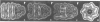Abstract
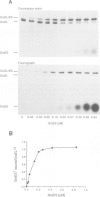
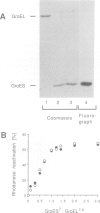
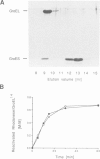
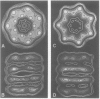
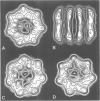
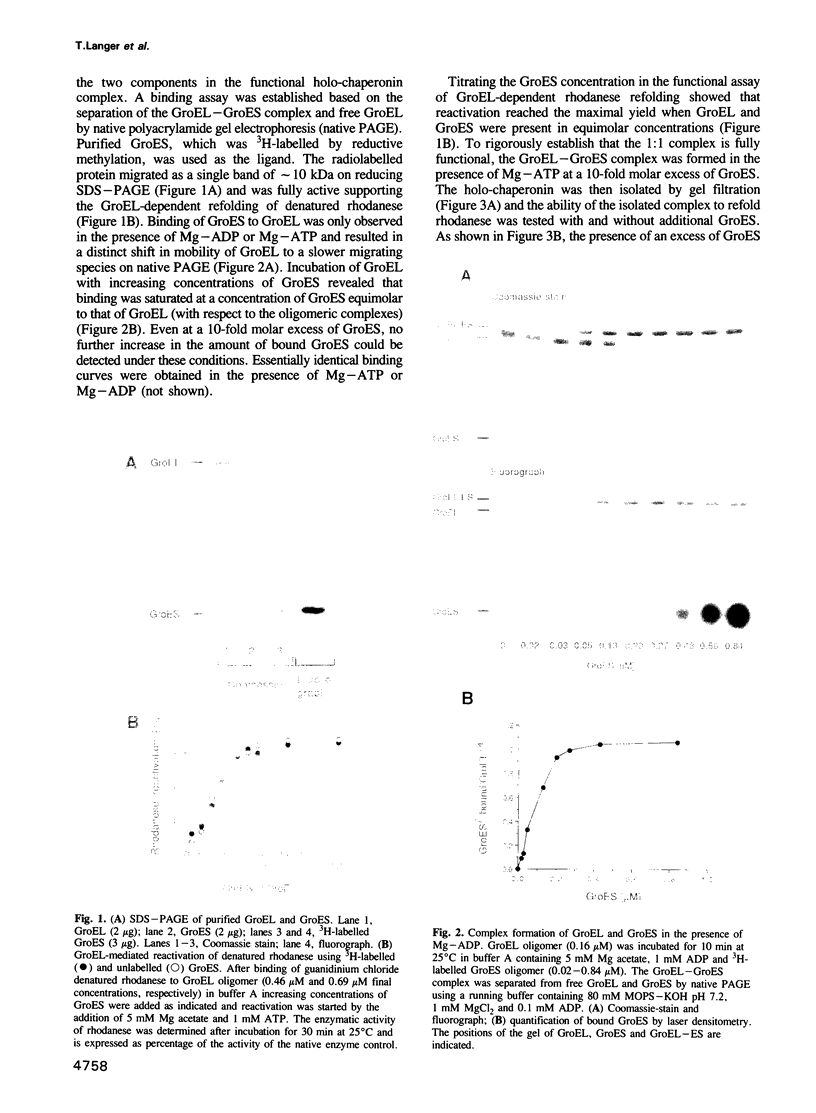
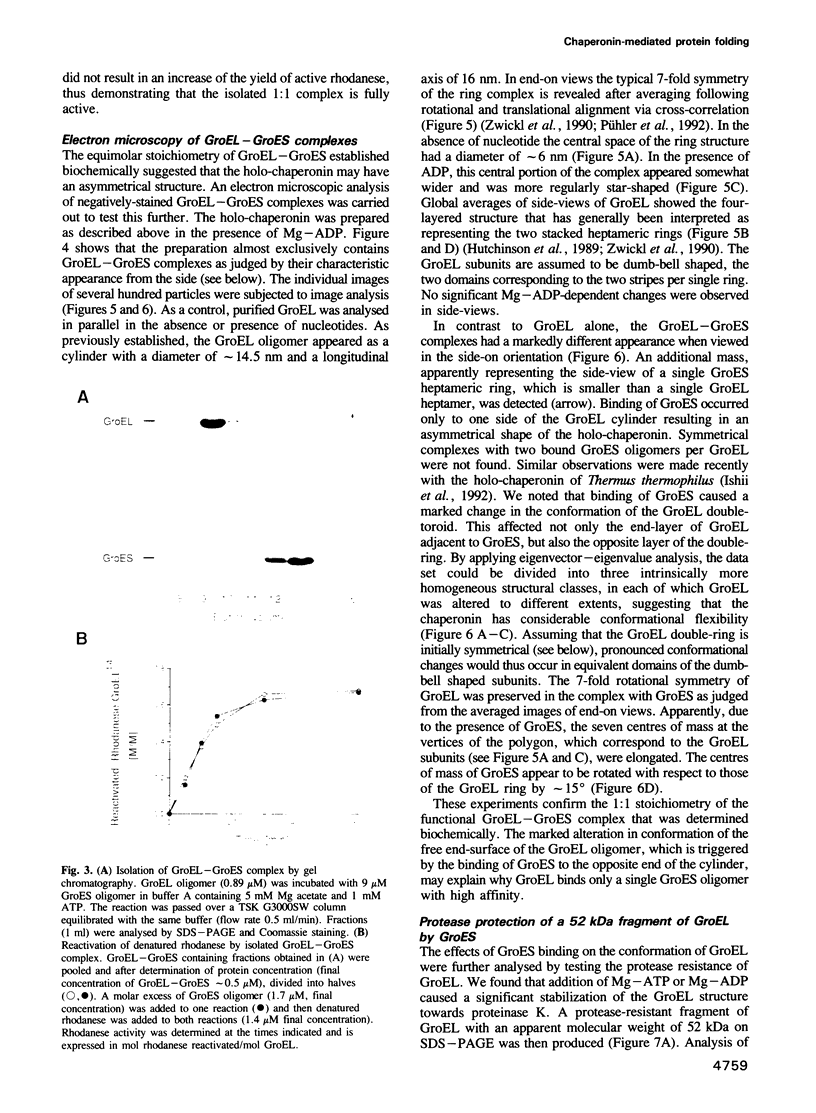
The mechanism of GroEL (chaperonin)-mediated protein folding is only partially understood. We have analysed structural and functional properties of the interaction between GroEL and the co-chaperonin GroES. The stoichiometry of the GroEL 14mer and the GroES 7mer in the functional holo-chaperonin is 1:1. GroES protects half of the GroEL subunits from proteolytic truncation of the approximately 50 C-terminal residues. Removal of this region results in an inhibition of the GroEL ATPase, mimicking the effect of GroES on full-length GroEL. Image analysis of electron micrographs revealed that GroES binding triggers conspicuous conformational changes both in the GroES adjacent end and at the opposite end of the GroEL cylinder. This apparently prohibits the association of a second GroES oligomer. Addition of denatured polypeptide leads to the appearance of irregularly shaped, stain-excluding masses within the GroEL double-ring, which are larger with bound alcohol oxidase (75 kDa) than with rhodanese (35 kDa). We conclude that the functional complex of GroEL and GroES is characterized by asymmetrical binding of GroES to one end of the GroEL cylinder and suggest that binding of the substrate protein occurs within the central cavity of GroEL.
Full text
PDF
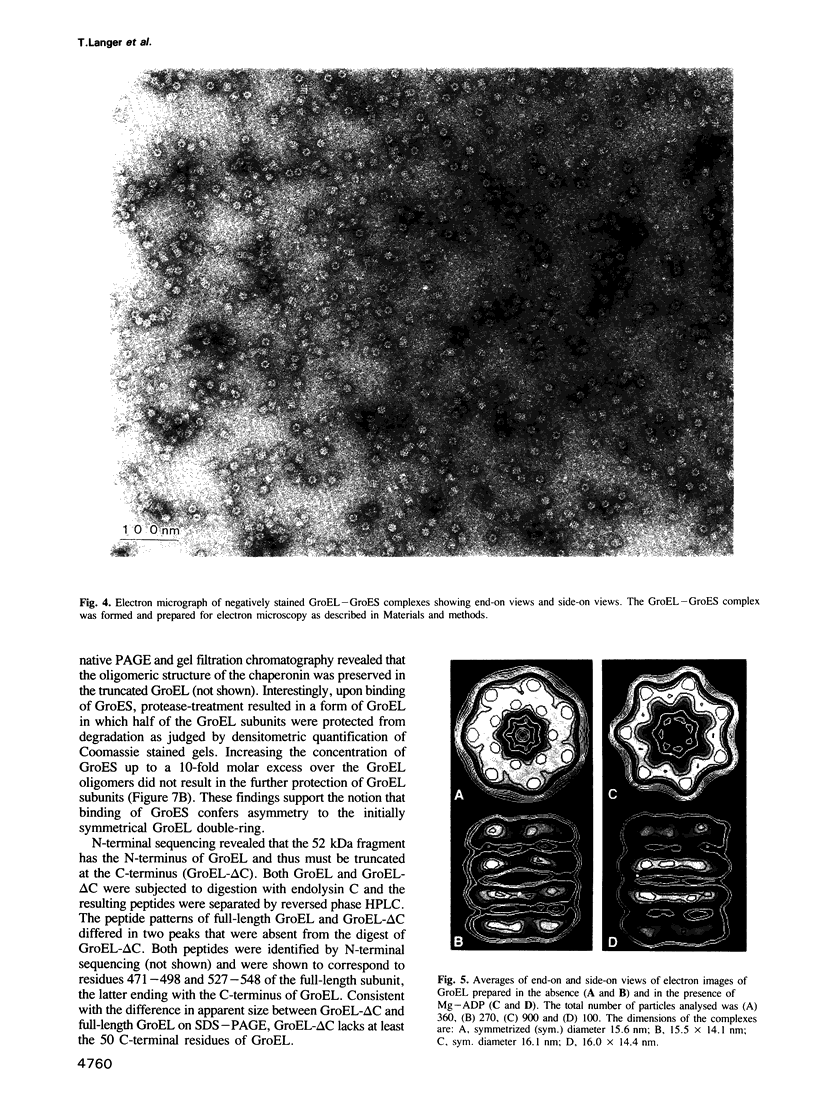
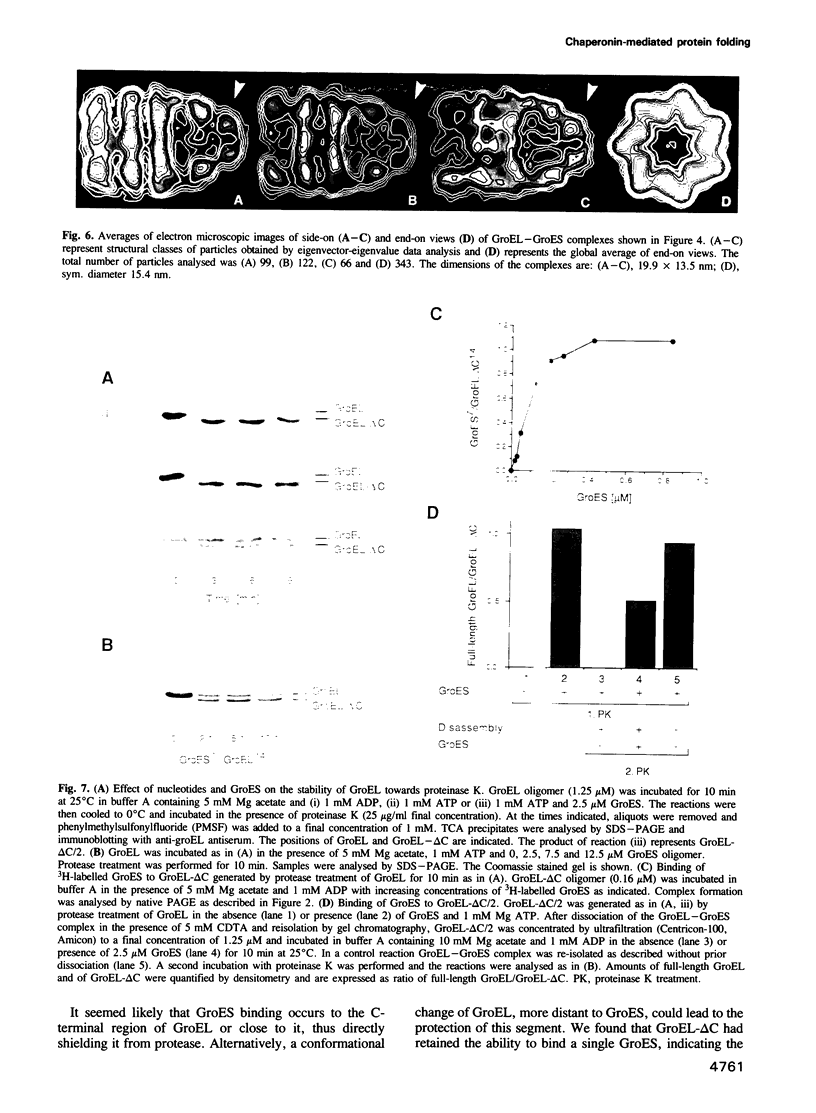
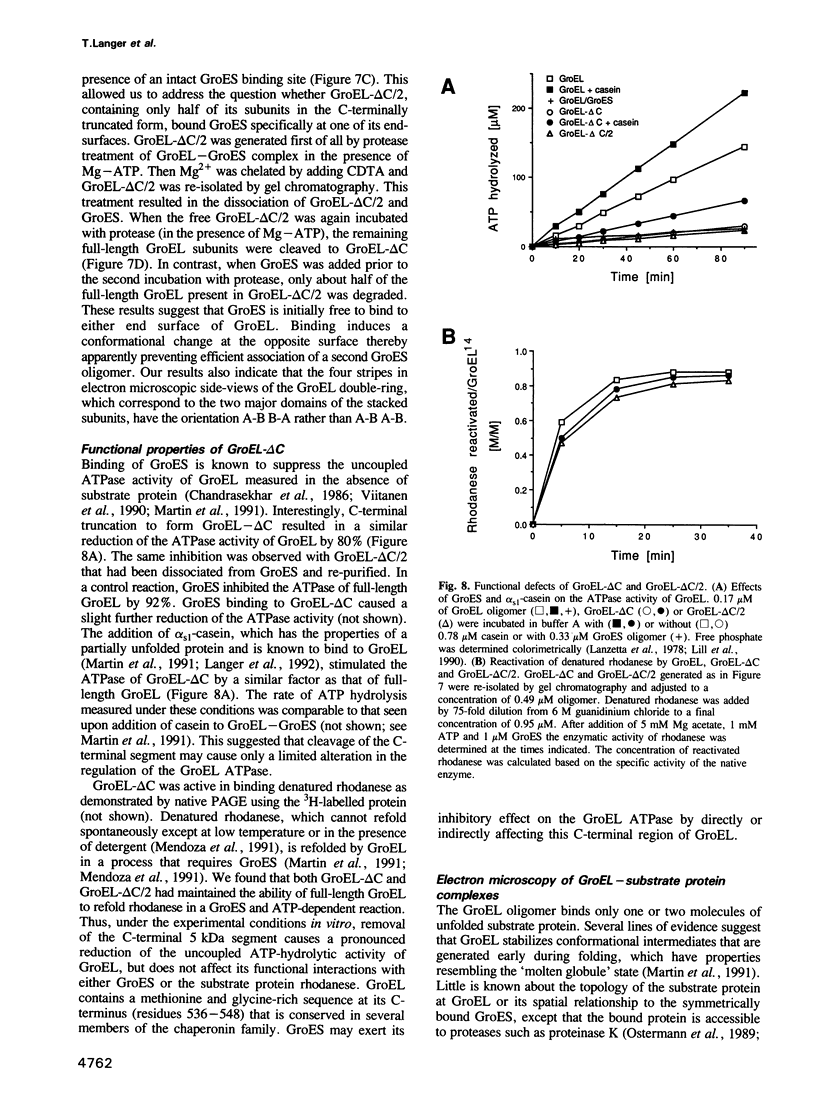
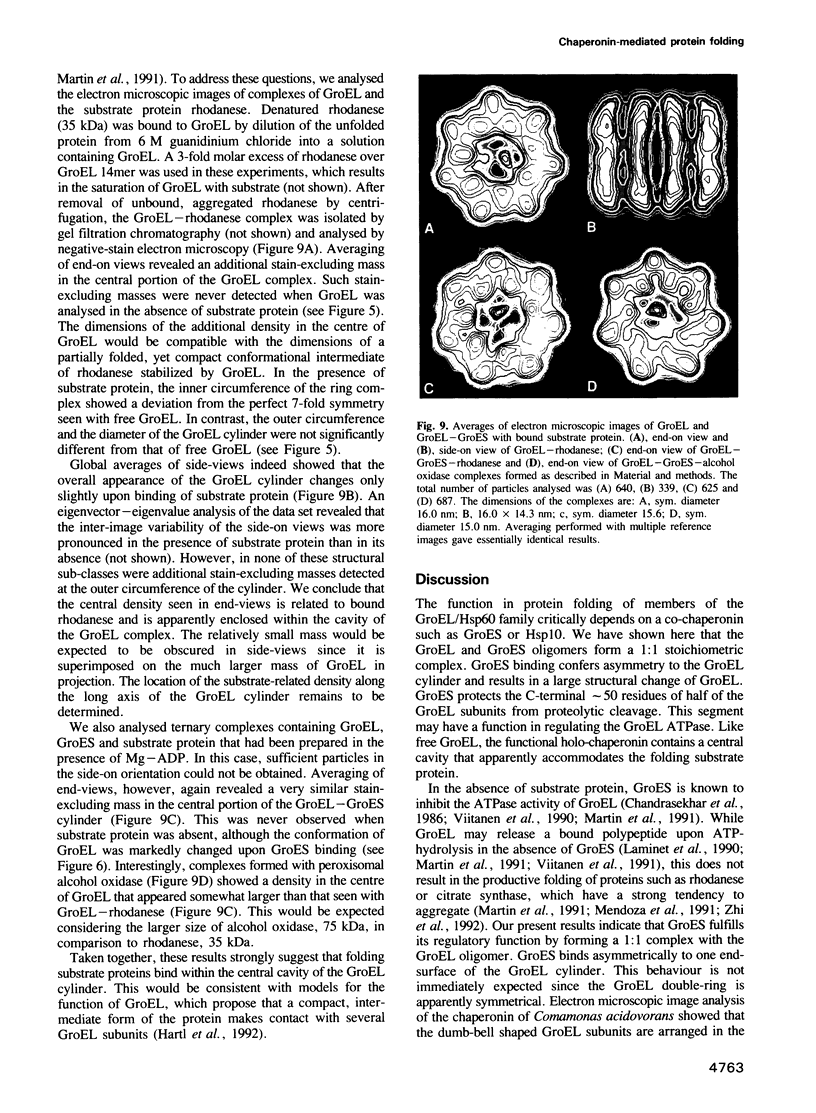


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Badcoe I. G., Smith C. J., Wood S., Halsall D. J., Holbrook J. J., Lund P., Clarke A. R. Binding of a chaperonin to the folding intermediates of lactate dehydrogenase. Biochemistry. 1991 Sep 24;30(38):9195–9200. doi: 10.1021/bi00102a010. [DOI] [PubMed] [Google Scholar]
- Bensadoun A., Weinstein D. Assay of proteins in the presence of interfering materials. Anal Biochem. 1976 Jan;70(1):241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- Bochkareva E. S., Lissin N. M., Flynn G. C., Rothman J. E., Girshovich A. S. Positive cooperativity in the functioning of molecular chaperone GroEL. J Biol Chem. 1992 Apr 5;267(10):6796–6800. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Buchner J., Schmidt M., Fuchs M., Jaenicke R., Rudolph R., Schmid F. X., Kiefhaber T. GroE facilitates refolding of citrate synthase by suppressing aggregation. Biochemistry. 1991 Feb 12;30(6):1586–1591. doi: 10.1021/bi00220a020. [DOI] [PubMed] [Google Scholar]
- Chandrasekhar G. N., Tilly K., Woolford C., Hendrix R., Georgopoulos C. Purification and properties of the groES morphogenetic protein of Escherichia coli. J Biol Chem. 1986 Sep 15;261(26):12414–12419. [PubMed] [Google Scholar]
- Creighton T. E. Molecular chaperones. Unfolding protein folding. Nature. 1991 Jul 4;352(6330):17–18. doi: 10.1038/352017a0. [DOI] [PubMed] [Google Scholar]
- Ellis J. Proteins as molecular chaperones. 1987 Jul 30-Aug 5Nature. 328(6129):378–379. doi: 10.1038/328378a0. [DOI] [PubMed] [Google Scholar]
- Ellis R. J., van der Vies S. M. Molecular chaperones. Annu Rev Biochem. 1991;60:321–347. doi: 10.1146/annurev.bi.60.070191.001541. [DOI] [PubMed] [Google Scholar]
- Evers M. E., Langer T., Harder W., Hartl F. U., Veenhuis M. Formation and quantification of protein complexes between peroxisomal alcohol oxidase and GroEL. FEBS Lett. 1992 Jun 22;305(1):51–54. doi: 10.1016/0014-5793(92)80653-x. [DOI] [PubMed] [Google Scholar]
- Fayet O., Louarn J. M., Georgopoulos C. Suppression of the Escherichia coli dnaA46 mutation by amplification of the groES and groEL genes. Mol Gen Genet. 1986 Mar;202(3):435–445. doi: 10.1007/BF00333274. [DOI] [PubMed] [Google Scholar]
- Gething M. J., Sambrook J. Protein folding in the cell. Nature. 1992 Jan 2;355(6355):33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Goloubinoff P., Christeller J. T., Gatenby A. A., Lorimer G. H. Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfoleded state depends on two chaperonin proteins and Mg-ATP. Nature. 1989 Dec 21;342(6252):884–889. doi: 10.1038/342884a0. [DOI] [PubMed] [Google Scholar]
- Gray T. E., Fersht A. R. Cooperativity in ATP hydrolysis by GroEL is increased by GroES. FEBS Lett. 1991 Nov 4;292(1-2):254–258. doi: 10.1016/0014-5793(91)80878-7. [DOI] [PubMed] [Google Scholar]
- Hartl F. U., Martin J., Neupert W. Protein folding in the cell: the role of molecular chaperones Hsp70 and Hsp60. Annu Rev Biophys Biomol Struct. 1992;21:293–322. doi: 10.1146/annurev.bb.21.060192.001453. [DOI] [PubMed] [Google Scholar]
- Hartman D. J., Hoogenraad N. J., Condron R., Høj P. B. Identification of a mammalian 10-kDa heat shock protein, a mitochondrial chaperonin 10 homologue essential for assisted folding of trimeric ornithine transcarbamoylase in vitro. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3394–3398. doi: 10.1073/pnas.89.8.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegerl R., Altbauer A. The "EM" program system. Ultramicroscopy. 1982;9(1-2):109–116. doi: 10.1016/0304-3991(82)90233-9. [DOI] [PubMed] [Google Scholar]
- Hemmingsen S. M., Woolford C., van der Vies S. M., Tilly K., Dennis D. T., Georgopoulos C. P., Hendrix R. W., Ellis R. J. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988 May 26;333(6171):330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- Hendrix R. W. Purification and properties of groE, a host protein involved in bacteriophage assembly. J Mol Biol. 1979 Apr 15;129(3):375–392. doi: 10.1016/0022-2836(79)90502-3. [DOI] [PubMed] [Google Scholar]
- Hohn T., Hohn B., Engel A., Wurtz M., Smith P. R. Isolation and characterization of the host protein groE involved in bacteriophage lambda assembly. J Mol Biol. 1979 Apr 15;129(3):359–373. doi: 10.1016/0022-2836(79)90501-1. [DOI] [PubMed] [Google Scholar]
- Hutchinson E. G., Tichelaar W., Hofhaus G., Weiss H., Leonard K. R. Identification and electron microscopic analysis of a chaperonin oligomer from Neurospora crassa mitochondria. EMBO J. 1989 May;8(5):1485–1490. doi: 10.1002/j.1460-2075.1989.tb03532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höll-Neugebauer B., Rudolph R., Schmidt M., Buchner J. Reconstitution of a heat shock effect in vitro: influence of GroE on the thermal aggregation of alpha-glucosidase from yeast. Biochemistry. 1991 Dec 17;30(50):11609–11614. doi: 10.1021/bi00114a001. [DOI] [PubMed] [Google Scholar]
- Ishii N., Taguchi H., Sumi M., Yoshida M. Structure of holo-chaperonin studied with electron microscopy. Oligomeric cpn10 on top of two layers of cpn60 rings with two stripes each. FEBS Lett. 1992 Mar 9;299(2):169–174. doi: 10.1016/0014-5793(92)80240-h. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laminet A. A., Ziegelhoffer T., Georgopoulos C., Plückthun A. The Escherichia coli heat shock proteins GroEL and GroES modulate the folding of the beta-lactamase precursor. EMBO J. 1990 Jul;9(7):2315–2319. doi: 10.1002/j.1460-2075.1990.tb07403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry S. J., Jordan R., McMacken R., Gierasch L. M. Different conformations for the same polypeptide bound to chaperones DnaK and GroEL. Nature. 1992 Jan 30;355(6359):455–457. doi: 10.1038/355455a0. [DOI] [PubMed] [Google Scholar]
- Langer T., Lu C., Echols H., Flanagan J., Hayer M. K., Hartl F. U. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992 Apr 23;356(6371):683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- Lanzetta P. A., Alvarez L. J., Reinach P. S., Candia O. A. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979 Nov 15;100(1):95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- Lill R., Dowhan W., Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990 Jan 26;60(2):271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- Lubben T. H., Gatenby A. A., Donaldson G. K., Lorimer G. H., Viitanen P. V. Identification of a groES-like chaperonin in mitochondria that facilitates protein folding. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7683–7687. doi: 10.1073/pnas.87.19.7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J., Langer T., Boteva R., Schramel A., Horwich A. L., Hartl F. U. Chaperonin-mediated protein folding at the surface of groEL through a 'molten globule'-like intermediate. Nature. 1991 Jul 4;352(6330):36–42. doi: 10.1038/352036a0. [DOI] [PubMed] [Google Scholar]
- Mendoza J. A., Rogers E., Lorimer G. H., Horowitz P. M. Chaperonins facilitate the in vitro folding of monomeric mitochondrial rhodanese. J Biol Chem. 1991 Jul 15;266(20):13044–13049. [PubMed] [Google Scholar]
- Ostermann J., Horwich A. L., Neupert W., Hartl F. U. Protein folding in mitochondria requires complex formation with hsp60 and ATP hydrolysis. Nature. 1989 Sep 14;341(6238):125–130. doi: 10.1038/341125a0. [DOI] [PubMed] [Google Scholar]
- Pühler G., Weinkauf S., Bachmann L., Müller S., Engel A., Hegerl R., Baumeister W. Subunit stoichiometry and three-dimensional arrangement in proteasomes from Thermoplasma acidophilum. EMBO J. 1992 Apr;11(4):1607–1616. doi: 10.1002/j.1460-2075.1992.tb05206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading D. S., Hallberg R. L., Myers A. M. Characterization of the yeast HSP60 gene coding for a mitochondrial assembly factor. Nature. 1989 Feb 16;337(6208):655–659. doi: 10.1038/337655a0. [DOI] [PubMed] [Google Scholar]
- Tandon S., Horowitz P. M. Reversible folding of rhodanese. Presence of intermediate(s) at equilibrium. J Biol Chem. 1989 Jun 15;264(17):9859–9866. [PubMed] [Google Scholar]
- Vachereau A. Luminescent immunodetection of western-blotted proteins from coomassie-stained polyacrylamide gel. Anal Biochem. 1989 May 15;179(1):206–208. doi: 10.1016/0003-2697(89)90227-3. [DOI] [PubMed] [Google Scholar]
- Viitanen P. V., Donaldson G. K., Lorimer G. H., Lubben T. H., Gatenby A. A. Complex interactions between the chaperonin 60 molecular chaperone and dihydrofolate reductase. Biochemistry. 1991 Oct 8;30(40):9716–9723. doi: 10.1021/bi00104a021. [DOI] [PubMed] [Google Scholar]
- Viitanen P. V., Lorimer G. H., Seetharam R., Gupta R. S., Oppenheim J., Thomas J. O., Cowan N. J. Mammalian mitochondrial chaperonin 60 functions as a single toroidal ring. J Biol Chem. 1992 Jan 15;267(2):695–698. [PubMed] [Google Scholar]
- Viitanen P. V., Lubben T. H., Reed J., Goloubinoff P., O'Keefe D. P., Lorimer G. H. Chaperonin-facilitated refolding of ribulosebisphosphate carboxylase and ATP hydrolysis by chaperonin 60 (groEL) are K+ dependent. Biochemistry. 1990 Jun 19;29(24):5665–5671. doi: 10.1021/bi00476a003. [DOI] [PubMed] [Google Scholar]
- Zahn R., Plückthun A. GroE prevents the accumulation of early folding intermediates of pre-beta-lactamase without changing the folding pathway. Biochemistry. 1992 Mar 31;31(12):3249–3255. doi: 10.1021/bi00127a029. [DOI] [PubMed] [Google Scholar]
- Zhi W., Landry S. J., Gierasch L. M., Srere P. A. Renaturation of citrate synthase: influence of denaturant and folding assistants. Protein Sci. 1992 Apr;1(4):522–529. doi: 10.1002/pro.5560010407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl P., Pfeifer G., Lottspeich F., Kopp F., Dahlmann B., Baumeister W. Electron microscopy and image analysis reveal common principles of organization in two large protein complexes: groEL-type proteins and proteasomes. J Struct Biol. 1990 May;103(3):197–203. doi: 10.1016/1047-8477(90)90037-d. [DOI] [PubMed] [Google Scholar]
- van Heel M., Frank J. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy. 1981;6(2):187–194. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]
- van der Vies S. M., Viitanen P. V., Gatenby A. A., Lorimer G. H., Jaenicke R. Conformational states of ribulosebisphosphate carboxylase and their interaction with chaperonin 60. Biochemistry. 1992 Apr 14;31(14):3635–3644. doi: 10.1021/bi00129a012. [DOI] [PubMed] [Google Scholar]