Abstract
Objective(s):
Tuberculosis (TB), a disease caused by Mycobacterium tuberculosis (Mtb), stayed a global health thread with high mortality rate. Since TB has a long-term treatment, it leads high risk of drug resistant development, and there is an urgent to find new drugs. The aim of this study was designing new inhibitors for a new drug target, iron dependent regulator, IdeR.
Materials and Methods:
Based on the interaction most populated amino acids of IdeR to the related gene operators, 50 short peptides were modeled. Bonding affinity of short peptides toward DNA were studied by docking. Top 10 best predicted bonding affinity were selected. DNA binding assay, microplate alamar blue assay, colony counting, quantitative real time- PCR (qRT-PCR) of IdeR corresponding genes, cell wall-associated mycobactin and whole-cell iron estimation were done to prove the predicted mechanism of in silico potent constructs.
Results:
Amongst the 10 synthesized short peptide candidates, glycine-valine-proline-glycine (GVPG) and arginine-proline-arginine (RPR) inhibited Mtb in vitro at 200 μM concentration. qRT-PCR showed mbtB 58-fold over expression that resulted in Mtb growth inhibition. Cell wall-associated mycobactin and whole-cell iron estimation confirmed the results of qRT-PCR.
Conclusion:
We introduced two new lead compounds to inhibit Mtb that are promising for the development of more potent anti-tubercular therapies.
Keywords: IdeR, Inhibitor, Modeling, Mycobacterium – Tuberculosis, RT-PCR
Introduction
Tuberculosis (TB), a disease caused by Myco-bacterium tuberculosis (Mtb), stayed a global health thread with high mortality rate. In 2016, it is estimated that 10.4 million new TB cases were developed and 1.4 million died from TB (1). High rate of TB in developing countries, low quality of education and long-term treatment are factors that lead risk of drug resistant Mtb. Indeed, 3.3% to new TB cases and 20% of treated cases have developed multi-drug resistant TB (MDR-TB). The problem has worsened with arising of extensively drug-resistant TB (XDR-TB) that presented in 9.7% of MDR-TB cases (1). Whereas there is an urgent need to find new drugs, it has only two new drugs, bedaquiline and delamanide, have been approved after five decades (2, 3).
Efforts to find new drug targets in Mtb have resulted in identifying key proteins that are promising to defeat TB (4). Global regulators that are central in biological metabolisms are of interest since
there are different genes regulated by them (5). Iron dependent regulator protein, IdeR, nominated Rv2711, as repressor/inducer controls 51 genes that are critical for Mtb metabolism, especially those that are related to iron-dependent genes expression, iron metabolism, and oxidative stress responses (5). Of which, mbtB and bfrB are more affected due to their straight role in uptake and storage of iron, respectively (5, 6).
Iron is involved in essential cellular functions, but very limited in the environment. However, due to generating toxic reactive oxygen species (ROS) in aerobic respiration in the Fenton reaction (7) controlling intracellular concentration is critical to the organism. IdeR and under-regulated genes play important roles in this issue. Since IdeR protein is crucial to Mtb, ideR mutation, non-functional protein and disordering function kills bacterium (6).
Crystallographic studies on IdeR revealed the protein-protein and protein-DNA interactions of this protein, in detail (8). In the case of monomer construction, three structural domains are considered, that could explain the biological function of this protein. Domains were assumed to be DNA-binding domain (DBD), includes amino acid no. 1 to 74, dimerization domain (DD), 75 to 140, and SH3-like domain, consisting of residues 151 to 230. However, complete functional activity of IdeR occurred when it forms dimers, but in the presence of iron, that interact to the conserved region on DNA (8). Therefore, successful inhibition of dimer formation or inhibiting the interaction of IdeR to related sequence of DNA may result in non-expression or over-expression of related genes. In this study, we have focused on the in silico designed competitors that intercept IdeR to DNA.
Materials and Methods
Designing constructs
The structures of short peptides were drawn by ChemDraw Ultra 8.0. The two dimensional (2D) structures of peptides were exported to HyperChem 8 software. To simulate the three dimensional (3D) structures, conformational analysis has been performed by MM+ (RMS gradient=0.05 kcal mol-1) and AM1 methods (convergence limit=0.01; Iteration limit=50; RMS gradient=0.05 kcal mol-1; Polak-Ribiere optimizer algorithm) (9).
For docking analysis, the 33-mer DNA sequence of iron box and constructs were submitted to AutoDock software (AutoDock Tools, V. 1.5.6).
The docking regions of the DNA were defined by considering Cartesian chart -5.534, -0.100, and 3.698 as the central of a grid size with 60, 60 and 60 points in X, Y and Z axis. Lamarckian genetic algorithm Parameters (GALS) were used for generating the docking parameter files. The number of generations and maximum number of energy evaluations was set to 200 and 2,500,000, respectively. The root-mean-square deviation tolerance (RMSD) of 2.0 Å was considered to cluster 200 docked complexes. Calculations were outputted as free energy of binding (ΔGb). Estimated dissociation constant (Kd) was calculated by using of this equation: ΔGb=2.3RTlogKd (Kd=1/Kb). Peptides were ordered with >95% purity (MIMOTOPES, Australia). Then, they were subjected for further investigations (Figure 1).
Figure 1.
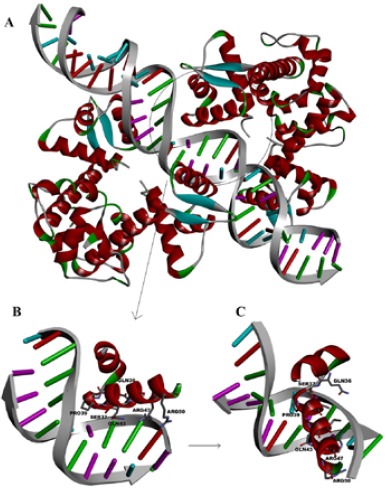
The interaction of IdeR with iron box of DNA. A) Double-dimer interaction of IdeR. The region of direct interaction of protein with DNA (PDB: 1U8R) is circled. B) Residues of IdeR that make hydrogen bonds are illustrated. C) The B picture rotated 90 ° to show better view of interacted residues with DNA
Modified DNA binding assay
To evaluate the affinity of constructs to DNA duplex, a competitive assay based on DNA/Methyl green displacement was performed (10).
A 33-mer oligonucleotide of both forward and reverse strands of the mbtA-mbtB operator sequences was obtained at the highest available molarity (Macrogene, Korea). Duplexes were generated through mixing equimolar amounts of both strands, by heating to 95 °C for 5 min, and slowly cooling. To confirm duplex formation a 7% polyacrylamide gel was used to run 1 μl of mixture.
Methyl green (MG) (Sigma), at the final concentration of 10 μM was mixed with 1 μM of DNA duplex and remained at least 15 min at room temperature before adding constructs. Constructs at 11 concentrations, 800, 400, 200, 100, 50, 25, 12, 6, 3, 1 and 0.5 μM, were added to the DNA/MG mixture and the volume adjusted to 100 μl with distilled water per well. DNA/MG, construct/DNA, construct/ MG and MG solely were separately embedded in distinct wells to unravel the discrepancies. Experiments were repeated at least in triplicate. A non-binding flat bottom clear ELISA microplate (Greiner Bio One, Austria) were used to spectrophoto-metric assay. Inoculated plates were left 24 hr at darkness and results were measured spectro-photometrically (BioTek, Winooski, VT, USA) at 630 nm wavelength. IC50 of each construct calculated based on the concentration that was required to decrease 50% of initial absorbance of DNA/MG complex by using of binding-competitive equation in PRISM 6.0. (11).
Bacterial strain and media
Mtb strain H37Rv ATCC 27294 was used in all of the experiments. Middlebrook 7H9 broth (Himedia, India) was used as high-iron medium (h7H9). To assay Mtb growth under low-iron conditions (1 μM), we reconstitute 7H9-based composition medium (r7H9) by adding 1 μM of iron in the form of FeCl3. The amount of iron was determined by atomic absorption spectroscopy. Both of media were supplemented by 10% ADC (Fluka), 0.2% glycerol, 0.05% Tween-80 and 0.085% NaCl. All chemicals were provided from Sigma.
Microplate alamar blue assay (MABA) and colony counting
The microplate alamar blue was used to assess sensitivity of Mtb to the constructs (12). Briefly, 106 colony forming units (CFUs) of Mtb cells were inoculated in 1 ml of h7H9 and r7H9 that at serial dilution manner, contained 800, 400, 200, 100, 50, 25, 12, 6, 3, 1 and 0.5μM concentration of constructs. After one week incubation, 100 μl of media transferred to 96-well U-bottom microplates (Greiner Bio One, Austria) and 20 μl of 0.01% resazurin was added to each well. After 24 hrs results were recorded. In addition, 100 μl of initial inoculated broth mediums were subjected to 10-3 dilution, and 50 μl of diluted bacteria were seeded into LJ (Löwenstein–Jensen) medium for colony counting. Experiments were done in triplicates. After three weeks incubation colony forming units of Mtb were counted. Construct-free bacterial inoculated broth media, h7H9 and r7H9, were considered as initial growth controls.
To analyze the exact function of IdeR-construct-DNA interaction in different iron-content media, we defined several processes (Table 1). Mtb cells were cultured in r7H9 and h7H9, with and without constructs. After 48 hrs of incubation in 37° C, cells were washed three times by phosphate buffer saline (PBS), and then transferred to the second medium.
Table 1.
Different processes to find out the interaction of IdeR-construct-DNA. In the processes of 9 and 10 Mtb cells were not transferred to the second medium, and cells remained in the initial medium to the end of incubation. Experiments were repeated three times. Abbreviations: r7H9, reconstituted 7H9; h7H9, hi-iron 7H9; compd., compound
| Process | Initial medium | Second transferred medium |
|---|---|---|
| 1 | r7H9 | h7H9-Compd. |
| 2 | r7H9 | r7H9-Compd. |
| 3 | h7H9 | h7H9-Compd. |
| 4 | h7H9 | r7H9-Compd. |
| 5 | r7H9-Compd. | r7H9 |
| 6 | r7H9-Compd. | h7H9 |
| 7 | h7H9-Compd. | r7H9 |
| 8 | h7H9-Compd. | h7H9 |
| 9 | r7H9-Compd. | - |
| 10 | h7H9-Compd. | - |
Quantitative real-time PCR
Mtb cells from different processes (Table 1) were subjected to qRT-PCR. Total RNA extraction, cDNA synthesis and quantitative reverse transcription-PCR (RT-PCR) were performed as previously described by Pandey et al. using primers targeting mbtB and bfrB of Mtb (6). qRT-PCR was performed by the Rotor-Gene 6000 instrument (Corbett Life Science, Valencia, CA) using Real-Time SYBR green 2x Master Mix (Parstous, Iran) in triplicates. The results were normalized to the amount of 16S rRNA and were calculated by 2-ΔΔCT Method (13).
Cell wall-associated mycobactin and whole-cell iron estimation
To assess inhibitory role of constructs on IdeR function, cell wall-associated mycobactin and whole cell iron were measured. The prior, as the consequence of mbtB expression, and the latter, as the outcome of bfrB, could explain the efficacy of each construct, respectively.
Cell wall-associated mycobactin was determined by a simple method, as described by Schwyn et al (14). Mtb cells from processes (Table 1) were washed three times by PBS, then, were subjected to overnight chloroform-ethanol (1:1) protein extraction. A drop of FeCl3 saturated in ethanol was added, and, the optical density at 450 nm was observed.
Intracellular iron content was measured as previously described by Dragset et al (15). Briefly, Mtb was cultured in h7H9 and r7H9 with construct. After 48 hr bacteria were centrifuged and three times washed with PBS. After acid nitric digestion, H2O2 was added to the mixture; then diluted with 2 ml water, metal content was scaled by atomic absorption spectroscopy (Analyst 300; PerkinElmer, Foster City, CA, USA).
Cellular toxicity assay
To found the cellular toxicity of constructs MDCK cell line was used. The number of viable cells was measured via 0.25% Trypan Blue on haemocytometer. Cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Sigma) culture medium which supplemented by 10% heat-inactivated fetal bovine serum (FBS) (Gibco), 100 mg/ml streptomycin (Sigma), 100 unit/ml penicillin (Sigma) and 2 mM L-glutamine (Sigma). A total of 5.0×103 cells were seeded in each well of 96-well microplate that contained 100 μL of medium and incubated overnight at 37 C in 5% CO2. Then, with a 100 μl of 0.25% FBS culture medium containing 800 μM of constructs the cell culture medium replaced, and incubated 24 hr at 37 °C in 5% CO2. About 20 μl of 0.01 % of resazurin was added to each well and incubated in the same conditions again. The results were recorded observationally 24 hr later. Experiments were done in triplicate for all constructs. Furthermore, bacterial strains including Escherichia coli, Staphylococcus aureus and Pseudomonas aeruginosa were studied to find the sensitivity of these strains to constructs at the concentration of 800 μM.
Results
Modeling of the constructs
According to the crystallographic analysis of IdeR (PDB entry: 1U8R), the interaction of the DBD of IdeR, few residues directly abutted on the iron box of DNA promoters. Predominant amino acids in this region are valine (V), alanine (A), glycine (G), threonine (T), proline (P), arginine (R), serine (S), glutamine (Q) and asparagine (N). Among them R47, R50, Q43, S37 and Q36 have hydrogen bonds with the nucleotides of DNA iron box (Figure 1).
According to these data, 50 constructs with three to four residues from the aforementioned amino acids were designed and their binding potency towards DBD were evaluated by AutoDocTools 4.2 software. The docking results were further analyzed to find a binding conformer from each cluster with the lowest Kd (estimated DNA-ligand dissociation constant), and finally, the average of the aforementioned Kd were calculated and named as Kd* for each of the synthetic peptides. Constructs that had Kd* lower than 100 μM were prepared and used for DNA binding assay. Among the proposed peptides, 10 cases had Kd* less than 100 μM. All of the aforementioned peptides were synthesized and their DNA binding potency were assayed.
Amongst the 10 peptides, GVPG was composed of four amino acids while the rest were triads. In these 10 selected compounds, proline was found in all except construct QTQ. Furthermore, PRP had the lowest Kd* among all constructs (Figure 2).
Figure 2.
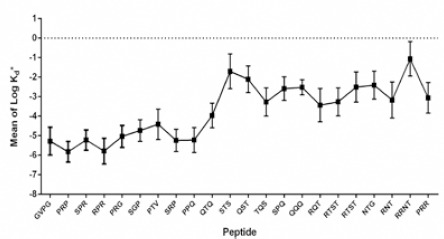
Some of the designed peptides based on IdeR sequence and DNA interacted amino acids. Peptides with mean of LogKd* less than -4 were considered for testing in vitro
Interestingly, it can be discussed that inserting of proline in the synthetic constructs form a pre-organized shape for binding to the major groove of the desired DNA moiety (Figure 3) as seen for proline-39 in the DBD of IdeR (Figure 1).
Figure 3.
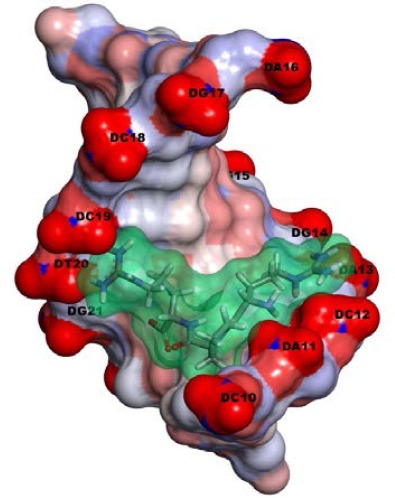
The solvent-surface presentation of the molecule interaction of RPR (a model with the best Kd) with DNA iron box. The RPR is distinguished by stick and transparent surface view
Modified DNA binding assay
To find the affinity of the constructs to the iron box of mbtB operator, competition assay with methyl green (MG) to attach double stranded DNA was performed. The IC50 calculated through spectrophotometric assay of the constructs, which are presented in detail in (Table 2). Amongst the constructs, RPR had the best IC50.
Table 2.
Characteristics comparison of construct’s in silico and in vitro. Bolded constructs showed anti-mycobacterial activity. Abbreviations: Kd*, average of lowest Kd; MABA, microplate alamar blue assay; MIC, minimum inhibitory constant
| Structure | Kd* | Best predicted Kd calculated by AutoDoc (μM) | DNA/MG spectrophotometric IC50 (μM) | MABA MIC (μM) | Colony counting |
|---|---|---|---|---|---|
| GVPG | -5.28 | 2.9 | 4 | 200 | 78.103 |
| SGP | -4.73 | 33 | 128 | >800 | >106 |
| PTV | -4.35 | 38.75 | 147 | >800 | >106 |
| PRP | -5.81 | 2.55 | 10.2 | >800 | >106 |
| SPR | -5.2 | 17.83 | 20 | >800 | >106 |
| SRP | -5.24 | 9.74 | 101 | >800 | >106 |
| RPR | -5.78 | 2.79 | 2.8 | 200 | 55.103 |
| PRG | -5.04 | 6.77 | 8 | >800 | >106 |
| QTQ | -4.00 | 60.72 | 316 | >800 | >106 |
| PPQ | -5.22 | 103.6 | 158 | >800 | >106 |
Linear regression was observed between Kd* and IC50 variations (Figure 4). Except two compounds, RPR and SPR, all of the rest showed linear correlation between logarithmic form of Kd* and IC50 with R-square value of 0.97. The plot shows that the computer based analysis can be responsible for in vitro prediction behavior of the designed compounds.
Figure 4.
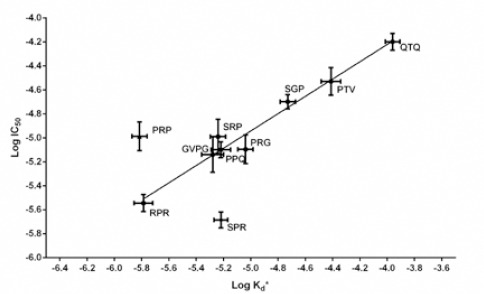
Linear correlation of LogIC50 and LogKd* of compounds (R-square = 0.97)
Microplate alamar blue assay (MABA) and colony counting
Constructs were tested in vitro to found the minimum inhibitory concentration (MIC) in practice. Constructs GVPG and RPR inhibited Mtb growth at 200 μM and the others were not effective at the highest concentration (800 μM). Colony forming units of Mtb cells were counted. Two effective constructs, GVPG and RPR, were in accordance with MIC definition, that of 90% of the inoculated cells inhibited by inhibitor. At the concentration of 400 μM no colony was seen for GVPG and RPR constructs.
qRT-PCR of mbtB and bfrB
Constructs were designed to interfere with iron hemostasis, and, they were expected to up or down-regulate related genes. Real time results showed that constructs GVPG and RPR at the concentration of 200 μM could affect the expression of mbtB genes, where Mtb cells were grown in r7H9 and h7H9 (Figure 5). A 41-fold increase in expression of mbtB was seen in the condition that Mtb was cultured in depleted iron medium, r7H9, followed by growth in constructs containing high iron medium, h7H9-compd. Constructs GVPG and RPR showed the same level of increase in expression of mtbB. Furthermore, 58-fold increase in mbtB expression were observed where Mbt cells were grown initially in r7H9 containing constructs (r7H9-compd) and then transferred to h7H9 medium. The expression level of bfrB was not affected by constructs, grown or transferred, neither r7H9 nor h7H9.
Figure 5.
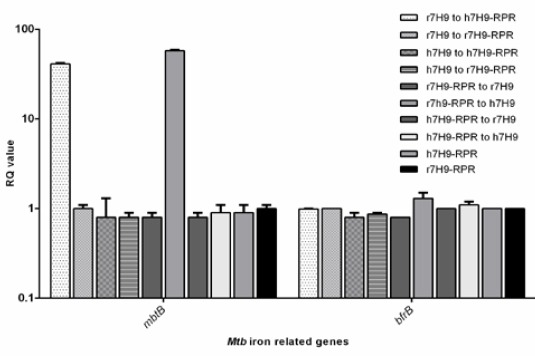
Changes of the expression level of iron-responsive genes, mbtB and bfrB, of Mbt cells grown in r7H9 and h7H9, with or without construct RPR. Construct GVPG had the same results (data not shown). The relative transcript levels of two iron-responsive genes (mbtB and bfrB) were measured by quantitative RT-PCR. Results from three experiments were calculated by 2-ΔΔCT. RQ (relative quantification) values are presented in Log10
Cell wallassociated mycobactin and wholecell iron estimation
To ascertain IdeR dysfunction, we analyzed the cell wallassociated mycobactin and wholecell iron content, as the result of bfrB and mbtB expression level.
As the same as real-time PCR results, constructs GVPG and RPR at the concentration of 200 μM could only change iron and mycobactin content of those Mtb cells that grown in r7H9 medium and transferred to h7H9. No differences were observed in iron and mycobactin content in Mtb cells grown under the other conditions (Figure 6).
Figure 6.
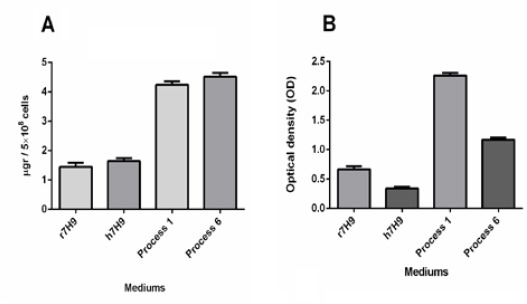
Wholecell iron and cellassociated mycobactin after treating with the 200 μM concentration of construct RPR. Construct GVPG had the same results (data not shown). Results were collected from 5×108 CFU of Mtb cells. A) Wholecell iron of Mtb cells in media, r7H9 and h7H9, processes 1 and 6. B) Cellassociated mycobactin harvested from Mtb cells cultured in media, r7H9 and h7H9, r7H9 and h7H9, processes 1 and 6
Cellular toxicity assay
MDCK cell line growth did not inhibited under 800 μM of constructs. As the constructs were indeed short peptides, it could be expected that, they did not affect mammalian cell growth. Additionally, bacterial strains were not susceptible to the constructs at the concentration of 800 μM (data not shown).
Discussion
Iron is crucial metal for Mtb survival, but excess amount is toxic. Hence, iron hemostasis is strongly controlled. Therefore, iron metabolism, and, in particular, IdeR has the potential to be considered as new drug target. In this study we targeted the binding site of IdeR holo-protein to DNA operator through mimicking DBD of IdeR to DNA, in the form of short peptides.
According to the processes presented in Table 1, only the processes were successful that Mtb cells were in the low-iron 7H9, r7H9, and then transferred to high-iron 7H9, h7H9. In iron limited conditions, like intracellular conditions, IdeR subunits cannot assemble. In the other hand, constructs entered, and when iron increases, assembled IdeR unable to regulate mbtB. Therefore, it up-regulates and cellwall associated mycobactin increased. Consequently, intracellular iron increased and overloaded that harms the cell through Fenton reaction. However, bfrB regulation, in comparison to the normal conditions, is not affected by the constructs (Figure 6). It can be explained in the way that, when cell is in iron deplete conditions, IdeR does not attached to the bfrB operator. But, when the iron increased in the presence of constructs, IdeR still cannot attach to the bfrB operator, and the expression level of this gene remained as the same as iron limited conditions. The lack of bacreioferritin, as the product of bfr gene, leads to lack of control over the iron storage, which leads to increasing free iron in the cell. Free oxygen radicals as the result of Fenton reaction damage biomolecules.
In the other study 92 components and 85 protein-protein interactions in iron hemostasis of Mtb, as a modeled system, were analyzed. They have found the importance of IdeR in Mtb as a key component that is critical for establishment of all interactions. This study only confirmed the key role of IdeR in Mtb as well as the importance of this protein as drug target, and no inhibitors were studied (16).
Due to production facility from point of view of the synthetic procedure (solid phase technique) and structure variation using of this kind of compounds for discovery of new drugs is common, in spite of instability of peptide compounds in biological media (17–19).
To assess the effectiveness of infected cells by Mtb, our constructs should be challenged in vivo. However, we defined two constructs that act as lead constructs which worth for more evaluations.
Conclusion
Whereas the ten of designed constructs showed potent results, in terms of computation and DNA binding energy, but only two of them could inhibit bacterial growth in the medium. Other factors that can affect the potency of drugs are cellwall transporting and chemical stability in the medium during culturing. The synthetic peptides are usually unstable in culture medium because of hydrolyzing of the amide bond. It can be accelerated by passing more time and temperature elevating (herein the culturing was down during one week at 37 °C). This could be a rational reason why the MIC of the synthetic amides is high.
The structure of the effective peptides (RPR and GVPG) can lead us to design new compounds by using of structure based searching method with high cell wall permeability and stability in biological medium.
Acknowledgment
The authors would thank Vice Chancellor of Health of Mashhad University of Medical Sciences for permission to use their laboratory in this study. The results described in this paper were part of PhD student thesis, Himen Salimizand, and supported by grant No 930710 from Mashhad University of Medical Sciences.
References
- 1.World Health Organization (WHO) Global Tuberculo-sis Report. 2015. Available at: http://www.who.int/tb/publications/global_report/en/)
- 2.Matsumoto M, Hashizume H, Tomishige T, Kawasaki M, Tsubouchi H, Sasaki H, et al. OPC-67683, a nitro-dihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med. 2006;3:e466. doi: 10.1371/journal.pmed.0030466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole ST, Alzari PM. Microbiology. TB- a new target, a new drug. Science. 2005;307:214–215. doi: 10.1126/science.1108379. [DOI] [PubMed] [Google Scholar]
- 4.Chung BK-S, Dick T, Lee D-Y. In silico analyses for the discovery of tuberculosis drug targets. J Antimicrob Chemother. 2013;68:2701–2709. doi: 10.1093/jac/dkt273. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. ideR, an essential gene in mycobacterium tuberculosis: role of ideR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immunol. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pandey R, Rodriguez GM. IdeR is required for iron homeostasis and virulence in Mycobacterium tuberculosis. Mol Microbiol. 2014;91:98–109. doi: 10.1111/mmi.12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keyer K, Imlay JA. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci U S A. 1996;93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wisedchaisri G, Chou CJ, Wu M, Roach C, Rice AE, Holmes RK. Crystal structures, metal activation, and DNA-Binding properties of two-domain ideR from Mycobacterium tuberculosis. Biochemistry. 2007;46:436–447. doi: 10.1021/bi0609826. [DOI] [PubMed] [Google Scholar]
- 9.El-Ayaan U, Abdel-Aziz AA, Al-Shihry S. Solvatochromism, DNA binding, antitumor activity and molecular modeling study of mixed-ligand copper(II) complexes containing the bulky ligand: Bis[N-(p-tolyl)imino] acenaphthene. Eur J Med Chem. 2007;42:1325–1233. doi: 10.1016/j.ejmech.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 10.Mcalpine B. A colorimetric microassay for the detection of agents that interact with DNA. 1992;55:1582–1587. doi: 10.1021/np50089a004. [DOI] [PubMed] [Google Scholar]
- 11.Bronstein JC, Weber PC. A Colorimetric Assay for high-throughput screening of inhibitors of herpes simplex virus Type 1 alkaline nuclease. Anal Biochem [Internet] 2001;293:239–245. doi: 10.1006/abio.2001.5144. [DOI] [PubMed] [Google Scholar]
- 12.Franzblau SG, Witzig RS, McLaughlin JC, Torres P, Madico G, Hernandez A, et al. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J Clin Microbiol. 1998;36:362–366. doi: 10.1128/jcm.36.2.362-366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 15.Dragset MS, Poce G, Alfonso S, Padilla-Benavides T, Ioerger TR, Kaneko T, et al. A novel antimycobacterial compound acts as an intracellular iron chelator. Antimicrob Agents Chemother. 2015;59:2256–2264. doi: 10.1128/AAC.05114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh S, Prasad KVS, Vishveshwara S, Chandra N. Rule-based modelling of iron homeostasis in tuberculosis. Mol Biosyst. 2011;7:2750–2768. doi: 10.1039/c1mb05093a. [DOI] [PubMed] [Google Scholar]
- 17.Hao G, Rongji D, Kui Q, Zhongqiu T, Heyao W. A synthetic peptide derived from NK-lysin with activity against mycobacterium tuberculosis and its structure-function relationship. Int J Pept Res Ther. 2011;17:301–306. [Google Scholar]
- 18.Flexner C. HIV drug development: the next 25 years. Nat Rev Drug Discov. 2007;6:959–966. doi: 10.1038/nrd2336. [DOI] [PubMed] [Google Scholar]
- 19.Lau QY, Choo XY, Lim ZX, Kong XN, Ng FM, Ang MJY, et al. A head-to-head comparison of the antimicrobial activities of 30 ultra-short antimicrobial peptides against staphylococcus aureus, pseudomonas aeruginosa and Candida albicans. Int J Pept Res Ther. 2015;21:21–28. [Google Scholar]


