Abstract
Objective(s):
MicroRNAs (miRNAs) are considered as powerful, post-transcriptional regulators of gene expression in hepatocellular carcinoma cells (HCC). However, the function of miR-92a is still unclear in HCC.
Materials and Methods:
Expression of miR-92a in human HCC cell lines was evaluated using qRT-PCR. MTT assay and transwell assay were used to examine the function of miR-92a in HepG2 and Huh7 cells. Bioinformatic analyses and luciferase reporter assays were used to validate FOXA2 as a direct target gene of miR-92a. Consistently, the biological outcome of miR-92a on regulating FOXA2 was examined by proliferation and invasion analysis in vitro.
Results:
Here, we detected the higher expression of miR-92a in human HCC cell lines, such as HepG2, Huh7 and Hep3B, compared with the normal human hepatocyte L02 cells. Overexpression of miR-92a significantly increased cell growth and invasion ability, while the knockdown of miR-92a could remarkably inhibit the growth and invasion possibility. We identified that miR-92a has specific targeting sites in the 3’-UTR of the FOXA2. By overexpressing miR-92a in HepG2 cells or Huh7 cells, the expression of FOXA2 was remarkably repressed.
Conclusion:
We demonstrated that miR-92a may play a critical role in HCC proliferation and invasion and may serve as a novel therapeutic target by the repression of FOXA2.
Keywords: FOXA2, Hepatocellular carcinoma, Invasion, miR-92a, Proliferation
Introduction
Hepatocellular carcinoma (HCC) is one of the most important malignant tumor, which causes highly cancer-related mortality all over the world (1), especially in China (2). During the development of HCC, multiple genes are deregulated, and oncogenes are activated, while tumor suppressor are inhibited (3, 4) Although numerous therapeutic strategies have been developed, the 5-year survival of HCC still remained low (5). Therefore, it is critical to understand the molecular mechanism of HCC and therefore to explore new potential therapeutic agents.
MiRNAs are a cluster of non-coding small RNAs, which approximately contains 22 to 25 nucleotide (6). Through the interaction with the 3’-untranslated regions (3’-UTRs) of target mRNAs, miRNA may inhibit expression of the targeted genes or induce their degradation (7). Findings have indicated that abnormal miRNA expression plays crucial roles in the development of cancer (8, 9).
Among those miRNAs, miR-92a generally plays a promoting role in several types of human cancers; for example, by inhibiting p21, miR-92a promotes cell proliferation in cervical cancer (10). By suppressing F-box and WD repeat-containing protein 7, miR-92a promotes osteosarcoma tumor growth (11). By inhibiting KLF4, miR-92a promotes colorectal cancer cell proliferation and migration (12). However, although it is reported that deregulation of miR-92a is implicated in HCC development (13), but the underlying mechanism in HCC remains unclear.
The forkhead box transcription factor A family contains FOXA1 (HNF3α), FOXA2 (HNF3β) and FOXA3 (HNF3γ) (14). Among them, FOXA2 belongs to the hepatocyte nuclear factor family and is responsible for liver glucose homeostasis (15). As reported, FOXA2 suppressed the HCC metastasis through inhibition of matrix metalloproteinase-9 (16), but the mechanism for the regulation of FOXA2 was still unclear.
Materials and Methods
Cell culture
The human HCC cell line HepG2, Huh7 and Hep3B were obtained from the American Type Tissue Culture Collection (ATCC). Normal human hepatocyte L02 cells were purchased from the Shanghai Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China).
The above cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS; Gibco, Uruguay), 100 mg/ml penicillin -Streptomycin (invitrogene), in a humidified atmosphere containing 5% CO2 at 37 °C.
Reagents and Transfection
Synthetic miR-92a and the scrambled negative control RNA (miR-control) were purchased from (Genechem, Shanghai, China). All cells were cultured in 6-well plates.
Small interfering RNA (siRNA) sequence targeting human Foxa2 cDNA was synthesized from GenePharma in the RNA interference assay (Shanghai, China). The scrambled siRNA was used as a negative control (NC). When the cells growth reached to 70% confluent, Lipofectamine 2000 (Invitrogen) was used for transfection. Equal amounts of miR-92a or miR-control or relative plasmids were added to each well, 48 hr after transfection, the cells were harvested for qRT-PCR and western blotting.
RNA isolation and qRT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions, and cDNA was synthesized using the Primescript RT reagent (TAKARA, Dalian, China). Using oligo dT as primers, 1 μg of total RNA was reverse-transcribed to cDNA. RNeasy MinElute Cleanup Kit (Qiagen) and miRNeasy Mini Kit were used to extract the miRNAs according to the manufacturer’s instructions.
Quantitative RT-PCR was performed on ABI 7500 Real-Time Sequence Detection System (Applied Biosystems) using SYBER Green Dye (Invitrogen), and GAPDH was used as an internal control using the 2-ΔΔCT method.
TaqMan miRNA (Applied Biosystems, Foster City, CA) probes were used to measure the miRNAs according to the manufacturer’s instructions. In quantitative PCR, the relative levels of miRNAs were normalized to U6.
Luciferase reporter assay
The Dual Luciferase Reporter Assay System (Promega, USA) was used for luciferase reporter assay, for searching the target gene for miR-92a. The entire 3’-UTR of human FOXA2 was amplified using PCR. The PCR product was inserted into the p-GL3 -reporter plasmid (Promega) and confirmed by sequencing. The mutant of FOXA2 3’-UTR (from CCCACC to CCCAGC) was used to test the binding specificity, and FOXA2 3’-UTR mutant form was also inserted into the equivalent luciferase reporter. HepG2 cells were cultured in 24-well plates. The plasmids and 100 pmol of miR-92a or miR-control were co-transfected with the Renilla plasmid using Lipofectamine 2000 (Invitrogen) into HepG2 cells, and 48 hr post-transfection, the cells were assayed. The promoter activity was calculated as firefly/Renilla.
Western blotting
All cells were lysed using RIPA Lysis buffer (Beyotime, China), which were supplemented with a protease inhibitor cocktail (Roche) on ice for 30 min. After centrifuged for 15 min at 4°C, (12000 g), the supernatant was collected. A BCA protein assay kit (Thermo Scientific, USA) was used to calculate the protein concentration. The membranes were blocked in 5% nonfat milk in TBST and incubated with the appropriate primary antibodies overnight at 4 °C. Followed by washing with TBST for 5 times, the membranes were incubated with goat anti-rabbit or goat anti-mouse secondary antibodies at room temperature. Finally, immunoreactive bands were measured using the Bandscan ImageJ software.
Cell proliferation assay
MTT assay was used to assess cell proliferation. Briefly, 2,500 transfected HepG2 cells were seeded in triplicate in 96-well plates per well in 100 μl of culture medium. Afterwards, 20 μl of MTT reagent was added to each well 12, 24, 36, 48, 72, and 96 hr after transfection and then incubated for 4 hr according to the manufacturer’s instructions. To terminate the reaction, 100 μl of DMSO was added in each well. The absorbance was measured at 570 nm on an ELISA plate reader (Biotek Instruments, Inc, USA). Each assay was performed in triplicate.
Cell cycle analysis
Cells were seeded into 6-cm diameter plates and treated with 10% FBS. 48 hr after transfection, 1 × 106 cells in each group were harvested, and after washing with phosphate buffered saline (PBS) for 3 times, the cells were fixed with 70% ice-cold ethanol for 24 hr at 4 °C. Then, a mixture containing 0.5 ml propidium iodide, 0.5 mg/ml ribonuclease A and 0.2% Triton X-100 were used to incubate with the cells for 30 min. Cell cycle distribution was measured using BD C6 flowcytometry (BD Biosciences, San Jose, CA, USA).
Cell invasion assay
Cell invasion was determined using 24-well transwell boyden chamber (8.0 μm pore size with polycarbonate membrane) (Costar, USA) according to the manufacturer’s instructions. The upper chamber was covered with 100 μl diluted Matrigel (1 mg/ml) (BD Company) and incubated at 37 °C in 5% CO2 for 1 hr. Cells transfected with relative constructs were suspended in 500 μl DMEM medium without FBS and added to the upper compartment (1 × 104 cells/well). Then, 600 μl of DMEM with 10% FBS was added to the lower chamber. Following 24 hr incubation, the non-invading cells were scraped off gently using cotton-tipped swabs. Cells that had entered the lower chamber were stained with 0.1% crystal violet and 2% ethanol for 15 min at room temperature and photographed using a photomicroscope (BX51 Olympus, Japan), 5 fields per chamber and then counted blindly.
Statistical analysis
SPSS version 17.0 software (SPSS, Inc. Chicago, IL, USA) was used for statistical analysis. Quantitative RT-PCR, cell proliferation assay, invasion assay and the luciferase reporter were performed in triplicate. The data were shown as the mean±SD, of at least three independent experiments. P<0.05 was considered as statistically significant. Student’s t test was used for determining statistical significance.
Results
The expression of miR-92a was up-regulated in human HCC cell lines
To investigate the possible role of miR-92a in HCC, qRT-PCR was used to analyze the expression of miR-92a in three HCC cell lines, HepG2, Huh7 and Hep3B. Compared with the normal human hepatocyte L02 cells, miR-92a expression was significantly up-regulated (Figure 1A). These data indicated that the higher expression of miR-92a may contribute to the HCC malignant phenotypes.
Figure 1.
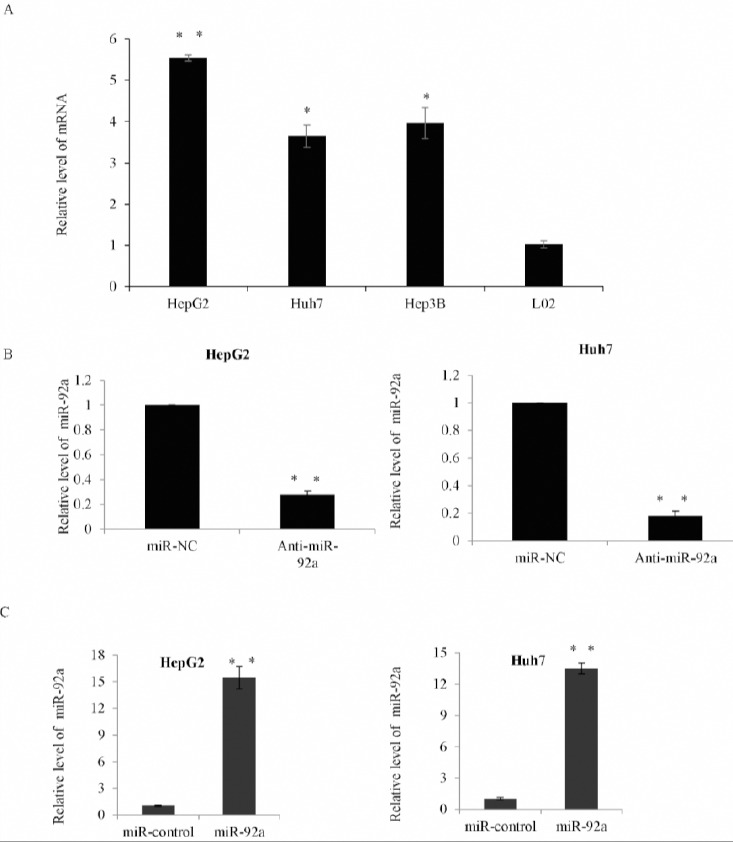
The expression of miR-92a was up-regulated in human hepatocellular carcinoma (HCC) cell lines
A) Quantitative RT-PCR analysis of the relative miR-92a levels in three hepatoma cells HepG2, Huh7 and Hep3B compared with the normal human hepatocyte L02 cells. Data was presented as the mean±SD, and was normalized to U6. *P<0.05; **P<0.01. B) The expression of miR-92a in HepG2 cells or Huh7 cells transfected with miR-92a antagomir (Anti-miR-92a) or negative control (miR-NC) was assessed by qRT-PCR. *P<0.05; **P<0.01. C) The expression of miR-92a in HepG2 cells or Huh7 cells transfected with miR-92a or the control mimics was assessed by qRT-PCR *P<0.05; **P<0.01
As a result, we performed miR-92a gain-of-function and loss–of-function studies in HepG2 cells and Huh7 cells in the following research. As shown in Figure 1B, qRT-PCR analysis showed that the expression level of miR-92a was obviously decreased in the HepG2 and Huh7 cells transfected with anti-miR-92a compared with miR-NC cells. Subsequently, the result of overexpression assay that was performed in HepG2 and Huh7 cells transfected with miR-92a or the control mimics showed that the expression of miR-92a was obviously increased in miR-92a mimic-transfected cells (Figure 1C).
miR-92a promoted cell growth and proliferation of HCC cells
We further investigated the role of miR-92a in the regulation of HCC cell growth and proliferation. MTT assay was used in the HepG2 cells transfected with the miR-92a or control mimics, or cells transfected with anti-miR-92a or miR-NC. The results showed that overexpression of miR-92a significantly increased HepG2 cell growth, while the knockdown of miR-92a could remarkably inhibit the growth rates (Figure 2A). Similar result was also shown in the Huh7 cells (Figure 2B). Further, cell cycle distribution was performed using flowcytometry in HepG2 and Huh7 cells (Figure 2C and 2D). The function of miR-92a was measured, and compared with the control group cells. In the miR-92a transfection cells, there was a substantial increase in S phase and a decrease in G1 phase populations, while knockdown of miR-92a blocked the progression of cell cycle from the G1 phase to S phase. The above data suggested that Foxk2 miR-92a promoted cell growth and proliferation of HCC cells through induction of G1 arrest.
Figure 2.
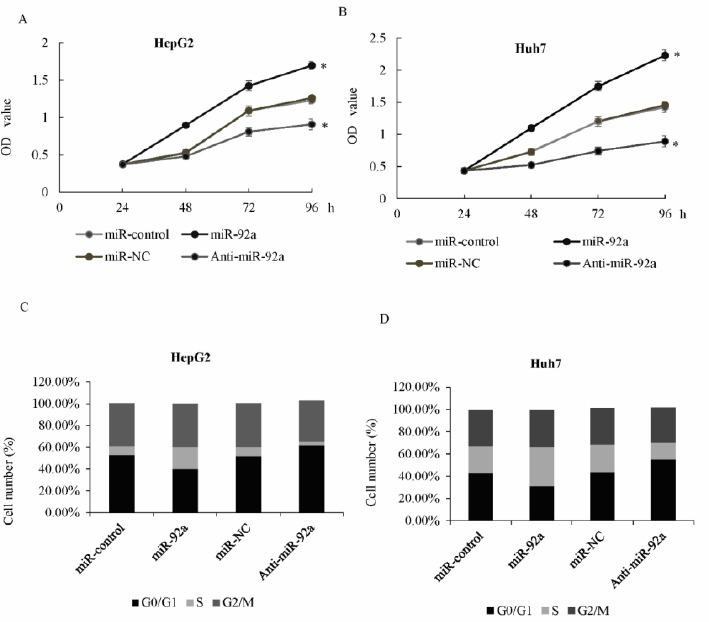
miR-92a promoted cell growth and proliferation of hepatocellular carcinoma (HCC) cells
A) MTT assay was performed in HepG2 cells transfected with miR-92a or control mimics, or transfected with anti-miR-92a or miR-NC. Optical density (OD) was measured 12, 24, 36, 48, 72, and 96 hr after transfection. B) MTT assay was performed in the Huh7 cells. C) Cell cycle distribution was performed using flowcytometry (FCM) in HepG2 cells transfected with anti-miR-92a or miR-NC, or miR-92a or control mimics. The percentage of cells in the G0/G1, S, and G2/M phase was shown, * P<0.05. D) Cell cycle distribution was performed in Huh7 cells
miR-92a promoted cell invasion of HCC cells
Transwell invasion assay was used to examine the effect of miR-92a on HepG2 cell motility. The invasion ability of HepG2 cells was significantly increased in the miR-92a transfection group compared with cells transfected with the miR-control, while in the cells transfected with anti-miR-92a, the invasion ability was obviously suppressed compared with the miR-NC (Figure 3A and 3B). Similarly, miR-92a was able to significantly promote Huh7 cells motility (Figure 3C). These above data suggested that miR-92a could remarkably promote both proliferation and invasion of HepG2 and Huh7 cells.
Figure 3.
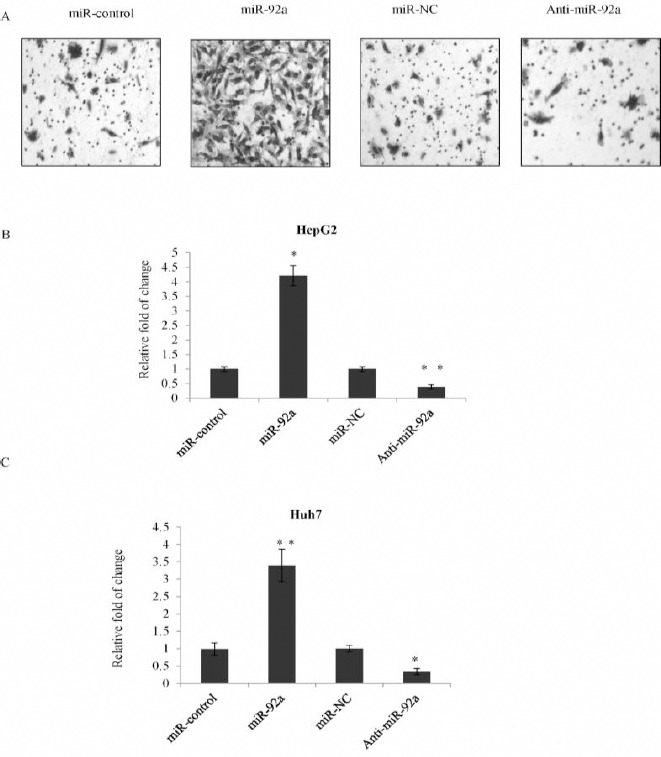
miR-92a promoted cell invasion of hepatocellular carcinoma (HCC) cells
A) Invasion analysis was performed in HepG2 cells transfected with miR-92a or the control mimics or anti-miR-92a or miR-NC. Representative photos were shown. B) Statistically analysis was represented as fold of change compared with vector. Error bars represented as the mean±SD for three different measurements. *P<0.05; **P<0.01. C) Statistically analysis was represented as fold of change compared with vector using transwell assay in Huh7 cells. *P<0.05; **P<0.01
Validation of FOXA2 as a direct target of miR-92a
TargetScan 6.2 (http://www.targetscan.org) was used to screen the potential target of miR-92a, and FOXA2 was considered as a target with potential miR-92a-binding sites. According to the bioinformatics data, several potential targets for miR-92a were identified. However, the binding of FOXA2 was with high confident scores. In light of the previous observation that FOXA2 was implicated in the development and progression of HCC, we focused our study on FOXA2 (Figure 4A). Luciferase activity assay was performed in HepG2 cells, and the results showed that miR-92a significantly inhibited the wild type 3’-UTR of FOXA2 luciferase activity, but not the mutant (Mut) type. The similar result was also shown in the Huh7 cells (Figure 4B and 4C). Furthermore, western blot was used to measure the protein level of FOXA2, and we found that the miR-92a significantly inhibited FOXA2 expression in HepG2 cell transfected with miR-92a compared with control mimic (Figure 4D). Consistently, overexpression of miR-92a significantly suppressed FOXA2 protein levels in Huh7 cells (Figure 4E). These data indicated that FOXA2 is a target of miR-92a.
Figure 4.
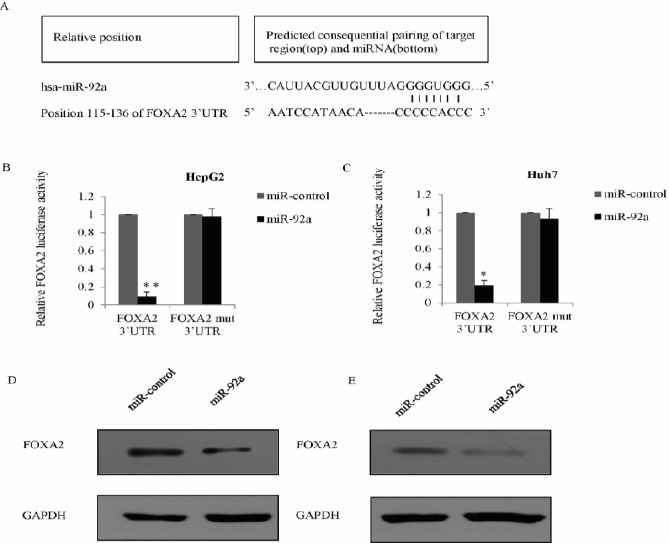
Validation of FOXA2 as a direct target of miR-92a
A) Schematic description of FOXA2 wild-type (WT) and FOXA2 mutant (Mut) 3’-UTR, which indicating the interactions between the FOXA2 3’-UTR binding site and miR-92a. B) HepG2 cells were co-transfected with luciferase gene driven WT or Mut UTR of FOXA2 and miR-92a or control mimics. The luciferase activity was examined 48 hr post-transfection. The normalized luciferase activity in the control group was used as a control. Each bar represented the mean±SD of triplicate experiments. *P<0.05, **P<0.01. C) The luciferase activity assay was performed in Huh7 cells. D) Western blot was used to measure the expression of FOXA2 in HepG2 cells transfected with miR-92a or the control mimics; experiments were performed in triplicate, and GAPDH was used as an internal control. E) The expression level of FOXA2 was measured in Huh7 cells transfected with miR-92a or the control mimics
MiR-92a induces HCC proliferation and invasion through the inhibition of FOXA2 expression
Since FOXA2 was a direct target of miR-92a, we further studied whether FOXA2 was involved in the function of miR-92a. We first established the FOXA2 overexpression plasmid and transfected into HepG2 or Huh7 cells, as measured by qRT-PCR. The FOXA2 mRNA level was significantly increased in FOXA2 overexpression groups compared with the vector groups (Figure 5A). MTT assay in HepG2 cells, co-transfected either with miR-92a and a FOXA2 overexpression plasmid, or control mimics, or miR-92a and pcDNA3.1, or pcDNA3.1-FOXA2 showed that FOXA2 restoration obviously reversed the promotion effects of miR-92a (Figure 5B); similar results were also observed in the Huh7 cells (Figure 5C). Transwell assay showed that overexpression of FOXA2 dramatically reversed the effects of miR-92a on HCC invasion ability (Figure 5D); similar results were also found in Huh7 cells (Figure 5E). The above data suggested that miR-92a functions as partially driving factor by targeting FOXA2.
Figure 5.
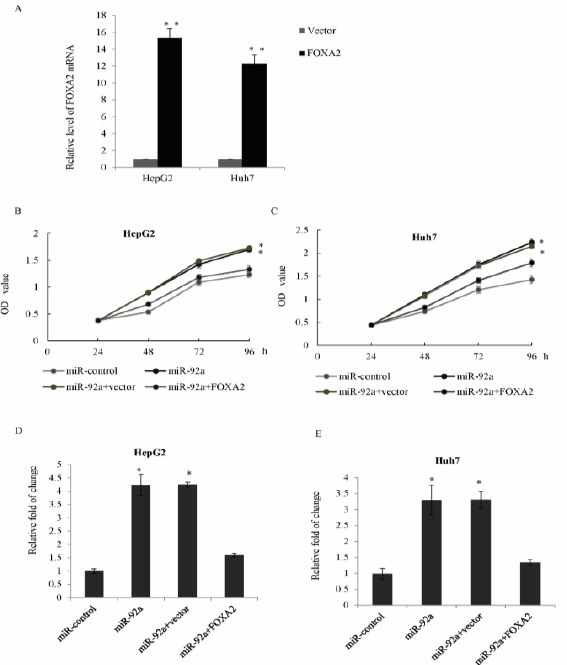
MiR-92a induces HCC proliferation and invasion through the inhibition of FOXA2 expression
A) HepG2 cells or Huh7 cells were transfected with FOXA2 plasmid, and the relative level of FOXA2 was examined by qRT-PCR. Each bar represented the mean±SD **P<0.01. B) HepG2 cells were co-transfected with miR-92a and a pcDNA3.1-FOXA2 overexpression plasmid, or control mimics, or miR-92a and pcDNA3.1, or pcDNA3.1-FOXA2. MTT assay was performed in triplicate experiments. *P<0.05. C) MTT assay was performed in Huh7 cells. *P<0.05. D) HepG2 cells were co-transfected with miR-92a and a pcDNA3.1-FOXA2 overexpression plasmid, or control mimics, or miR-92a and pcDNA3.1, or pcDNA3.1-FOXA2, transwell assay was performed, relative statistically analyses were represented. E) The above transwell assay was performed in Huh7 cells
Discussion
Increasing evidence shows that miRNAs play essential roles during cancer progression, such as proliferation, metastasis, and angiogenesis (17, 18). HCC, the most common liver cancer, has been reported to have a variety of aberrantly altered miRNAs, many of which function as oncogenes or tumor suppressors (19), (20).
As previously (21) reported, microRNA-24 level is increased in HCC and is positively correlated with HCC prognosis and tumorigenesis. As well, it is reported that microRNA-30d promoted cancer invasion and metastasis by targeting Galphai2 in HCC (22), while microRNA-422a was significantly down-regulated in HCC samples via targeting forkhead box G1, FOXQ1, and FOXE1 in the regulation of HCC development (23). Also, it is reported that miR-135b was usually amplified and up-regulated in HCC tissues and promoted HCC cells invasion and metastasis in vitro and in vivo (24). Moreover, findings have shown that miR-522 contributed to HCC cells proliferation by regulation of DKK1 and SFRP2 (25), while microRNA-141 inhibited HCC cells proliferation and invasion and promoted apoptosis by targeting hepatocyte nuclear factor-3β (26). But, the function of miR-92a in hepatocellular carcinoma is still unclear.
In this study, we found that miR-92a was usually up-regulated in the HCC cell lines. Knockdown of miR-92a suppressed HCC cell proliferation and invasion, while overexpression of miR-92a showed the opposite tendency. FOXA2 was identified as a target of miR-92a in HCC cells. As shown from the luciferase reporter assay, miR-92a could bind to the 3’UTR of FOXA2. RT-PCR and western blot showed that miR-92a regulates FOXA2 expression through leading to the mRNA degradation. Restoration of FOXA2 could partially reverse the oncogenic effects of miR-92a. The above data suggest that miR-92a might function as an oncogene through targeting FOXA2. As known, HCC should be characterized as a multi-stage process, which contains numerous gene expression alterations (27, 28). Discovery of critical miRNAs in modulating gene expression has not only changed our concept of tumorigenesis regulation, but has also offered opportunities for designing new anticancer strategies and therapies (29). In the future, more targets of miR-92a should be found and more evidence should be provided. The underlying on the molecular mechanisms may provide potential targets for HCC treatment.
Conclusion
We demonstrated that miR-92a may play a critical role in HCC proliferation and invasion. By regulating the repression of FOXA2, miR-92a may serve as a novel therapeutic target.
Acknowledgment
This study was supported by a grant from the Scientific Research Foundation of the Health Department of Zhejiang Province (2011KYA069). Zhejiang Provincial Natural Science Foundation of China (No. LY14C070003)
Conflicts of interest
The authors declare that no conflict of interest exists.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Ke X, Li L, Dong HL, Chen ZN. Acquisition of anoikis resistance through CD147 upregulation: A new mechanism underlying metastasis of hepatocellular carcinoma cells. Oncol Lett. 2012;3:1249–1254. doi: 10.3892/ol.2012.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen RX, Xia YH, Xue TC, Zhang H, Ye SL. Down-regulation of osteopontin inhibits metastasis of hepatocellular carcinoma cells via a mechanism involving MMP-2 and uPA. Oncol Rep. 2011;25:803–808. doi: 10.3892/or.2010.1116. [DOI] [PubMed] [Google Scholar]
- 5.Tang ZY. Hepatocellular carcinoma--cause, treatment and metastasis. World J Gastroenterol. 2001;7:445–454. doi: 10.3748/wjg.v7.i4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 2010;24:1339–1344. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’UTR as in the 3’UTR. Proc Natl Acad Sci U S A. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allegra A, Alonci A, Campo S, Penna G, Petrungaro A, Gerace D, et al. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (review) Int J Oncol. 2012;41:1897–1912. doi: 10.3892/ijo.2012.1647. [DOI] [PubMed] [Google Scholar]
- 9.Kunej T, Godnic I, Ferdin J, Horvat S, Dovc P, Calin GA. Epigenetic regulation of microRNAs in cancer: an integrated review of literature. Mutat Res. 2011;717:77–84. doi: 10.1016/j.mrfmmm.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Su Z, Yang H, Zhao M, Wang Y, Deng G, Chen R. MicroRNA-92a promotes cell proliferation in Cervical Cancer via Inhibiting p21 Expression and Promoting Cell cCycle Progression. Oncol Res. 2017;25:137–145. doi: 10.3727/096504016X14732772150262. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Jiang X, Li X, Wu F, Gao H, Wang G, Zheng H, et al. Overexpression of miR-92a promotes the tumor growth of osteosarcoma by suppressing F-box and WD repeat-containing protein 7. Gene. 2017;606:10–16. doi: 10.1016/j.gene.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Lv H, Zhang Z, Wang Y, Li C, Gong W, Wang X. MicroRNA-92a promotes colorectal cancer cell growth and migration by inhibiting KLF4. Oncol Res. 2016;23:283–290. doi: 10.3727/096504016X14562725373833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shigoka M, Tsuchida A, Matsudo T, Nagakawa Y, Saito H, Suzuki Y, et al. Deregulation of miR-92a expression is implicated in hepatocellular carcinoma development. Pathol Int. 2010;60:351–357. doi: 10.1111/j.1440-1827.2010.02526.x. [DOI] [PubMed] [Google Scholar]
- 14.Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 15.Lantz KA, Vatamaniuk MZ, Brestelli JE, Friedman JR, Matschinsky FM, Kaestner KH. Foxa2 regulates multiple pathways of insulin secretion. J Clin Invest. 2004;114:512–520. doi: 10.1172/JCI21149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Zhu CP, Hu PF, Qian H, Ning BF, Zhang Q, et al. FOXA2 suppresses the metastasis of hepatocellular carcinoma partially through matrix metalloproteinase-9 inhibition. Carcinogenesis. 2014;35:2576–2583. doi: 10.1093/carcin/bgu180. [DOI] [PubMed] [Google Scholar]
- 17.Fiorino S, Bacchi-Reggiani ML, Visani M, Acquaviva G, Fornelli A, Masetti M, et al. MicroRNAs as possible biomarkers for diagnosis and prognosis of hepatitis B- and C-related-hepatocellular-carcinoma. World J Gastroenterol. 2016;22:3907–3936. doi: 10.3748/wjg.v22.i15.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes CN, Chayama K. MicroRNAs as biomarkers for liver disease and hepatocellular carcinoma. Int J Mol Sci. 2016;17:280. doi: 10.3390/ijms17030280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao B, Wang G. MicroRNAs involved with hepato-cellular carcinoma (Review) Oncol Rep. 2015;34:2811–2820. doi: 10.3892/or.2015.4275. [DOI] [PubMed] [Google Scholar]
- 20.Gnoni A, Santini D, Scartozzi M, Russo A, Licchetta A, Palmieri V, et al. Hepatocellular carcinoma treatment over sorafenib: epigenetics, microRNAs and microenvironment. Is there a light at the end of the tunnel? Expert Opin Ther Targets. 2015;19:1623–1635. doi: 10.1517/14728222.2015.1071354. [DOI] [PubMed] [Google Scholar]
- 21.Liu YX, Long XD, Xi ZF, Ma Y, Huang XY, Yao JG, et al. MicroRNA-24 modulates aflatoxin B1-related hepatocellular carcinoma prognosis and tumorigenesis. Biomed Res Int. 2014;2014:482926. doi: 10.1155/2014/482926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao J, Liang L, Huang S, Ding J, Tan N, Zhao Y, et al. MicroRNA-30d promotes tumor invasion and metastasis by targeting Galphai2 in hepatocellular carcinoma. Hepatology. 2010;51:846–856. doi: 10.1002/hep.23443. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Yang Y, Yang T, Yuan S, Wang R, Pan Z, et al. Double-negative feedback loop between microRNA-422a and forkhead box (FOX)G1/Q1/E1 regulates hepatocellular carcinoma tumor growth and metastasis. Hepatology. 2015;61:561–573. doi: 10.1002/hep.27491. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Xu D, Bao C, Zhang Y, Chen D, Zhao F, et al. MicroRNA-135b, a HSF1 target, promotes tumor invasion and metastasis by regulating RECK and EVI5 in hepatocellular carcinoma. Oncotarget. 2015;6:2421–2433. doi: 10.18632/oncotarget.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Yu C, Chen M, Li Z, Tian S, Jiang J, et al. miR-522 contributes to cell proliferation of hepatocellular carcinoma by targeting DKK1 and SFRP2. Tumour Biol. 2016;37:11321–11329. doi: 10.1007/s13277-016-4995-0. [DOI] [PubMed] [Google Scholar]
- 26.Lin L, Liang H, Wang Y, Yin X, Hu Y, Huang J, et al. microRNA-141 inhibits cell proliferation and invasion and promotes apoptosis by targeting hepatocyte nuclear factor-3beta in hepatocellular carcinoma cells. BMC Cancer. 2014;14:879. doi: 10.1186/1471-2407-14-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyra-Gonzalez I, Flores-Fong LE, Gonzalez-Garcia I, Medina-Preciado D, Armendariz-Borunda J. MicroRNAs dysregulation in hepatocellular carcinoma: Insights in genomic medicine. World J Hepatol. 2015;7:1530–1540. doi: 10.4254/wjh.v7.i11.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu LN, Li DD, Xu HX, Zheng SG, Zhang XP. Role of microRNAs in hepatocellular carcinoma. Front Biosci (Landmark Ed) 2015;20:1056–1067. doi: 10.2741/4358. [DOI] [PubMed] [Google Scholar]
- 29.Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840–847. doi: 10.1002/hep.26095. [DOI] [PubMed] [Google Scholar]


