Supplemental Digital Content is available in the text
Keywords: biomarker, herpes zoster, postherpetic neuralgia, Serum microRNAs
Abstract
Postherpetic neuralgia (PHN) is commonly defined as pain persisting for at least 3 months after acute herpes zoster (AHZ) rash presentation. About one-tenth of all acute herpes zoster patients develop PHN. Circulating microRNAs (miRNAs) are promising biomarkers for infectious diseases; however, there has been no relationship established between circulating miRNAs and PHN to date; the aim of the present investigation was to elucidate this relationship.
We compared serum levels of miRNA in PHN and AHZ patients. Twenty-nine patients with PHN and 37 patients with AHZ participated. MiRNA serum levels were determined via TaqMan Low Density Array (TLDA) and confirmed individually by RT-qPCR.
TLDA results showed that the expression levels of 157 serum miRNAs in PHN patients were distinct from those in AHZ patients. Among these PHN patient serum miRNAs, 17 were upregulated and 139 were downregulated in contrast to those in AHZ patients. Receiver operational characteristic (ROC) curve analysis and RT-qPCR results altogether confirmed that the levels of miR-34c-5p, miR-107, miR-892b, miR-486–3p, and miR-127–5p were notably increased in PHN patients in comparison with those of AHZ patients. These miRNAs in circulation may regulate numerous relevant pathways. A few likely participate in the nervous system and inflammatory reactions.
This study is the first to show that the expression profiles of numerous miRNAs vary in the PHN process. Among these, 5 types of serum miRNAs are very likely related to PHN development.
1. Introduction
The varicella-zoster virus (VZV), which results in AHZ as well as varicella, is a type of the herpes virus which exists ubiquitously in human beings.[1] In prosperous countries and regions, the sero-prevalence of VZV antibodies is above 95%; that is to say, almost all of the world's adult population is at risk for AHZ.[2] The virus initially infects human beings and then hides itself in sensory cranial nerve ganglia and spinal dorsal root ganglia, potentially laying dormant (asymptomatic) for many years.[3] The immunity mediated by cells diminishes due to aging, stress, or illness, and thus, the risk of AHZ recurrence increases over time.[4] Though AHZ symptoms usually subside in 15 to 30 days, nearly one-tenth of the affected population suffers from PHN, commonly known as pain sustained over 3 months or more at the site of the AHZ rash.[5,6] The risk of developing PHN likewise increases with age. Pain caused by PHN is attributed to damage to the sensory nerves and exhibits intermittency, spontaneity, and chronic characteristics.[7] Although intervention is possible, PHN-related pain is well capable of interfering with the patient's daily activities and sleep quality.
MicroRNAs (miRNAs) are a group of noncoding RNAs which have low molecular weight and single-strand structure.[8] They play a critical role in regulating various diseases, and are abundant in human body fluids. Studies have shown that they inhibit abundant downstream genes through binding specific target loci of mRNA transcripts.[9] Circulating miRNAs are considered to be more robust to degradation, particularly in contrast to mRNA, which makes them well-suited to indicating potential diseases.[10–12] A growing body of evidence suggests that miRNAs take part in maintaining normal cell physiological function, relate to host-virus interactions, and play a role in limiting the replication of certain kinds of viruses.[13]
The primary aim of this investigation was to elucidate the relationship between circulating miRNAs and PHN by comparing serum levels of miRNA in PHN versus AHZ patients. The expression levels of various serum miRNAs in AHZ and PHN groups were also determined for the sake of comparison and to investigate their potential biological functions.
2. Methodology
2.1. Patient selection
A total of 66 participants were enrolled between June 2013 and April 2016, including 29 PHN patients and 37 AHZ patients, all from Nanjing Gulou Hospital. Patients showed no signs of apparent secondary bacterial infection on affected areas or any other inflammatory reactions which would potentially affect the results. No patients were given immunosuppression therapy or steroids prior to or during the study. Serum samples were obtained from the enrolled AHZ and PHN patients (N = 37, N = 29, respectively) after they showed clinical manifestations. Blood samples were collected from the AHZ patients at the beginning of the acute phase and from PHN patients who suffered from pain for 3 months or more after vesicle eruption. All obtained samples were preserved in the laboratory at −80°C within 4 hours of sampling. The research ethics committee board of the Nanjing Gulou Hospital approved this protocol and all patient participants submitted written informed consent forms. Demographic information was obtained through verifying electronic medical records or abstracting charts.
2.2. Methods
In order to analyze serum samples via TLDA, control and PHN patient pools were built by pooling isovolumetric serum from each patient individually; the volume of each pool was 20 mL. Total RNAs of each pool were extracted with the TRIzol reagent (Invitrogen), and then, 20 μL RNase-free H2O was used to dissolve each RNA pellet. The total RNA samples as-prepared were then preserved at −80°C until use.
To conduct the RT-qPCR analysis, a mixture of 100 μL serum, 300 μL RNase-free H2O, 200 μL acid phenol, and 200 μL chloroform was prepared and shaken vigorously with a vortex device to guarantee its uniformity. The mixture was then centrifuged under ambient temperature for 15 minutes. After the phrases were separated, the water phase was collected and 0.1 volume of 3 mol/L sodium acetate (pH 5.3) plus 1.5 volumes of isopropyl alcohol were added. The solution was then chilled at −20°C for 1 hour and centrifuged at 4°C and 16,000g for 20 minutes. The resulting pellet was washed in 750 ml/L ethanol once and dried at ambient temperature for 10 minutes. Finally, 20 μL RNase-free H2O was applied to dissolve the pellet and the solution was preserved at −80°C until use.
Megaplex RT Primers and a TaqMan miRNA Reverse Transcription Kit were used to perform reverse transcription. Extracted RNA (50 ng) was used as the template and mixed with reverse transcription mixture containing 10× Megaplex RT primers, 100 mmol dTTP and dNTPs, 50 U/μL multiscribe reverse transcriptase, 10× reverse transcription buffer, 25 mmol MgCl2, 20 U/μL RNase inhibitor, and nuclease-free water. After incubating the mixture with ice for 5 minutes, a thermal cycler was used to perform reverse transcription followed by a pre-amplification employed to improve TLDA sensibility; TLDA was then applied to analyze the expression profiles of 768 individual microRNAs. All procedures were conducted according to the manufacturers’ recommendations with a 7900 HT Fast Real-Time PCR System (Applied Biosystems). The comparative Ct method (2−ΔΔCt) was applied to calculate the relative expression. The mean Ct of U6 snRNA in each pool (ΔCt) was used to normalize the results.
The serum samples were analyzed with a hydrolysis probe-based RT-qPCR method per the manufacturer's instructions (Applied Biosystems). The RT reaction was conducted in a 10-μL reaction system containing 2 μL total RNA, 3.5 μL DEPC water, 2 μL RT buffer, 1 μL stem loop RT primer (Applied Biosystems), 0.5 μL AMV reverse transcriptase (TaKaRa), and 1 μL 10 mmol/L dNTPs. The technological process of cDNA synthesis involved incubating the reaction mixture at 16°C for 30 minutes, then at 42°C for 30 minutes, at 85°C for 5 minutes, and finally holding it at 4°C. Pre-amplification was conducted in order to improve the RT-qPCR sensitivity. The reaction system included 2 μL cDNA from the reverse-transcription mixed with 0.3 μL Taq, 0.33 μL miRNA specifically stem-loop primer (Applied Biosystems), 2 μL 10 × PCR buffer, 0.4 μL 10 mmol/L dNTPs, 2 μL of 25 mmol/L MgCl2, and 13.77 μL DEPC water. The technological process of reamplification included a 5-minutes first cycle of incubating the reaction mixture at 95°C, 12 cycles of 15 seconds at 95°C, 1 minute at 60°C with the Applied Biosystems PCR instrument, and finally holding the mixture at 4°C. After the pre-amplification was complete, the PCR product was diluted 20 times before use as cDNA for the RT-qPCR assay. An Applied Biosystems 7900 Sequence Detection System was used to perform the RT-PCR. The technological process included 1 cycle at 95°C for 5 minutes, then 40 cycles at 95°C for 1 minutes and another 1 minutes at 60°C. The reaction system was a 20-μL mixture containing 1 μL cDNA, 2 μL 10 × PCR buffer, 0.4 μL 10 mmol/L dNTPs, 1.2 μL 25 mmol/L MgCl2, 0.33 μL hydrolysis probe (Applied Biosystems), 0.3 μL Taq, and 14.77 μL DEPC water. All examinations were conducted in triplicate including the control group, which had no template. Ct was defined as the number of fractional cycles when the fluorescence intensity exceeded the threshold value. Default settings were used to assign the base line and analyze the data; U6 snRNA and the ΔΔCt method were utilized to normalize and calculate the expression levels of miRNAs.
Target genes which could be regulated by candidate miRNAs were predicted with TargetScan[14] and analyzed on the DAVID server using the default setup to determine the physiological functions or signal pathways in which the genes were likely involved.
2.3. Statistical analysis
The relative expression levels of miRNA in qRT-PCR results transformed into Log2 values were calculated in light of the differences in Ct between target miRNAs and U6 snRNA (ΔCt). All analyses above were performed with the use of SPSS statistical software (V18.0). Any P lower than 0.05 was considered under the threshold of statistical significance. ROC curves were established to determine the sensitivity and specificity of PHN prediction on each individual miRNA and the combinations of different miRNAs.
3. Results
3.1. Participant clinical characteristics
Sixty-six participants in total, among which 37 were AHZ patients and 29 were PHN patients, participated in this study with a median age of over 60 years. Most participants were examined in their first visit to the clinic for any conditions which could have decreased their immunity in the 6 months before their visit including aging, mechanical injury, serious psychological stress, chronic kidney disease, malignant tumors, or type II diabetes. The age and sex distribution of PHN patients was not significantly different from that of AHZ patients (P>0.05). The sensory nerve branches that innervated dermatomes in which most skin lesions occurred were from C2 to L5. The patient characteristics are presented in Table 1.
Table 1.
The patients’ characteristic.
3.2. Serum miRNA screening by TLDA
The expression profiles of miRNAs in 2 serum sample pools (29 PHN patients, 37 AHZ patients) were investigated using TLDA. For any miRNA in the PHN pool, if Ct value was >35 and the concentration was at least twice that of its counterpart in the AHZ pool, it was considered upregulated. Class-comparison analysis of all 768 miRNAs revealed that 17 miRNAs were upregulated in the PHN sample (Supplemental Table 1), 5 of the upregulated miRNAs were subsequently validated in the individual patients using RT-qPCR.
3.3. Increases in serum miRNA concentrations confirmed by RT-qPCR
The expression levels of 17 miRNAs as-determined by TLDA were verified again by RT-qPCR for each serum sample. Five miRNAs, including miR-34c-5p, miR-107, miR-892b, miR-486-3p, and miR-127-5p were found to be markedly upregulated in sera from PHN patients (P < 0.05). Conversely, the expression levels of miR-30d, miR-518b, miR-376c, miR-200b, miR-487b, miR-642, miR-98, miR-519d, miR-517c, miR-301b, miR-34a, and miR-184 in PHN serum samples were not significantly different from those in AHZ samples (P > 0.05) (Fig. 1).
Figure 1.

In total 17 serum miRNA levels in PHN patients and AHZ patients were selected for verification using real-time qRT-PCR in individual PHN patients (N = 29) and AHZ controls (N = 37). Serum levels of miR-34c-5p, miR-107, miR-892b, miR-486–3p, and miR-127–5p were significantly higher in PHN patients compared with those in the AHZ group (∗∗, P < 0.01). But miR-642 was not significantly higher in PHN patients compared with that in the AHZ group (ns, P>0.05). Expression levels of the miRNAs are normalized to U6 snRNA (Log2 relative level). AHZ = acute herpes zoster, miRNA = microRNAs, PHN = postherpetic neuralgia.
3.4. Evaluating miRNAs in PHN and analyzing variables with ROC curves
Next, we conducted ROC curve analyses on each of the individual 5 serum miRNAs to assess the value for discriminating between PHN and AHZ (Fig. 2, Table 2). The area values under curves of miR-34c-5p, miR-107, miR-892b, miR-486–3p, and miR-127–5p were 0.847 (95% CI 0.751–0.943), 0.883 (95% CI 0.799–0.967), 0.788 (95% CI 0.679–0.898), 0.872 (95% CI 0.786–959), and 0.797 (95% CI 0.619–0.915), respectively, which indicated that the distinguishing efficiency of the proposed technique is reasonable. The AUC value of miR-107 was 0.883, which indicated that miR-107 is capable of distinguishing PNH infection. The ROC curves showed that the proposed diagnosis technique was remarkably accurate. The combined set AUC value of 0.978 (95% CI 0.950–1.0; P < 0.001) was much higher than those of the 5 miRNAs.
Figure 2.
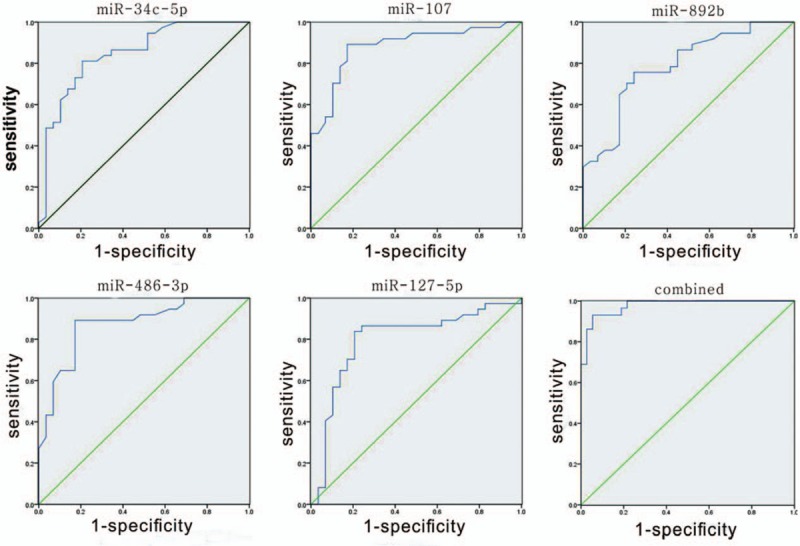
Receiver operating characteristic (ROC) curves of differentially expressed miRNAs between PHN patients and AHZ controls. ROC curves of miR-34c-5p, miR-107, miR-892b, miR-486–3p, and miR-127–5p showed a moderate distinguishing efficiency. The combination of the 5 miRNAs showed a slightly higher AUC value of 0.883. AHZ = acute herpes zoster, miRNA = microRNAs, ROC = receiver operating characteristic, PHN = postherpetic neuralgia.
Table 2.
Areas under the curve and the asymptotic 95% confidence intervals of the individual miRNA in ROC curves.

3.5. Target gene prediction
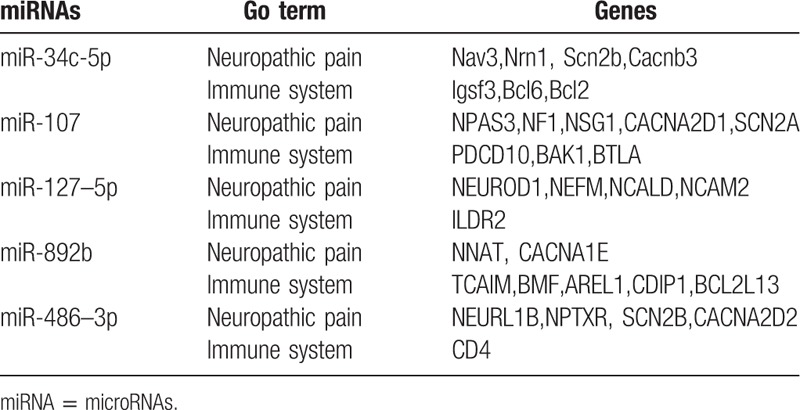
In recent years, the effects of miRNA on host-virus interactions have garnered significant research attention. Previous scholars have hypothesized that human miRNA plays an essential role in promoting cell growth, inhibiting apoptosis, controlling anti-viral reactions, and virus replication.[15] Research also shows that miRNAs are related to inflammatory reaction and immunoreactionduring bacterial infection.[16] Here, the TargetScan program was utilized to predict the targets of miR-34c-5p, miR-107, miR-892b, miR-486–3p, and miR-127–5p in order to reveal their potential functions. Gene ontology (GO) analysis results suggested that a portion of these genes were indeed involved in neuropathic pain and immune system development in the study participants. For example, miR-127–5p coordinated the gene expression of neurofilament medium polypeptide (NEFM), a member of the neuro-filament triplet protein family that is the main constituent of the CNS neuron cytoskeleton.[17] NEFM exists abundantly in axons and endows them with robustness and mechanical strength; it also localizes and transports constituents in the axons.[18] The results of this study suggest that NEFM may be related to the neuropathy pain in PHN, though the mechanism remains unclear. Bcl-2, the target of miR-34c-5p, may inhibit the apoptosis of various kinds of cells including lymphocytes.[19] The inflammatory reaction in PHN could be related to this, but the mechanism again remains unclear (Table 3).
Table 3.
The list of genes predicted to be targeted by the candidated miRNAs.
4. Discussion
AHZ manifests as pain and rash with blisters caused by reactivated VZV, a herpes virus that is ubiquitous in human beings.[1] Though AHZ symptoms typically subside within 15 to 30 days, nearly one-tenth of the affected population suffers from PHN, that is, pain continuing for at least 3 months after AHZ presentation. The risk of developing PHN increases with age. The pain caused by PHN is attributed to injuries to sensory nerves and exhibits intermittency, spontaneity, and chronic characteristics.[7] Allodynia is usually included in the symptoms of PHN – even ordinary touches such as brushing the skin gently can evoke considerable pain. Although intervention is possible, pain in PHN can certainly interfere with the patient's quality of life.
To the best of our knowledge, this study is the first to assess the miRNA profiles in PHN patients. The calculation results based on the ROC curves indicated that PHN patient miRNAs (miR-34c-5p, miR-107, miR-892b, miR-486–3p, and miR-127–5p) are significantly elevated in the serum compared to AHZ patients. Further research with larger numbers of patients and healthy volunteers is required to fully validate these findings.
Our results suggest that these miRNAs may exert substantial influence on the interaction between neuropathic pain and the immunological responses though pathogenic mechanisms for PHN remain elusive. MiR-107 regulates the levels of dicers in differentiating neurons, which is essential for maintaining the homeostasis of specified miRNAs;[20] the accurate accumulated quantity of these miRNAs is indispensable for neurogenesis. The results of this study showed that the miR-107 level in PHN patients was higher than that in AHZ patients. Previous studies have demonstrated that miR-892b[21] and miR-127–5p[22] directly target and suppress many mediators of NF-κB such as TAB3, TAK1, and TRAF2 in addition to weakening the NF-κB signal. MiR-34c-5p[23] also may impact the invasion, vascular repair, and adhesion of anti-inflammatory cells. Further investigation is necessary to confirm their roles in PHN. More studies are needed to reveal the biological mechanism of differently expressed miRNAs in PHN and to comprehensively evaluate the capacity of these miRNAs for diagnosis.
New interactions between miRNA and herpes zoster were clarified here, but this study was not without limitations. First, the quantity of participants was relatively small and thus not able to provide definitive conclusions. Larger-scale studies are necessary to confirm these findings. The degrees of distinction in serum miRNAs between PHN and AHZ groups were reasonable but only mid-level, likely because the quantity of up-regulated miRNAs under evaluation in this study was insufficient. A greater number of combinations of miRNAs should be tested in the future to determine more effective biomarkers and further support the conclusions discussed here. In short, further studies on larger scales and including both ill and healthy people are yet needed to verify our conclusions.
In this study, TLDA analysis results confirmed that the expression profiles of serum miRNAs in PHN patients differ markedly from those in AHZ patients. The assembly of miR-34c-5p, miR-107, miR-892b, miR-486–3p, and miR-127–5p showed potential application as a biomarker for diagnosing PHN noninvasively.
Supplementary Material
Footnotes
Abbreviations: AHZ = acute herpes zoster, GO = gene ontology, NEFM = neurofilament medium polypeptide, PHN = postherpetic neuralgia, ROC = receiver operational characteristic, TLDA = TaqMan Low Density Array, VZV = varicella–zoster virus.
YH and XL contributed equally to this work.
YH devised the research, conducted the statistical analysis, wrote the manuscript, and checked and submitted the ultimate manuscript. XL conducted the statistical analysis, modified the manuscript, and checked and proofed the ultimate manuscript. GT collected the clinical data and checked and proofed the ultimate manuscript. TZ collected the clinical data and checked and proofed the ultimate manuscript. JL designed the research, obtained the manuscript, and checked and wrote the ultimate version. All authors participated in the discussion of findings and left comments on the manuscript.
Funding: This work was supported by the National Natural Science Foundation of China (grant no. 31500125).
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Kang CI, Choi CM, Park TS, et al. Incidence of herpes zoster and seroprevalence of varicella-zoster virus in young adults of South Korea. Int J Infect Dis 2008;12:245–7. [DOI] [PubMed] [Google Scholar]
- [2].Johnson RW. Herpes zoster and postherpetic neuralgia. Expert Rev Vaccines 2010;9(3 suppl):21–6. [DOI] [PubMed] [Google Scholar]
- [3].Sampathkumar P, Drage LA, Martin DP. Herpes zoster (shingles) and postherpetic neuralgia. Mayo Clinic Proc 2009;84:274–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nalamachu S, Morley-Forster P. Diagnosing and managing postherpetic neuralgia. Drugs Aging 2012;29:863–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Argoff CE. Review of current guidelines on the care of postherpetic neuralgia. Postgrad Med 2011;123:134–42. [DOI] [PubMed] [Google Scholar]
- [6].Ragozzino MW, Melton LJ, 3rd, et al. Population-based study of herpes zoster and its sequelae. Medicine 1982;61:310–6. [DOI] [PubMed] [Google Scholar]
- [7].Yawn BP, Saddier P, Wollan PC, et al. A population-based study of the incidence and complication rates of herpes zoster before zoster vaccine introduction. Mayo Clinic Proc 2007;82:1341–9. [DOI] [PubMed] [Google Scholar]
- [8].Ristori E, Lopez-Ramirez MA, Narayanan A, et al. A dicer-miR-107 interaction regulates biogenesis of specific miRNAs crucial for neurogenesis. Dev Cell 2015;32:546–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Irmak MK, Erdem U, Kubar A. Antiviral activity of salivary microRNAs for ophthalmic herpes zoster. Theor Biol Med Model 2012;9:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hennessey PT, Sanford T, Choudhary A, et al. Serum microRNA biomarkers for detection of non-small cell lung cancer. PloS One 2012;7:e32307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sun Y, Wang M, Lin G, et al. Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PloS One 2012;7:e47003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zampetaki A, Kiechl S, Drozdov I, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 2010;107:810–7. [DOI] [PubMed] [Google Scholar]
- [13].Michael A, Bajracharya SD, Yuen PS, et al. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis 2010;16:34–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hsu PW, Lin LZ, Hsu SD, et al. ViTa: prediction of host microRNAs targets on viruses. Nucleic Acids Res 2007;35:D381–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Grassmann R, Jeang KT. The roles of microRNAs in mammalian virus infection. Biochim Biophys Acta 2008;1779:706–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ma F, Xu S, Liu X, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol 2011;12:861–9. [DOI] [PubMed] [Google Scholar]
- [17].Wu J, Pan Z, Cheng M, et al. Ginsenoside Rg1 facilitates neural differentiation of mouse embryonic stem cells via GR-dependent signaling pathway. Neurochem Int 2013;62:92–102. [DOI] [PubMed] [Google Scholar]
- [18].Tzekov R, Dawson C, Orlando M, et al. Sub-chronic neuropathological and biochemical changes in mouse visual system after repetitive mild traumatic brain injury. PloS One 2016;11:e0153608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Koh YW, Park C, Yoon DH, et al. Prognostic significance of COX-2 expression and correlation with Bcl-2 and VEGF expression, microvessel density, and clinical variables in classical Hodgkin lymphoma. Am J Surg Pathol 2013;37:1242–51. [DOI] [PubMed] [Google Scholar]
- [20].Wang C, Ding M, Xia M, et al. A five-miRNA panel identified from a multicentric case-control study serves as a novel diagnostic tool for ethnically diverse non-small-cell lung cancer patients. EBioMedicine 2015;2:1377–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Belleannee C, Calvo E, Thimon V, et al. Role of microRNAs in controlling gene expression in different segments of the human epididymis. PloS One 2012;7:e34996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Siebert M, Westbroek W, Chen YC, et al. Identification of miRNAs that modulate glucocerebrosidase activity in Gaucher disease cells. RNA Biol 2014;11:1291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Xu X, Wang S, Liu J, et al. Hypoxia induces downregulation of soluble guanylyl cyclase beta1 by miR-34c-5p. J Cell Sci 2012;125(pt 24):6117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


