with the IM on the outside defining its ∼50 nm width, and two layers of developing PG cell wall in the middle (Fig. 5B). The OM remained on the outside of the septum at the top of the V. Burdette and Murray suggested that the conventional fixatives did not preserve the short septum, allowing it to be pulled apart into an expanded V-shaped cleft [38]. When the septum was preserved it was always at the bottom of the V-shaped cleft, in contrast to the septa in Gram-positive bacteria, which extend all the way to the cylindrical cell wall. We will refer to this mechanism as “septation-constriction.”
Burdette and Murray [38] also obtained beautiful images of septa in the E. coli mutant CRT 97. In this strain, which may be defective in one or more murein hydrolases, septa were easily preserved by almost any fixative (Fig. 5C,D). Note that these septa also extend from the base of a V-shaped constriction, similar to the septation-constriction division seen in wild type E. coli fixed by acrolein-glutaraldehyde. A later study of Heidrich et al. [41] created mutants defective in various combinations of the 18 known murein hydrolases. Some of these grew as chains of cells, with septa spanning the full diameter, i.e. they had no V-shaped constriction. E. coli thus has the capability of forming clear plate-like septa, suggesting that this is the basic mechanism for its division.
Overall, these early studies of Murray and colleagues set the stage for understanding constrictive division as a modification of septation. We will pursue this idea below.
Super resolution light microscopy provides evidence for septal division in E. coli
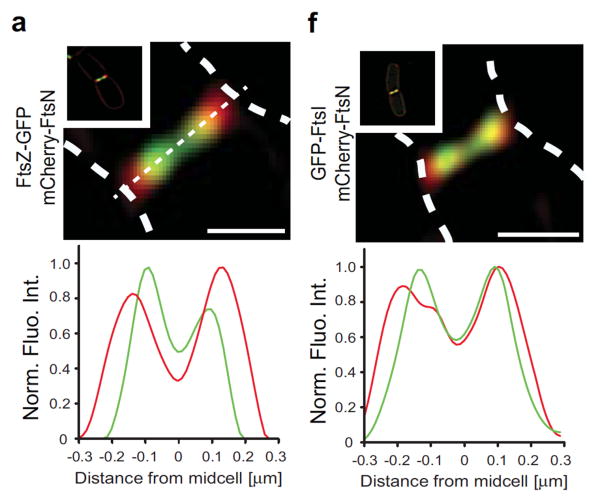
Söderstrom et al. [42] recently used GFP- and mCherry-labeled division proteins to determine their time of exit from the Z ring at the end of division. FtsZ departed first, followed by FtsA and ZipA. FtsI and FtsN remained at the division site longer. Their images also provided surprising new information on the radial localization of the division proteins. FtsZ, FtsA and ZipA co-localized in a ring throughout division, consistent with the direct binding of FtsA and ZipA to the C-terminal peptide of FtsZ. FtsN, FtsI, FtsL and FtsQ were mostly co-localized with each other. Surprisingly, the ring of FtsZ moved ahead of the ring of FtsN in constricting cells. The structured illumination microscopy (SIM) used for these images has a resolution of about 100 nm, but the centroid of the green and red labeled proteins could be localized much more precisely, to about 10 nm. As shown in Fig. 6a, the FtsZ ring was significantly inside FtsN ring. The rings differed in diameter by 105 nm over repeated measurements. Fig. 6f showed that FtsI almost co-localized with FtsN; repeated measurements gave a 26 nm smaller diameter for FtsI. Combining these two measurements and converting to radius, the FtsZ ring is about 40 nm closer to the cell center than FtsI.
Figure 6.
Localization of FtsZ, FtsN and FtsI by SIM superresolution microscopy. Dashed white lines indicate the cell boundaries. Panel a shows that FtsZ (green) is significantly toward the center of the cell relative to FtsN (red). Panel f shows that FtsI is almost co-localized with FtsN, but slightly toward the center. Reprinted from [42].
The images of Söderstrom et al. [42] can be further interpreted to provide evidence for a septal mechanism of division, based in particular on the width of the FtsI and FtsZ rings. These appear to be diffraction limited (100 nm in the SIM images) in both radial and axial directions. If the division were purely constrictive, with sides of the V at 45 degrees, the FtsN would have to expand axially as it moved radially outward from FtsZ. The 50 nm radial separation would convert the FtsN ring axially to a double ring 100 nm apart. This would not be resolved by SIM, but it should produce an obvious axial broadening. This was not seen. The images are consistent with a septal width of 50 nm or less for both FtsZ and FtsN.
Hypothesis – constrictive division in Gram-negative bacteria is a variation of septation
The study of Burdett and Murray [38] provided convincing images of septa in wild type E. coli fixed with acrolein-glutaraldehyde, and in the mutant CRT 97 fixed with conventional glutaraldehyde plus OsO4. The earlier study of Steed and Murray [36] found septa in E. coli preserved by conventional fixation in certain growth conditions. These studies clearly show that E. coli has the ability to divide by formation of septa. In most cases the septa of E. coli invaginate from the bottom of a V-shaped constriction. In some cases, especially early in division, the V-shaped constriction dominated [37,39].
These structures can be resolved by postulating septation as the universal division mechanism, but recognizing three variations in how the PG wall is hydrolyzed. These are diagrammed in Fig. 7. (A) At one extreme are Gram-positive bacteria, which divide by a full septation mechanism. Here the bridge PG of the mother cell is maintained in place until the septum has completed its invagination. The bridge PG is hydrolyzed and/or mechanically broken only after the IM and the PG plates have completed their fusion for the daughter cells. The E. coli hydrolase mutants of Heidrich et al. [41], which formed septa spanning the full diameter of the mother cell, suggest that E. coli has the potential for similar extreme septation. (B) An intermediate mechanism, which we term septation-constriction, has a short septum extending inward, but it splits into a V-shaped constriction on the outside. This could be formed if hydrolysis of the outer layers of PG begins while the septum is still advancing. This would permit the forming septum to separate into a V-shaped constriction above the site of hydrolysis, while the septum advances below. This is apparently the state captured in the images in Figs. 5B-D, 4E. (C) Finally, at the other extreme, are images showing a V-shaped constriction, with no obvious septum (Fig. 4A-D). We suggest these are produced when hydrolysis follows so closely after the advancing septum that it virtually obliterates the septal structure. Importantly however, even these images retained a hint of septation. The IM appeared to advance ahead of the OM-PG, leading to an extended periplasmic space and a sharper V for the IM (Fig. 4B, D).
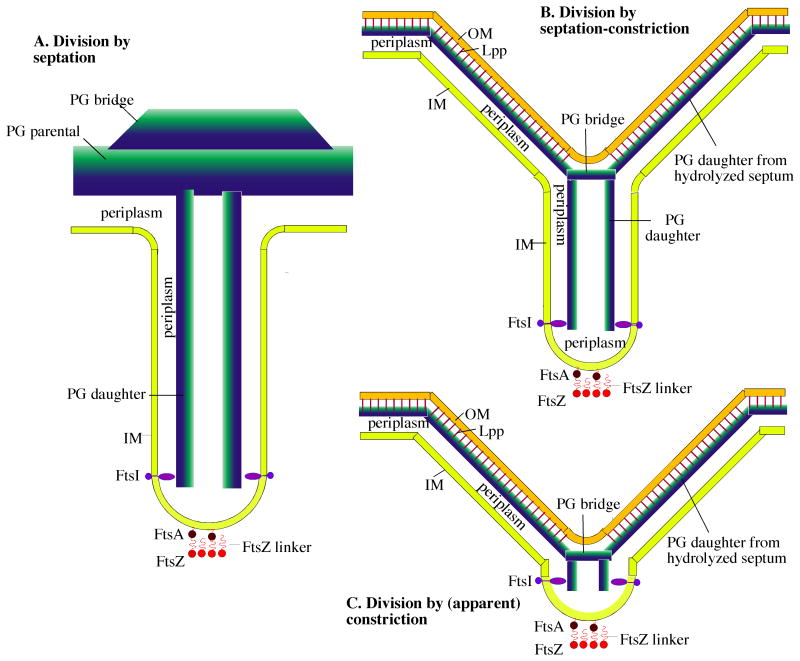
Figure 7.
Three variations on a general model of bacterial division by septation. A: The Gram-positive septum. The components are: FtsZ, a 5 nm diameter sphere with a 10 nm peptide linker that connects to FtsA, a 5 nm sphere with an amphipathic helix that inserts into the IM. The Z ring shown here in cross-section is a ribbon of four protofilaments, consistent with recent cryoEM [39]; see also [50] for arguments supporting this narrow width. The IM, a 4 nm thick lipid bilayer that defines the two sides of the septum ∼70 nm apart; PG parental, a 30 nm wide sheet represented as a blue to green gradient inside to outside; PG bridge, a thickening of the parental PG at the septum [33]; periplasm, the 20 nm space between the IM and PG; FtsI (PBP3 in E. coli and PBP2B in B. subtilis), a transmembrane protein with an elongated periplasmic domain [51] that can bridge the 10 nm wide periplasmic space in the septum. FtsI is shown here spaced 40 nm above the FtsZ ring [42]). This septum remains connected to the outer wall PG of the mother cell until invagination is complete. B: The Gram-negative “septation-constriction” mechanism. Most components are the same as in the Gram-positive septum with different dimensions: The periplasm is 10 nm and the PG is 6 nm thick. In addition the Gram-negative bacteria have an OM, a 4 nm thick bilayer (ignoring here the oligosaccharide projections) covalently attached to the PG by the abundant Lpp lipoprotein rods [20]. In this septation-constriction mechanism the outer layers of the septum have split to generate the V-shaped constriction. (C) In the (apparent) constriction-only mechanism, the splitting of the septum on the outside follows very closely its invagination toward the center, so that the septum is hardly visible.
An essential part of the constriction-septation mechanism is the PG bridge, at the point of the V, which must maintain a seal with the OM-PG on the sides of the V. This bridge must also somehow be attached to the daughter PG in the septum. Its growth on the inside and hydrolysis on the outside would need to be coupled to avoid a rupture. In this scenario the major difference between Gram-positive and Gram-negative division is the timing of murein hydrolases digesting the old PG wall. In Gram-positive division this occurs only after the septum is complete, while in Gram-negative it occurs while the septum is still advancing.
The structure of the V raises the question, why doesn't the turgor pressure collapse it outward? Probably because the PG is rigid not just to stretching, but also to compression and bending. An example of this rigidity is the structure of the cell walls of S. aureus immediately following the popping separation (Fig. 6C). Here they are abruptly exposed to turgor pressure, but they maintain their flat shape. Rigidity of the PG layer is also suggested by the observations that E. coli cells forced to grow into odd shapes retain their shapes when released, and only reform after one or more cell cycles [43,44].
Another point in favor of a common septal mechanism is based on the argument of Gupta that the earliest bacteria were the simpler Gram-positive “monoderms,” and that the Gram-negative “diderms,” with the extra outer membrane, evolved from them [45]. In this case the septal mechanism would have been inherited by the earliest Gram-negative bacteria. Consistent with this, Deinococcus species, which appear to be a primitive Gram-negative form, divide by a distinctly septal mechanism [46]. The V-shaped constrictive division would then be a later modification on the septal mechanism. This is supported by the observation that E. coli still retains the possibility of dividing by a septal mechanism (Fig. 5B-D).
This overall hypothesis is not really new. Steed and Murray [36] concluded in 1966: “These electron microscope observations indicate that it is possible to regularly demonstrate an incontrovertible septum in these organisms [Gram-negative], which show only constrictive divisions [when grown and fixed under certain conditions]. There is no escaping the suspicion that septum formation is the normal prelude to cell division in E. coli.”
Conclusions and outlook - What generates the constriction force?
Coltharp et al. [13] suggested that FtsZ pf bending would not be able to generate a force sufficient to overcome the turgor pressure, and proposed PG wall growth as the primary constriction force. However, if septation takes place within the high osmolar environment of the cytoplasm-periplasm, weaker forces would suffice. It is possible that both FtsZ pf bending and PG ingrowth contribute. Two recent studies have shown that FtsI moves in circular paths around the Z ring, driven by patches of treadmilling FtsZ [47,48]. Bisson-Filho et al. [48] suggested that FtsZ filaments in the treadmilling patch may deform the membrane, and PG synthesis could reinforce the deformation on the other side. “Thus multiple sites of local deformation and coupled reinforcing synthesis moving around the division site would iteratively build the invaginating septum.”
Acknowledgments
I thank Dr. Masaki Osawa for many informative and challenging discussions. Supported by NIH grant GM066014. I declare no conflicts of interest.
Abbreviations
- EM
electron microscopy
- IM
inner membrane
- MDO
membrane-derived oligosaccharide
- OM
outer membrane
- PG
peptidoglycan
References
- 1.Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–4. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 2.RayChaudhuri D, Park JT. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992;359:251–4. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- 3.de Boer P, Crossley R, Rothfield L. The essential bacterial cell division protein FtsZ is a GTPase. Nature. 1992;359:254–6. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 4.Mukherjee A, Dai K, Lutkenhaus J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci USA. 1993;90:1053–7. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erickson HP. The discovery of the prokaryotic cytoskeleton: 25th anniversary. Mol Biol Cell. 2017;28:357–8. doi: 10.1091/mbc.E16-03-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–4. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erickson HP, Taylor DW, Taylor KA, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93:519–23. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu C, Reedy M, Erickson HP. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J Bacteriol. 2000;182:164–70. doi: 10.1128/jb.182.1.164-170.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osawa M, Anderson DE, Erickson HP. Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 2009;28:3476–84. doi: 10.1038/emboj.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson HP, Anderson DE, Osawa M. FtsZ in Bacterial Cytokinesis: Cytoskeleton and Force Generator All in One. Microbiol Mol Biol Rev. 2010;74:504–28. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osawa M, Erickson HP. Inside-out Z rings - constriction with and without GTP hydrolysis. Mol Microbiol. 2011;81:571–9. doi: 10.1111/j.1365-2958.2011.07716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Housman M, Milam SL, Moore DA, Osawa M, et al. FtsZ Protofilament Curvature Is the Opposite of Tubulin Rings. Biochemistry. 2016;55:4085–91. doi: 10.1021/acs.biochem.6b00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coltharp C, Buss J, Plumer TM, Xiao J. Defining the rate-limiting processes of bacterial cytokinesis. Proc Natl Acad Sci USA. 2016;113:E1044–53. doi: 10.1073/pnas.1514296113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coltharp C, Xiao J. Beyond force generation: Why is a dynamic ring of FtsZ polymers essential for bacterial cytokinesis? BioEssays. 2017;39:1–11. doi: 10.1002/bies.201600179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonald KL. Out with the old and in with the new: rapid specimen preparation procedures for electron microscopy of sectioned biological material. Protoplasma. 2014;251:429–48. doi: 10.1007/s00709-013-0575-y. [DOI] [PubMed] [Google Scholar]
- 16.Matias VR, Al-Amoudi A, Dubochet J, Beveridge TJ. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J Bacteriol. 2003;185:6112–8. doi: 10.1128/JB.185.20.6112-6118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stock JB, Rauch B, Roseman S. Periplasmic space in Salmonella typhimurium and Escherichia coli. J Biol Chem. 1977;252:7850–61. [PubMed] [Google Scholar]
- 18.Matias VR, Beveridge TJ. Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol Microbiol. 2005;56:240–51. doi: 10.1111/j.1365-2958.2005.04535.x. [DOI] [PubMed] [Google Scholar]
- 19.Zuber B, Chami M, Houssin C, Dubochet J, et al. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bacteriol. 2008;190:5672–80. doi: 10.1128/JB.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shu W, Liu J, Ji H, Lu M. Core structure of the outer membrane lipoprotein from Escherichia coli at 1.9 A resolution. J Mol Biol. 2000;299:1101–12. doi: 10.1006/jmbi.2000.3776. [DOI] [PubMed] [Google Scholar]
- 21.Decad GM, Nikaido H. Outer membrane of gram-negative bacteria. XII. Molecular-sieving function of cell wall. J Bacteriol. 1976;128:325–36. doi: 10.1128/jb.128.1.325-336.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarz H, Koch AL. Phase and electron microscopic observations of osmotically induced wrinkling and the role of endocytotic vesicles in the plasmolysis of the Gram-negative cell wall. Microbiology. 1995;141(Pt 12):3161–70. doi: 10.1099/13500872-141-12-3161. [DOI] [PubMed] [Google Scholar]
- 23.Sochacki KA, Shkel IA, Record MT, Weisshaar JC. Protein diffusion in the periplasm of E. coli under osmotic stress. Biophys J. 2011;100:22–31. doi: 10.1016/j.bpj.2010.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy EP. Osmotic regulation and the biosynthesis of membrane-derived oligosaccharides in Escherichia coli. Proc Natl Acad Sci USA. 1982;79:1092–5. doi: 10.1073/pnas.79.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller KJ, Kennedy EP, Reinhold VN. Osmotic adaptation by gram-negative bacteria: possible role for periplasmic oligosaccharides. Science. 1986;231:48–51. doi: 10.1126/science.3941890. [DOI] [PubMed] [Google Scholar]
- 26.Cayley DS, Guttman HJ, Record MT., Jr Biophysical characterization of changes in amounts and activity of Escherichia coli cell and compartment water and turgor pressure in response to osmotic stress. Biophys J. 2000;78:1748–64. doi: 10.1016/s0006-3495(00)76726-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scherrer R, Gerhardt P. Molecular sieving by the Bacillus megaterium cell wall and protoplast. J Bacteriol. 1971;107:718–35. doi: 10.1128/jb.107.3.718-735.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Demchick P, Koch AL. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J Bacteriol. 1996;178:768–73. doi: 10.1128/jb.178.3.768-773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasupuleti M, Schmidtchen A, Malmsten M. Antimicrobial peptides: key components of the innate immune system. Crit Rev Biotechnol. 2012;32:143–71. doi: 10.3109/07388551.2011.594423. [DOI] [PubMed] [Google Scholar]
- 30.Foreman-Wykert AK, Weinrauch Y, Elsbach P, Weiss J. Cell-wall determinants of the bactericidal action of group IIA phospholipase A2 against Gram-positive bacteria. J Clin Invest. 1999;103:715–21. doi: 10.1172/JCI5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matias VR, Beveridge TJ. Lipoteichoic acid is a major component of the Bacillus subtilis periplasm. J Bacteriol. 2008;190:7414–8. doi: 10.1128/JB.00581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhavsar AP, Brown ED. Cell wall assembly in Bacillus subtilis: how spirals and spaces challenge paradigms. Mol Microbiol. 2006;60:1077–90. doi: 10.1111/j.1365-2958.2006.05169.x. [DOI] [PubMed] [Google Scholar]
- 33.Matias VR, Beveridge TJ. Cryo-electron microscopy of cell division in Staphylococcus aureus reveals a mid-zone between nascent cross walls. Mol Microbiol. 2007;64:195–206. doi: 10.1111/j.1365-2958.2007.05634.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhou X, Halladin DK, Rojas ER, Koslover EF, et al. Bacterial division. Mechanical crack propagation drives millisecond daughter cell separation in Staphylococcus aureus. Science. 2015;348:574–8. doi: 10.1126/science.aaa1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuber B, Haenni M, Ribeiro T, Minnig K, et al. Granular layer in the periplasmic space of gram-positive bacteria and fine structures of Enterococcus gallinarum and Streptococcus gordonii septa revealed by cryo-electron microscopy of vitreous sections. J Bacteriol. 2006;188:6652–60. doi: 10.1128/JB.00391-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steed P, Murray RG. The cell wall and cell division of gram-negative bacteria. Can J Microbiol. 1966;12:263–70. doi: 10.1139/m66-036. [DOI] [PubMed] [Google Scholar]
- 37.Judd EM, Comolli LR, Chen JC, Downing KH, et al. Distinct constrictive processes, separated in time and space, divide caulobacter inner and outer membranes. J Bacteriol. 2005;187:6874–82. doi: 10.1128/JB.187.20.6874-6882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burdett ID, Murray RG. Septum formation in Escherichia coli: characterization of septal structure and the effects of antibiotics on cell division. J Bacteriol. 1974;119:303–24. doi: 10.1128/jb.119.1.303-324.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szwedziak P, Wang Q, Bharat TA, Tsim M, et al. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife. 2014;3:e04601. doi: 10.7554/eLife.04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berezuk AM, Goodyear M, Khursigara CM. Site-directed fluorescence labeling reveals a revised N-terminal membrane topology and functional periplasmic residues in the Escherichia coli cell division protein FtsK. J Biol Chem. 2014;289:23287–301. doi: 10.1074/jbc.M114.569624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heidrich C, Ursinus A, Berger J, Schwarz H, et al. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J Bacteriol. 2002;184:6093–9. doi: 10.1128/JB.184.22.6093-6099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soderstrom B, Mirzadeh K, Toddo S, von Heijne G, et al. Coordinated disassembly of the divisome complex in Escherichia coli. Mol Microbiol. 2016;101:425–38. doi: 10.1111/mmi.13400. [DOI] [PubMed] [Google Scholar]
- 43.Takeuchi S, DiLuzio WR, Weibel DB, Whitesides GM. Controlling the shape of filamentous cells of Escherichia coli. Nano Lett. 2005;5:1819–23. doi: 10.1021/nl0507360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mannik J, Driessen R, Galajda P, Keymer JE, et al. Bacterial growth and motility in sub-micron constrictions. Proc Natl Acad Sci USA. 2009;106:14861–6. doi: 10.1073/pnas.0907542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta RS. Origin of diderm (Gram-negative) bacteria: antibiotic selection pressure rather than endosymbiosis likely led to the evolution of bacterial cells with two membranes. Antonie Van Leeuwenhoek. 2011;100:171–82. doi: 10.1007/s10482-011-9616-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murray RG, Hall M, Thompson BG. Cell division in Deinococcus radiodurans and a method for displaying septa. Can J Microbiol. 1983;29:1412–23. doi: 10.1139/m83-217. [DOI] [PubMed] [Google Scholar]
- 47.Yang X, Lyu Z, Miguel A, McQuillen R, et al. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science. 2017;355:744–7. doi: 10.1126/science.aak9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bisson-Filho AW, Hsu YP, Squyres GR, Kuru E, et al. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science. 2017;355:739–43. doi: 10.1126/science.aak9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matias VR, Beveridge TJ. Native cell wall organization shown by cryo-electron microscopy confirms the existence of a periplasmic space in Staphylococcus aureus. J Bacteriol. 2006;188:1011–21. doi: 10.1128/JB.188.3.1011-1021.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erickson HP, Osawa M. FtsZ constriction force – curved protofilaments bending membranes. In: Amos LA, Löwe J, editors. Prokaryotic Cytoskeletons: Filamentous Protein Polymers Active in the Cytoplasm of Bacterial and Archaeal Cells. Springer; 2017. in press. [Google Scholar]
- 51.Han S, Zaniewski RP, Marr ES, Lacey BM, et al. Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2010;107:22002–7. doi: 10.1073/pnas.1013092107. [DOI] [PMC free article] [PubMed] [Google Scholar]