Abstract
Reactive oxygen species generate some 20,000 base lesions per human cell per day. The vast majority of these potentially mutagenic or cytotoxic lesions are subject to base excision repair (BER). Although chromatin remodelers have been shown to enhance the excision of oxidized bases from nucleosomes in vitro, it is not clear that they are recruited to and act at sites of BER in vivo. To test the hypothesis that cells possess factors that enhance BER in chromatin, we assessed the capacity of nuclear extracts from human cells to excise thymine glycol (Tg) lesions from exogenously added, model nucleosomes. The DNA glycosylase NTHL1 in these extracts was able to excise Tg from both naked DNA and sites in nucleosomes that earlier studies had shown to be sterically accessible. However, the same extracts were able to excise lesions from sterically-occluded sites in nucleosomes only after the addition of Mg2+/ATP. Gel mobility shift assays indicated that nucleosomes remain largely intact following the Mg2+/ATP -dependent excision reaction. Size exclusion chromatography indicated that the NTHL1-stimulating activity has a relatively low molecular weight, close to that of NTHL1 and other BER glycosylases; column fractions that contained the very large chromatin remodeling complexes did not exhibit this same stimulatory activity. These results indicate that cells possess a factor(s) that promotes the initiation of BER in chromatin, but differs from most known chromatin remodeling complexes.
Keywords: base excision repair (BER), NTHL1, nucleosomes, chromatin remodelers, thymine glycol (Tg)
Introduction
Reactive oxygen species, produced during oxidative metabolism or as a result of collisions between ionizing radiation and water, can react with and damage DNA. Oxidatively damaged bases can stall replication or mispair during replication, thereby increasing the risk of mutation and cell death. Nearly all oxidized bases are removed and replaced via base excision repair (BER)[1–3], which begins with their recognition and excision by bifunctional DNA glycosylases. These enzymes cleave the glycosidic bond between a base and its sugar moiety, and then introduce a nick into the DNA backbone. This leaves a sugar moiety attached to the 3′ side of the DNA nick, which is removed by apurinic / apyrimidinic endonuclease 1 (APE1). The resulting DNA gap is then filled by DNA polymerase and sealed by DNA ligase.
DNA in eukaryotes is packaged in chromatin, which consists in large part of tandemly arrayed nucleosomes. The DNA in nucleosomes is wrapped around a histone octamer [4, 5], which generally inhibits enzymatic processing of DNA. Damage that generates large structural perturbations in DNA, such as stand breaks and bulky lesions, trigger a DNA damage Response (DDR), which includes histone modifications and changes in chromatin structure that facilitate repair [6–8]. However, most of the damages subject to BER have relatively small effects on DNA structure, and may not trigger a DDR. Consistent with this, multiple BER enzymes can initiate BER of oxidized bases in nucleosomes in vitro, in the absence of chromatin remodeling complexes [9–19]. This is possible because BER enzymes exploit structural and dynamic properties of nucleosomes. For example, at certain sites in nucleosomes, oxidized bases are able to rotate through the major groove and into the glycosylase active site without steric hindrance. These ‘outward-facing’ lesions are processed fairly readily. Inward-facing lesions are also processed, but with a much reduced efficiency. Specifically, inward facing lesions appear to be processed only during brief intervals when the lesions are exposed, by periodic, spontaneous partial unwrapping of DNA from the histone octamer [9, 20]. Such DNA unwrapping events begin at the edge of the nucleosome and vary in extent [21, 22]. Thus, bases closer to the edge are generally exposed more frequently and for longer intervals. Our measurements suggested that the frequency with which inward-facing lesions near the center of nucleosomes are exposed by spontaneous unwrapping is too low to account for the relatively efficient BER of damaged bases in vivo [20, 23]. Thus, although partial unwrapping of DNA from nucleosomes likely occurs in vivo [24], it is likely that cells contain additional factors that promote BER of lesions in chromatin. The most obvious candidates are the chromatin remodeling complexes, and previous studies indicated that such complexes can facilitate BER of nucleosomal substrates in vitro [25, 26]. As well, the depletion of a critical subunit in the chromatin remodeling complex RSC decreased the efficiency of BER in yeast [27]. However, because the RSC-depleted cells also developed many other defects, it is not clear that RSC acts directly at sites of oxidative damage.
To further investigate the possibility that chromatin remodelers, modifiers or other factors act at sites of oxidative damage to promote BER in cells, we took advantage of the fact that DNA glycosylases do not require Mg2+or ATP to function, whereas chromatin-remodeling factors do. This enabled us to determine if nuclear extracts from human cells contain Mg2+/ATP -dependent activities that promote the glycosylase-dependent excision of oxidized bases from nucleosomes. We report here that cells do in fact contain a factor (or factors) that promotes excision of oxidized bases from occluded sites in nucleosomes, by the endogenous glycosylase NTHL1. Interestingly, this stimulatory activity does not co-fractionate with most high molecular weight chromatin remodeling complexes, but instead with smaller proteins (or protein complexes). These results provide evidence that cells do possess factors that facilitate BER in chromatin, and that they may differ from factors that facilitate other DNA repair pathways.
Materials and Methods
Cell culture
HEK293T cells, used as packaging cells, were obtained from ATCC and maintained in DMEM medium (Corning, Cellgro), supplemented with 10% Fetal Bovine Serum and 1% penicillin-streptomycin (Gibco). MCF10A cells, an immortalized, non-transformed mammary epithelial cell line, were also obtained from ATCC, and maintained in DMEM/F12 medium (Corning, Cellgro), supplemented with 5% horse serum (HyClone), 1% penicillin-streptomycin (Gibco), epidermal growth factor (EGF; 20 ng/mL, Peprotech), hydrocortisone (0.5 μg/mL, Sigma-Aldrich), insulin (10 μg/mL, Invitrogen), and Cholera Toxin (100 ng/mL, Sigma-Aldrich). All cells were maintained at 37°C in a humidified 5% CO2 incubator. To generate MCF10A control and hNTHL1 knockdown cell lines (MCF10A shGFP andMCF10A shNTHL1, respectively), we transfected HEK293T cells with 6 ug of pLKO.1 shGFP (Addgene) or pLKO.1 NTHL1 shRNA (Dharmacon clone IDs TRCN0000007915 and TRCN0000007916), together with 6 ug each of pRGR, pRSV, and pVSV-G, which code for proteins that packaged the pLKO.1 constructs into viral particles. After 72 hours, the virus-containing media was collected and replaced with fresh media. The virus-containing media was filtered through a 0.45 uM pore filter and then added to MCF10A cells that had been pre-grown for 24 hours in 6-well plates. The plates were centrifuged at 2300 RPM for 1.5 hrs, and then returned to the incubator. 24 hrs later, virus-containing media from the transfected HEK293T cells was collected once again, filtered, and added to the MCF10A cells for a second round of infection. Cell transformants stably expressing the shRNA constructs (shGFP or shNTHL1) were selected in MCF10A complete media containing 1ug/mL puromycin; expression was maintained by growing cells in complete media containing 75ng/mL puromycin.
Western Blotting
To compare the abundance of hNTH1 before and after its expression was suppressed by shRNA, cells were collected, suspended in modified RIPA buffer (150 mM NaCl, 50 mM Tris pH 7.8, 1% NP-40, 0.25% sodium deoxycholate), and then centrifuged at 10,000 × g for 10 minutes. The resulting cleared cell lysates were mixed with 6x Laemmli buffer (375 mM Tris-HCl, 9% SDS, 50% glycerol, bromophenol blue), heated at 95°C for 5 min, and subjected to 10% SDS-PAGE. The fractionated proteins were transferred to polyvinylidene difluoride (PVDF-FL) membranes (Millipore, Billerica, MA, USA). Membranes were blocked in Odyssey blocking buffer (PBS) (Millipore) for 1hr at room temperature (RT) with gentle shaking, and then incubated with a mouse monoclonal anti-HA primary antibody (1/1000) (ThermoScientific, 2–2.2.14), a rabbit monoclonal anti-NTHL1 primary antibody (1/1000) (Abcam, EPR15930) and a mouse monoclonal tubulin primary antibody (1/10000) (Abcam, DM1A), overnight at 4°C with gentle shaking. The membranes were washed three times for 5 min each in PBS/0.1% Tween, and then incubated with IRDye® 800CW goat anti-mouse IgG (H+L) (1/20,000) and IRDye® 680RD goat anti-rabbit IgG (H+L) (1/20,000), for 1 hr at RT. The membranes were then washed twice for 5 min each with PBS/0.1% Tween, and protein-antibody complexes visualized using an Odyssey® CLx Infrared Imaging System. Expression levels were quantified using Image Studio Software version 2.1.10. To track selected proteins and protein complexes during size exclusion chromatography (Figure 4B), 4 ug of the unfractionated extract and 1 ug of protein from pools 1–3 were subjected to 10% SDS-PAGE. Fractionated proteins were transferred to PVDF-FL membranes as before. Membranes were then incubated with mouse monoclonal anti-human NTHL1 primary antibody (1/1000; Abcam ab191413) and goat polyclonal anti-human Brg1 primary antibody (2 ug/ml; R and D systems AF5738), overnight at 4°C with gentle shaking. The membranes were washed as above, and then incubated with IRDye® 800CW donkey anti-goat IgG (H+L) (1/20,000) and IRDye® 680RD donkey anti-rabbit IgG (H+L) (1/20,000), for 1 hr at RT. The membranes were then washed, and the protein-antibody complexes were visualized as before.
Figure 4. The Mg2+/ATP-dependent, NTHL1-promoting activity does not co-fractionate with the large chromatin-remodeling complexes.
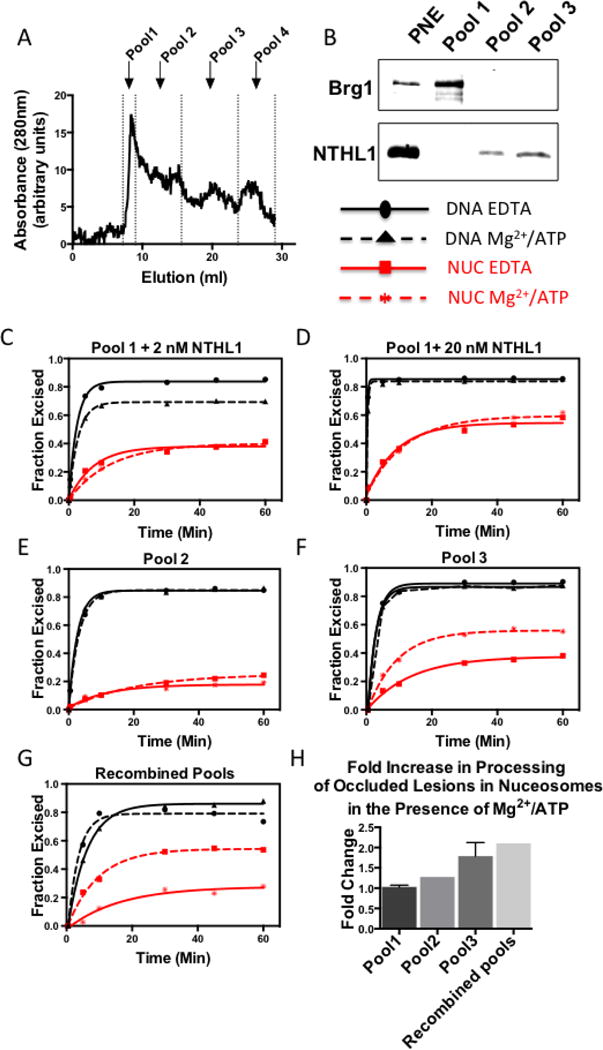
(A) Nuclear extracts were subjected to S200 superdex gel filtration and the elution was pooled as shown. (B)Western blots containing 4μg of the total (parental) nuclear extract (PNE) and 1ug from the pools containing most of the protein, were prepared and probed for the 34 kDa NTHL1 and the BAF-associated, 220 kDa protein, Brg1, as described in the Methods. Brg1 (220kDa) was detected in the parent nuclear extract and Pool 1, indicating that remodeling complexes remained intact during fractionation. As expected, Pool 1 was devoid of NTHL1 activity. Therefore, to determine if Pool 1 contained factors that would enhance Tg excision under permissive buffer conditions, we added either 2 nM (C) or 20nM (D) recombinant human NTHL1 protein to reactions containing 0.7 ug Pool 1 and 2.25 nM Tg-in nucleosomes. Although the exogenously added NTHL1 exhibited relatively high Tg-excision activity, this was not enhanced by addition of Mg2+/ATP. (E) Pool 2 (4ug) did contain endogenous hNTHL1 activity however excision of occluded lesion (2.25nM Tg-in) was not enhanced by addition of Mg2+/ATP. (F) Pool 3 (1.6ug) activity toward Tg-in nucleosomes (2.25nM) was substantially enhanced by the addition of Mg2+/ATP (G), Pools were recombined in an equivolume fashion and the Mg2+/ATP-dependent enhancement in Pool 3 remained, but its magnitude did not increase suggesting that the enhancing factor was present entirely within Pool 3. (H) A histogram comparing the relative enhancement provided by the addition of Mg2+/ATP. Total amplitudes of EDTA reactions for each pool were set to 1 and the fold-enhancement provided by the Mg2+/ATP condition was calculated. Error bars represent standard deviations of 2 independent assays.
Preparation and fractionation of nuclear extracts
MCF10A cells grown in 150mm tissue culture plates to 75–80% confluence were washed with phosphate buffered saline, suspended using a cell scraper, and pelleted by centrifugation at 1850 × g for 10 minutes at 4°C. Cells and reagents were kept on ice for the remainder of the procedure, and Roche complete protease inhibitor cocktail was added to all buffers according to the manufacturers instructions. Cells were resuspended in 5 packed cell volumes of buffer A10 (25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)-pH 8, 0.2 mM ethylene glycol tetraacetic acid (EGTA), 1.5 mM magnesium acetate (MgOAc2), 1 mM dithiothreitol (DTT), containing 10 mM Potassium acetate (KOAc)), and collected by centrifugation at 1850 × g for 5 minutes. Cells were then resuspended in 3 packed cell volumes of buffer A10, incubated on ice for 10 min, and lysed using a dounce homogenizer with a type B pestle. Nuclei were collected by centrifuged at 3300 × g for 15 min, and suspended in ½ volume of buffer A10 containing 25% glycerol. To this suspension, we added dropwise with gentle magnetic stirring, ½ packed nuclear volume of buffer A1200 (buffer A with 1.2 M KOAc) containing 25% glycerol. After 30 min of further incubation, the nuclear extract was cleared by centrifugation at 25,000 × g for 30 min., aliquoted, and stored at -80°C [28, 29]. A portion of the nuclear extracts was further fractionated using a Superdex 200GL (10/300 GE) size exclusion column in buffer A300 (buffer A with 0.3 M KOAc) containing 25% glycerol. Adjacent column fractions were combined into four size pools. These were aliquoted and stored at -80°. Protein concentrations were determined using the Bradford assay (Thermo).
DNA and nucleosome substrates
Rotationally positioned nucleosomes were assembled using a 184 bp blunt-ended DNA fragment containing the 5S rDNA sequence from L. variegatus, modified to include a single Tg residue [9]. As discussed in [20] most histone octamers assemble with 5S rDNA in one of three registers. Because these registers differ from one another by one-two helical turns, the rotational orientation of the Tg residue was the same in all nucleosomes, facing either away or toward the histone octamer (outward- vs. inward-facing). Our previous studies indicated that most inward- and outward-facing lesions would be located, respectively, at 46 and 51 nucleotides from the central dyad axis (36 and 41 nucleotides, and 56 and 61 nucleotides in the two other translational variants) [9]. Thus in all cases, the Tg lesions lay within the nucleosome proper. To assemble nucleosomes, we mixed Tg-containing DNAs, constructed and end-labeled with 32P, as in [10], with purified histone octamers, that we had assembled with recombinant Xenopus laevis histones, as described in [10, 30, 31]. To ensure nucleosome stability, we also added to the reconstitution reactions unlabeled, mono- and di-nucleosome length DNA (prepared from chicken chromatin as described in [9]), in amounts sufficient to ensure a total nucleosome concentration in later reactions of 40 nM (with 2.25 nM Tg-containing nucleosomes). Nucleosomes were assembled via dialysis to low salt, and then assayed using non-denaturing gels [9] (Figure 3A). Reconstitution efficiencies were calculated to allow for computational correction for small amounts of contaminating free DNA, as described in [9]. Substrates for naked DNA controls were identical except for the absence of histone octamers.
Figure 3. Tg-containing nucleosomes are not detectably altered by exposure to nuclear extract in the presence or absence of Mg2+/ATP. (A).
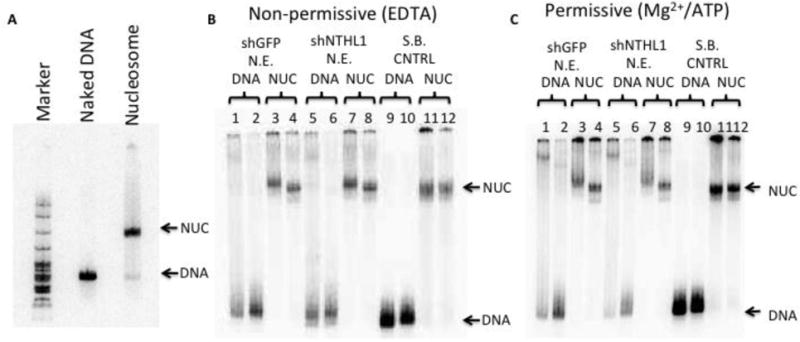
Tg-in DNA was reconstituted into a nucleosome and quality was checked using a 5% non-denaturing gel. DNA and nucleosome (NUC) bands are indicated and a pBR322/MspI digest size marker was used. Nucleosomes containing occluded Tg residues were incubated in storage buffer (S.B. CNTRL) or in nuclear extracts from either cells expressing shRNA against NTHL1 (N.E. shNTHL1), or cells expressing an off-target control shRNA (N.E. shGFP); buffers included either EDTA (B) or Mg2+/ATP (C). After one hour at 37°C, reactions were mixed with gel loading buffer, containing or lacking excess naked DNA (even and odd numbered lanes, respectively), and electrophoresed through non-denaturing gels. Lanes 3,4,7, and 8 indicate that nucleosomes are largely intact after exposure to the extracts. The super-shifted complexes that form with nucleosomes in extracts (lanes 3 and 7) are not altered by buffer conditions or knockdown of NTHL1, which suggests they are not BER-specific. The complexes decay with the addition of a naked DNA trap (lanes 4 and 8), leaving nucleosomes that migrate normally; no free DNA is released. (A small fraction of the nucleosomal DNA can be seen to migrate just ahead of the nucleosome band in some lanes but this occurs independently of buffer conditions, knockdown of NTHL1, or exposure to nuclear extract.) Collectively, these observations suggest that the NTHL1-enhancing factor does not irrevocably disrupt or alter nucleosome structure.
Tg excision assays
Tg excision reactions were conducted at 37°C, and initiated by the addition of DNA or nucleosome substrates to varying amounts of nuclear extract in 25 mM HEPES-pH 8, 100 mM KOAc, 5% glycerol, 250 uM each deoxynucleotide triphosphates (NEB), and 0.5 ug/ul bovine serum albumin (NEB). Buffers also contained either 2.5 mM adenosine triphosphate (ATP) and 2.5 mM Mg2+ (remodeler permissive conditions) or 5 mM EDTA (remodeler non-permissive conditions), and 155 nM mono- and di-nucleosome length, dsDNA (prepared from chicken chromatin, as above), which helped suppress nonspecific DNA binding to Tg-containing substrates. To monitor DNA glycosylase activities, aliquots taken at varying times were quenched in 0.5% sodium dodecyl sulfate, 20 mM EDTA, 1.4 mg/ml proteinase K. After 20 min of incubation at 37°C, abasic sites were converted to strand breaks by adding sodium hydroxide to 333 mM, and heating samples at 95°C for 2 minutes. Samples were then mixed with 2 volumes of formamide/20mM EDTA, heated again to 95° C for 2 minutes, and fractionated using 8% sequencing gels. Substrate and product bands were visualized and quantified using phosphoimaging (Bio-rad). Fraction excised was determined by normalizing the substrate band at any given time to the substrate band at 0.5 min, and graphed using Prism (Graphpad). Control experiments conducted with non-damaged DNA show that activity was Tg dependent (Supplementary Figure 1). Reactions in which nuclear extract was supplemented with recombinant NTHL1, reactions were conducted exactly as described above but Tg excision activity was calculated as the intensity of the product band divided by the combined intensity of the substrate and product bands. Errors bars represent standard deviations of at least 2 assays. Assays using a 35 base pair naked DNA substrate were conducted as above under the non-permissive conditions. Aliquots were taken at various timepoints and quenched in 2 volumes of formamide/20 mM EDTA. Product was separated from substrate using 12% polyacrylamide gels, and bands were visualized using phosphoimagery. Recombinant human NTHL1 and giant DNA mimivirus Nei used in the above assays were expressed in, and purified from, E.coli as described [32–34].
Gel mobility shift assays
Reactions were prepared just as for the excision assays, and incubated at 37°C for 60 minutes. Aliquots were added to a gel-loading buffer (50 mM HEPES-pH 8, 5 mM EDTA, 5% glycerol) containing or lacking 1 ug of a dsDNA trap. The samples were then heated for 2 minutes at 42°C to allow proteins that might be non-specifically bound to the damaged DNA to migrate to the DNA trap. Samples were loaded and run into an 8% non-denaturing polyacrylamide gels. Gels were dried and visualized via phosphoimaging.
Results
The DNA glycosylase NTHL1 present in nuclear extracts can excise Tg from exogenously added DNA substrates
To determine if cells contain factors that promote excision of oxidized bases from nucleosomes, we prepared nuclear extracts from cultured MCF10A cells, as described in the Methods. The extracts were largely free of protease activity, as judged by the fact that recombinant hNTHL1 remained intact when incubated with the extracts and then analyzed by western blotting (data not shown). As well, gel mobility shift assays indicated that nucleosomes remained intact when incubated with the extracts (c.f. Figure 3 below). The extracts did however contain a significant amount of non-specific DNA binding activity, which required the addition of undamaged double stranded DNA, prior to conducting DNA glycosylase assays with Tg-containing substrates. To determine if endogenous DNA glycosylases were present and active in the nuclear extracts, we added double stranded DNA containing a single centrally positioned Tg residue, and examined excision activity in the nuclear extracts, in the presence of EDTA, as described in the Methods. Figure 1A shows strand scission products, indicating that the extracts contained active glycosylase. NTHL1 is a member of the HhH superfamily and is of two bifunctional DNA glycosylases responsible for most of the Tg excision activity in human cells; it generates a β-elimination product [35]. The second glycosylase, NEIL1, is a member of the NEI family and generates β-δ-elimination products [36, 37]. Inspection of Figure 1A shows that nuclear extracts produced exclusively hNTHL1-like products.
Figure 1. Identification of NTHL1 as the enzyme in nuclear extracts responsible for excision of thymine glycol (Tg) from DNA. (A).
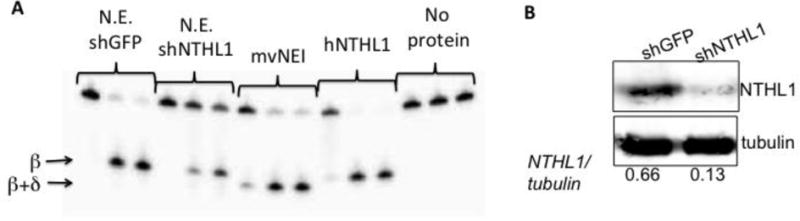
Nuclear extracts were prepared from cells expressing shRNA against NTHL1 (N.E. shNTHL1), and from cells expressing an off-target control shRNA (N.E. shGFP). 5.9 ug of each extract was incubated with 5 nM of 35 base pair Tg containing DNA fragment in an EDTA-containing buffer. Reactions were quenched after 0.5, 30 or 60 min. in formamide/20 mM EDTA, and products were fractionated using 12% polyacrylamide gels. Parallel reactions using 80 nM giant mimivirus NEI protein (MV NEI) and 20 nM human NTHL1 demonstrate that the NEI-generated β-δ-elimination product can be readily distinguished from the NTHL1-generated β-elimination product. (B) Western blot showing a ~5-fold decrease in NTHL1 abundance in MCF10A cells expressing shRNA against endogenous NTHL1 (shNTHL1) as compared to MCF10A cells expressing an off-target control shRNA (shGFP).
To confirm that hNTHL1 was responsible for the observed Tg excision activity in nuclear extracts, we generated stable MCF10A cell lines expressing shRNA against either GFP (as a control) or NTHL1. Quantitative western blotting indicated that NTHL1 levels in cells expressing the anti-NTHL1 shRNA were about five-fold lower than normal (Figure 1B). The Tg excision activity in nuclear extracts from these cells was also markedly lower than in extracts from the GFP knockdown control cells. As before, we did not observe any excision activity that could be attributed to NEIL1 or other NEI glycosylases. This may be because NEIL1 is expressed predominately during S-phase and associates with replication factors whereas NTHL1 is constitutively expressed [38–43]. NEIL1 has a higher affinity for undamaged DNA than does NTHL1[44]. Thus, its activity in extracts may also have been suppressed by the DNA we had added to sequester non-specific binding proteins. The extracts did, however, contain tetrahydrofuran incision and 8-oxoguanine excision activities (data not shown).
A factor in nuclear extracts facilitates the NTHL1-mediated excision of occluded (inward facing) lesions from nucleosomes
To test the hypothesis that cells possess (Mg2+/ATP-dependent) factors that enhance NTHL1-mediated excision of oxidized bases in nucleosomes, we added extracts to nucleosomes that contained either inward- or outward-facing (occluded or accessible) Tg lesions. Figure 2A shows Tg excision activity of the extracts in chromatin remodeling permissive and non-permissive buffers (“Mg2+/ATP” and “EDTA”). Endogenous NTHL1 rapidly excised Tg residues from naked DNA in either buffer. Nucleosomes containing an outward-facing Tg were also processed similarly under the two assay conditions, albeit with diminished amplitude when compared to the naked DNA. It is important here to note that our nuclear extracts contained numerous factors that might influence the kinetics of lesion processing. Thus, we focused primarily (both here and in later reactions) on the fraction of substrate processed in permissive and non-permissive buffers. With that caveat in mind, Figure 2A suggests that addition of Mg2+/ATP increased the rates of lesion processing for both substrates. The most likely explanation for this is that activation of APE1 (which follows NTHL1 during BER) by the addition of Mg2+ would reduce product inhibition of NTHL1, thereby increasing its efficiency in multi-turnover reactions.
Figure 2. The capacity of endogenous NTHL1 to excise Tg lesions from nucleosomes in nuclear extracts depends on lesion orientation when assayed in non-permissive (EDTA-containing) buffers but not in Mg2+/ATP-containing buffers.
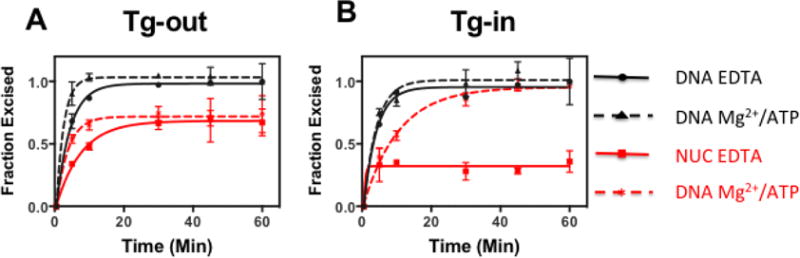
Tg excision capacity was measured in reactions with 5.9 ug nuclear extract and 2.25 nM nucleosomes containing either outward-facing (sterically accessible) Tg lesions (A) or inward-facing (occluded) lesions (B). Reactions were conducted in permissive or non-permissive buffers (dashed and solid lines, respectively). Parallel reactions were conducted with naked DNA’s identical to those used to assemble test nucleosomes. Note that Mg2+/ATP had little impact on excision activity measured for naked DNAs and nucleosomes with outward facing lesions, but substantially stimulated excision of Tg residues at occluded sites in nucleosomes. Error bars represent standard deviation of 2 independent assays for each trace.
In contrast to the results for the processing of sterically-accessible lesions (Figure 2A), addition of Mg2+/ATP to reactions with nucleosomes containing an occluded, inward-facing Tg residues dramatically increased the fraction of lesion processed (Figure 2B). As before, the naked DNA controls show similar rapid Tg excision in either buffer, but the significantly enhanced processing of occluded Tg residues in nucleosomes in remodeler permissive buffers strongly support the hypothesis that cells possess factors that enhance BER in chromatin.
The NTHL1 enhancing activity does not irrevocably alter nucleosome integrity
The enhancement of Tg excision activity shown in Figure 2 might be due to an ATP- and/or Mg2+-dependent factor that acts with NTHL1, allowing it to more efficiently find and bind occluded substrates in nucleosomes. Alternatively, the factor may modify nucleosome structure, either transiently or permanently, to render the substrate more accessible. To distinguish between these two possibilities, we incubated Tg-containing nucleosomes or naked DNA with nuclear extract for 60 minutes, in either permissive or non-permissive buffers. We then split the reactions, adding gel-loading buffer and 1ug of dsDNA to half and gel-loading buffer without DNA to the other half. All samples were then heated to 42°C for 2 minutes to promote migration of weakly or nonspecifically bound proteins from the substrates to the excess trap dsDNA. Samples were then electrophoresed into 8% non-denaturing polyacrylamide gels. In the presence of the added DNA trap, nucleosomes exposed to nuclear extract in either assay buffer (Figures 3B and 3C, lanes 4 and 8) exhibited a migration behavior nearly identical to that of nucleosomes incubated in storage buffer for 60 min at 37°C (Figures 3B and 3C, lanes 11 and 12). Although this assay may not detect minor structural perturbations, exposure of nucleosomes to nuclear extracts clearly did not lead to release of naked DNA. This indicates that the NTHL1 enhancing activity does not irrevocably disrupt Tg-containing nucleosomes. In the absence of added trap DNA, factors present in the extract did influence nucleosome mobility; however, this mobility shift was also evident in the EDTA containing reactions (Figure 3 B and C, lanes 3 and 7). As well, knockdown of NTHL1 did not alter the magnitude of the shift. This suggests that the mobility-shifted complexes do not contain NTHL1. We cannot, however, rule out the possibility that the NTHL1-stimulatory factor is able to bind substrate in EDTA-containing buffers but requires Mg2+/ATP to act. It is also worth noting that detecting interactions between chromatin remodeling factors and their substrates commonly requires extensive cross-linking (c.f. [45]). Thus, while these results indicate that the increased excision activity seen for the inward facing lesion is not due to an irrevocable disruption of the host nucleosome, they did not rule out the possible transient involvement of a chromatin remodeling activity.
Fractionation of the nuclear extract
Nucleosome remodelers are large multi-subunit complexes with molecular weights greater than 1 MDa [46]. To determine if one or more of these complexes might be responsible for the enhanced Tg excision activity, we fractionated nuclear extracts, using size exclusion chromatography, and assessed the Tg excision activity in each of four size pools, as indicated in Figure 4A and Table 1. Western blot analyses in Figure 4B show that Pool 1 contained Brg1, the 220 kDa catalytic subunit of the ~1 MDa chromatin remodeling complex, Brg Associated Factors (BAF). Thus, other large remodeling complexes were most likely also present in this pool. Endogenous NTHL1 (34 kDa) was present in both Pools 3 and 2 (Figure 4B). To determine if the NTHL1-stimulatory activity was present in Pool 1 (as seemed likely), we conducted Tg-excision reactions with Pools 1–3. In comparing fractionated extracts with the parent extract, it is important to recall that certain fractions will be enriched for specific factors and depleted of others. Thus, in assaying selected fractions, we used less total protein than in reactions with whole extract. As expected, Pool 1 did not exhibit any detectable Tg excision activity. Therefore, to determine if Pool 1 contained a factor that would stimulate the NTHL1-mediated excision of occluded lesions from nucleosomes, we added 2nM or 20nM of human recombinant NTHL1. Figures 4C and 4D show that addition of Mg2+/ATP to Pool 1 failed to increase the processing of occluded lesions by recombinant NTHL1. This result suggested the stimulatory factor is either not present in a large molecular weight complex, or that its activity requires the addition of a factor from a different size pool.
Table 1.
Molecular size fractionation of the nuclear extract
| Nuclear extract pool # | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Approx. M.W. range* | <750,000 | 750,000–45,000 | 45,000–6,500 | > 6,500 |
| Tg excision activity | − | + | + | − |
approx M.W.range is based on data available from the column manufacturer
One ug of Pool 2 protein exhibited only a limited amount of Tg excision activity when tested with naked DNA substrates and virtually no activity on Tg-containing nucleosomes (data not shown). A four-fold increase in the amount of Pool 2 protein did give detectable levels of excision activity, but this was not enhanced by Mg2+/ATP (Figure 4E).
Pool 3 exhibited the greatest amount of Tg excision activity/mg protein. Moreover, as shown in Figure 4F, the addition of Mg2+/ATP further enhanced the excision of lesions from occluded sites in nucleosomes. These results indicate that the NTHL1 enhancing effect evident in Figure 2 is due not to a large molecular weight complex but rather to a smaller factor whose size is similar to that of the individual DNA glycosylases. The stimulatory effect seen in Pool 3 was somewhat less than that in the unfractionated extract, which could be due to partial loss of activity during fractionation. Alternatively, the size fractionation may have altered the relative abundance of various factors, such that the molar ratio of NTHL1 to the stimulatory factor was no longer optimal. To help assess these possibilities we recombined the pools in an equi-volume fashion, and repeated the Tg excision assays as before. Figure 4G shows that the recombined nuclear extract did provide some stimulation under the permissive conditions but it was markedly reduced when compared to unfractionated extract (c.f. Fig 2). This result suggested that the activity of the stimulatory factor in Pool 3 is not enhanced by factors from other size pools. A summation of the enhancement of nucleosome excision provided addition of Mg2+/ATP for each pool is provided in Figure 4H.
Discussion
It was reported previously that the large chromatin remodeling complex SWI/SNF stimulated the activity of the BER glycosylase OGG1 on nucleosome substrates in vitro, and also the activities of the downstream BER enzymes APE1 and Polymerase β [25]. As well, Saccharomyces cerevesiae ISW1 and ISW2 were reported to stimulate pol β activity on nucleosome arrays in vitro [26]. These studies demonstrated that nucleosome remodeling can facilitate BER in vitro, but did not address the question of whether remodelers are recruited to and act at sites of BER in vivo. Depletion of Sth1p, the ATPase subunit of the RSC complex in S. cerevisiae, caused cells to become more sensitive to methyl methane sulfonate, which generates alkylated bases that normally are removed by BER [27]. However, since chromatin remodelers and modifiers are involved in many fundamental cellular pathways, depletion of such factors invariably results in multiple complex phenotypes, which makes interpretation of such experiments difficult. In this report, cells containing functional RSC exhibited significantly more BER but also more “open” chromatin, as assessed with MNase. Thus, RSC may enhance BER in cells due to its effect on general chromatin architecture rather than as a specific responder to oxidative damage in DNA.
To circumvent the above-described experimental challenges, and determine if human cells contain factors that act directly on chromatin substrates to promote BER, we examined the capacity of nuclear extracts to excise Tg bases from discrete sites within exogenously added nucleosomes. We determined that hNTHL1 was responsible for the base excision observed (Figure 1A), and discovered that the extracts also contain a Mg2+/ATP-dependent factor or effector that enables hNTHL1 to excise Tg from sites in nucleosomes are normally occluded (Figure 2). Gel mobility shift assays (Figure 3) indicated that any changes in nucleosome structure that allow for this enhanced excision activity are subtle or transient. Specifically, there was no indication that the histones are irrevocably displaced from host nucleosomes.
Recently, CHO-K1 nuclear extracts were reported to contain a factor that stimulates APE1-mediated incision at a tetrahydrofuran residue, located at a partially occluded site in nucleosomes, under conditions similar to our permissive conditions [47]. However, the size and identity of this factor, and its effect on nucleosome structure, is unknown. Thus, additional work will be required to determine if the factor(s) that promote APE1 incision is related to the NTHL1-enhancing factor reported here.
Our fractionation studies (Figure 4) yielded the surprising result that our NTHL1-enhancing activity does not reside in a large chromatin-remodeling complex but instead co-fractionates with smaller proteins, similar in size to NTHL1 itself. Thus it could be an accessory factor that acts through NTHL1, or, conceivably, it is a post-translationally modified version of NTHL1 itself. The most likely alternative is that the NTHL1-enhancing factor alters nucleosomes, either transiently, or subtly enough to preclude detectable differences in gel mobility shift assays. Should this prove true, it would be important to determine if the factor acts only on nucleosomes containing oxidized bases. This would suggest that it is recruited by DNA glycosylases. It is possible as well that the factor/effector acts in a more general fashion. Nuclear extracts may contain many factors that can ‘interrogate’ nucleosomes in a manner that allows DNA glycosylases better access to substrates in nucleosomes. This effect may be more pronounced under the permissive conditions as some DNA binding and modifying factors require Mg2+ or Mg2+/ATP. This sort of cooperative nucleosome interrogation has been seen for nucleosomes with binding sites for DNA sequence-specific binding factors, where those factors facilitate the binding (and thus the activity) of other factors [16, 48]. What is clear from this work is that enhancement of BER of substrates in nucleosomes involves a factor(s) that differ from the large remodeling complexes that help promote transcription and act in other DNA repair pathways.
Supplementary Material
Highlights.
Nucleosomes suppress the base excision repair (BER) of oxidatively damaged DNA.
We found in human cell extracts a factor that promotes BER in nucleosomes.
This factor promotes excision of oxidized bases from occluded sites in nucleosomes.
The factor requires Mg2+/ATP but acts without disrupting the host nucleosome.
The factor is smaller than known chromatin remodeling complexes.
Acknowledgments
We thank Drs. Aishwarya Prakash and Sylvie Doublie for a gift of the mimivirus Nei protein, and Drs. Joyce Heckman and Wendy Cannan for comments on the manuscript. Phosphorimaging was performed in the University of Vermont Cancer Center Advanced Genome Technologies Core and was supported by the University of Vermont Cancer Center, Lake Champlain Cancer Research Organization, and the University of Vermont College of Medicine.
Funding: Major funding for this study was from an NCI program project grant (P01-CA098993) The funders have no role in the study design, data analysis/interpretation or preparation of this manuscript. The choice to publish this manuscript was at the sole discretion of the authors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement:
The authors declare that there are no conflicts of interest.
Contributor Information
R.L. Maher, Department of Microbiology and Molecular Genetics, and The Markey Center for Molecular Genetics, University of Vermont, Burlington, Vermont, 05405, U.S.A
C.G. Marsden, Department of Microbiology and Molecular Genetics, and The Markey Center for Molecular Genetics, University of Vermont, Burlington, Vermont, 05405, U.S.A
A.M. Averill, Department of Microbiology and Molecular Genetics, and The Markey Center for Molecular Genetics, University of Vermont, Burlington, Vermont, 05405, U.S.A
S.S. Wallace, Department of Microbiology and Molecular Genetics, and The Markey Center for Molecular Genetics, University of Vermont, Burlington, Vermont, 05405, U.S.A
D.S. Pederson, Department of Microbiology and Molecular Genetics, and The Markey Center for Molecular Genetics, University of Vermont, Burlington, Vermont, 05405, U.S.A
J.B. Sweasy, Department of Therapeutic Radiology and Human Genetics, Yale University School of Medicine, New Haven, Connecticut 06520, U.S.A
References
- 1.Jacobs AL, Schar P. DNA glycosylases: in DNA repair and beyond. Chromosoma. 2012;121:1–20. doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 3.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 5.Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair?, Nature reviews. Molecular cell biology. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldecott KW. Single-strand break repair and genetic disease, Nature reviews. Genetics. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 7.Gong F, Kwon Y, Smerdon MJ. Nucleotide excision repair in chromatin and the right of entry. DNA repair. 2005;4:884–896. doi: 10.1016/j.dnarep.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prasad A, Wallace SS, Pederson DS. Initiation of base excision repair of oxidative lesions in nucleosomes by the human, bifunctional DNA glycosylase NTH1. Molecular and cellular biology. 2007;27:8442–8453. doi: 10.1128/MCB.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odell ID, Barbour JE, Murphy DL, Della-Maria JA, Sweasy JB, Tomkinson AE, Wallace SS, Pederson DS. Nucleosome disruption by DNA ligase III-XRCC1 promotes efficient base excision repair. Molecular and cellular biology. 2011;31:4623–4632. doi: 10.1128/MCB.05715-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinz JM. Impact of abasic site orientation within nucleosomes on human APE1 endonuclease activity. Mutation research. 2014;766–767:19–24. doi: 10.1016/j.mrfmmm.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez Y, Smerdon MJ. The structural location of DNA lesions in nucleosome core particles determines accessibility by base excision repair enzymes. The Journal of biological chemistry. 2013;288:13863–13875. doi: 10.1074/jbc.M112.441444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menoni H, Shukla MS, Gerson V, Dimitrov S, Angelov D. Base excision repair of 8-oxoG in dinucleosomes. Nucleic acids research. 2012;40:692–700. doi: 10.1093/nar/gkr761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye Y, Stahley MR, Xu J, Friedman JI, Sun Y, McKnight JN, Gray JJ, Bowman GD, Stivers JT. Enzymatic excision of uracil residues in nucleosomes depends on the local DNA structure and dynamics. Biochemistry. 2012;51:6028–6038. doi: 10.1021/bi3006412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinz JM, Mao P, McNeill DR, Wilson DM., 3rd Reduced Nuclease Activity of Apurinic/Apyrimidinic Endonuclease (APE1) Variants on Nucleosomes: IDENTIFICATION OF ACCESS RESIDUES. The Journal of biological chemistry. 2015;290:21067–21075. doi: 10.1074/jbc.M115.665547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannan WJ, Tsang BP, Wallace SS, Pederson DS. Nucleosomes suppress the formation of double-strand DNA breaks during attempted base excision repair of clustered oxidative damages. The Journal of biological chemistry. 2014;289:19881–19893. doi: 10.1074/jbc.M114.571588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez Y, Hinz JM, Smerdon MJ. Accessing DNA damage in chromatin: Preparing the chromatin landscape for base excision repair. DNA repair. 2015;32:113–119. doi: 10.1016/j.dnarep.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balliano AJ, Hayes JJ. Base excision repair in chromatin: Insights from reconstituted systems. DNA repair. 2015;36:77–85. doi: 10.1016/j.dnarep.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Odell ID, Wallace SS, Pederson DS. Rules of engagement for base excision repair in chromatin. Journal of cellular physiology. 2013;228:258–266. doi: 10.1002/jcp.24134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maher RL, Prasad A, Rizvanova O, Wallace SS, Pederson DS. Contribution of DNA unwrapping from histone octamers to the repair of oxidatively damaged DNA in nucleosomes. DNA repair. 2013;12:964–971. doi: 10.1016/j.dnarep.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nature structural & molecular biology. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 22.Li G, Widom J. Nucleosomes facilitate their own invasion. Nature structural & molecular biology. 2004;11:763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 23.Nyaga SG, Jaruga P, Lohani A, Dizdaroglu M, Evans MK. Accumulation of oxidatively induced DNA damage in human breast cancer cell lines following treatment with hydrogen peroxide. Cell Cycle. 2007;6:1472–1478. [PubMed] [Google Scholar]
- 24.Geraghty DS, Sucic HB, Chen J, Pederson DS. Evidence that partial unwrapping of DNA from nucleosomes facilitates the binding of heat shock factor following DNA replication in yeast. The Journal of biological chemistry. 1998;273:20463–20472. doi: 10.1074/jbc.273.32.20463. [DOI] [PubMed] [Google Scholar]
- 25.Menoni H, Gasparutto D, Hamiche A, Cadet J, Dimitrov S, Bouvet P, Angelov D. ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A.Bbd nucleosomes. Molecular and cellular biology. 2007;27:5949–5956. doi: 10.1128/MCB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakanishi S, Prasad R, Wilson SH, Smerdon M. Different structural states in oligonucleosomes are required for early versus late steps of base excision repair. Nucleic acids research. 2007;35:4313–4321. doi: 10.1093/nar/gkm436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czaja W, Mao P, Smerdon MJ. Chromatin remodelling complex RSC promotes base excision repair in chromatin of Saccharomyces cerevisiae. DNA repair. 2014;16:35–43. doi: 10.1016/j.dnarep.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abmayr SM, Yao T, Parmely T, Workman JL. Preparation of nuclear and cytoplasmic extracts from mammalian cells. In: Ausubel Frederick M, et al., editors. Current protocols in molecular biology. 2006. Chapter 12. Unit 12 11. [DOI] [PubMed] [Google Scholar]
- 29.Akbari M, Krokan HE. Base excision repair efficiency and mechanism in nuclear extracts are influenced by the ratio between volume of nuclear extraction buffer and nuclei-implications for comparative studies. Mutation research. 2012;736:33–38. doi: 10.1016/j.mrfmmm.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Methods in molecular biology. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 31.Lee JY, Greene EC. Assembly of recombinant nucleosomes on nanofabricated DNA curtains for single-molecule imaging. Methods in molecular biology. 2011;778:243–258. doi: 10.1007/978-1-61779-261-8_16. [DOI] [PubMed] [Google Scholar]
- 32.Galick HA, Kathe S, Liu M, Robey-Bond S, Kidane D, Wallace SS, Sweasy JB. Germ-line variant of human NTH1 DNA glycosylase induces genomic instability and cellular transformation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14314–14319. doi: 10.1073/pnas.1306752110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bandaru V, Zhao X, Newton MR, Burrows CJ, Wallace SS. Human endonuclease VIII-like (NEIL) proteins in the giant DNA Mimivirus. DNA repair. 2007;6:1629–1641. doi: 10.1016/j.dnarep.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandaru V, Blaisdell JO, Wallace SS. Oxidative DNA glycosylases: recipes from cloning to characterization. Methods in enzymology. 2006;408:15–33. doi: 10.1016/S0076-6879(06)08002-5. [DOI] [PubMed] [Google Scholar]
- 35.Ikeda S, Biswas T, Roy R, Izumi T, Boldogh I, Kurosky A, Sarker AH, Seki S, Mitra S. Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys-212 as the active nucleophilic residue. The Journal of biological chemistry. 1998;273:21585–21593. doi: 10.1074/jbc.273.34.21585. [DOI] [PubMed] [Google Scholar]
- 36.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA repair. 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 37.Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra S, Izumi T. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. The Journal of biological chemistry. 2002;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 38.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA repair. 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dou H, Theriot CA, Das A, Hegde ML, Matsumoto Y, Boldogh I, Hazra TK, Bhakat KK, Mitra S. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. The Journal of biological chemistry. 2008;283:3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- 41.Theriot CA, Hegde ML, Hazra TK, Mitra S. RPA physically interacts with the human DNA glycosylase NEIL1 to regulate excision of oxidative DNA base damage in primer-template structures. DNA repair. 2010;9:643–652. doi: 10.1016/j.dnarep.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prakash A, Moharana K, Wallace SS, Doublie S. Destabilization of the PCNA trimer mediated by its interaction with the NEIL1 DNA glycosylase. Nucleic acids research, ( 2016 doi: 10.1093/nar/gkw1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang C, Sengupta S, Hegde PM, Mitra J, Jiang S, Holey B, Sarker AH, Tsai MS, Hegde ML, Mitra S. Regulation of oxidized base damage repair by chromatin assembly factor 1 subunit A. Nucleic acids research. 2017;45:739–748. doi: 10.1093/nar/gkw1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Odell ID, Newick K, Heintz NH, Wallace SS, Pederson DS. Non-specific DNA binding interferes with the efficient excision of oxidative lesions from chromatin by the human DNA glycosylase, NEIL1. DNA repair. 2010;9:134–143. doi: 10.1016/j.dnarep.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annual review of biochemistry. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 47.Eccles LJ, Menoni H, Angelov D, Lomax ME, O’Neill P. Efficient cleavage of single and clustered AP site lesions within mono-nucleosome templates by CHO-K1 nuclear extract contrasts with retardation of incision by purified APE1. DNA repair. 2015;35:27–36. doi: 10.1016/j.dnarep.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polach KJ, Widom J. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. Journal of molecular biology. 1996;258:800–812. doi: 10.1006/jmbi.1996.0288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


