Abstract
The low molecular weight fraction of tomato plants inoculated with potato spindle tuber viroid (PSTVd) contains a population of short PSTVd-specific RNAs of either polarity. The main constituents were RNAs of 22 and 23 nt representing different domains of the viroid genome. The occurrence of such distinct RNA species indicated that the nuclear replicating PSTVd RNA induces post-transcriptional gene silencing. The short RNAs were slightly more abundant at 30 days post-inoculation than at later stages and were present in plants infected with a mild, severe or lethal isolate of PSTVd. There was no apparent correlation between the quantity of small PSTVd-specific RNAs and the degree of virulence of the viroid isolate.
INTRODUCTION
Post-transcriptional gene silencing (PTGS) results in the sequence-specific degradation of single-stranded RNAs. The phenomenon was first described in plants as ‘co-suppression’ when transgenes with highly active promoters were analysed (1,2). In many examples the affected RNA transcripts, including those expressed from viruses and recombinant viruses (reviewed in 3,4), were transcribed at high levels. However, poorly transcribed RNAs (reviewed in 5,6) or even ectopic promoterless DNA constructs have been shown to initiate PTGS (7). Once PTGS is induced it spreads systemically and homologous RNAs are degraded regardless of whether they had been expressed from nuclear genes, transgenes or cytoplasmic RNA viruses. It is likely that PTGS represents a host defence system against viruses (8), transposons (9) and potentially other forms of invading RNA. Transgenes or double-stranded (ds) RNA, also called interfering RNA (RNAi), can initiate similar processes in Neurospora crassa (10), Caenorhabditis elegans (11), Drosophila melanogaster (12) and vertebrates (13,14), including mice (15).
According to current models of PTGS, an endogenous RNA-dependent RNA polymerase or a virus-encoded RNA polymerase (16) synthesises a long dsRNA that is processed into 21–25 nt RNA fragments of both polarities. Such RNAs were first described in plants (17) and have recently been identified in Drosophila cells treated with RNAi (18,19). Current evidence suggests that the short RNAs are incorporated into a ribonuclease and that they confer sequence-specificity upon this enzyme (18,20–22).
Hamilton and Baulcombe (17) detected the characteristic RNAs in plants exhibiting PTGS that had been initiated in each of three different ways, including transgene-induced PTGS in the presence or absence of a homologous endogenous gene and systemic PTGS induced by local infiltration with Agrobacterium tumefaciens carrying a transformation vector. Since the size of the small RNA was identical in these examples of PTGS it was concluded that the occurrence of such RNA species can be used to diagnose PTGS and related processes in situations where it had not previously been suspected or confirmed. For example, the presence of potato virus X (PVX)-specific RNAs in PVX-infected plants was used as supporting evidence for the proposed role of PTGS as a natural anti-viral protection mechanism. Further examples of the occurrence of PTGS-related short RNAs in plants have been reported recently (23–25).
Viroids represent a distinct form of an RNA that can invade and infect some higher plant hosts. Like viruses they are pathogenic, autonomously replicating RNAs. However, their genomic RNA is unique. It consists of a single-stranded, covalently closed and thus circular RNA molecule ranging from 246 to 375 nt (reviewed in 26). Unlike RNA viruses, the viroid RNA is devoid of any protecting coat protein, does not use a pathogen-encoded replicase, and does not encode any protein. Potato spindle tuber viroid (PSTVd), which is the type-member of the largest groups of viroids, the pospiviroids, is replicated in the nucleus by the DNA-dependent RNA polymerase II (27,28) which also synthesises RNA transcripts from endogenous genes or transgenes. However, unlike a typical mRNA, PSTVd accumulates primarily in the nucleolus (29). In this report we show that PSTVd can induce the PTGS-typical response of its host as monitored by the occurrence of small RNAs.
MATERIALS AND METHODS
Infections with PSTVd
The tomato cultivar ‘Rentita’ was inoculated with in vitro synthesised RNA transcripts as previously described (30). Three different PSTVd isolates were used: (i) the mild isolate KF5 (accession number S54933; 31), (ii) the severe isolate KF440-2 (accession number X58388; 32,33) and (iii) the lethal isolate RG1 (accession number U23058; 34). Photographs showing the symptoms obtained with different viroid isolates have been published previously (32). Plants were kept under greenhouse conditions at ∼20°C.
Extraction of plant materials
RNA extraction shown in Figure 1 was carried out according to the method of Hamilton and Baulcombe (17). For extractions shown in Figures 2–4, ∼2–3 g of leaf material was harvested, frozen in liquid nitrogen and homogenised in a mortar. To the frozen powder, 7 ml of TEMS buffer (100 mM Tris–HCl pH 7.5, 100 mM NaCl, 10 mM EDTA) supplemented with 100 mM 2-mercaptoethanol prior to usage, was added. This was followed immediately by the addition of 10 ml extraction phenol (1 kg phenol, 300 ml TEMS buffer, 1 g 8-hydroxyquinoline and 250 ml chloroform). The mixture was vortexed and centrifuged at 4°C, 4000 g for 30 min. The aqueous phase was extracted once more with phenol then once with chloroform–isoamyl alcohol (24:1) (v/v), in each case followed by a further centrifugation step. After increasing the sodium concentration to 200 mM by the addition of sodium acetate pH 5.6, the mixture was precipitated with 2.5 vol ethanol. After collecting the precipitate by centrifugation the samples were washed with 70% ethanol and dried.
Figure 1.
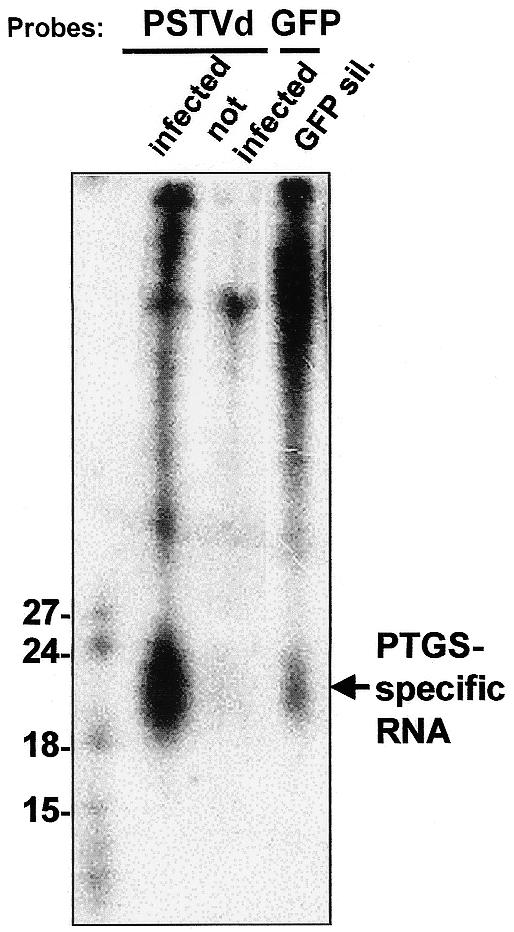
Comparison of low molecular weight RNA from PSTVd-infected and non-infected tomato and from Nicotiana benthamiana transformed with a [35S]GFP transgene that is undergoing PTGS. All samples were run on the same gel and blotted onto the same membrane. The lanes containing tomato RNA were hybridised with a full length PSTVd(–) RNA transcript. The uninfected control contains an unspecific cross hybridisation signal detected under these low stringency conditions. The lane containing N.benthamiana RNA was hybridised with a full length GFP(–) RNA transcript. Markers were 32P-labelled mGFP5 RNA, transcribed in vitro and digested with RNase T1. Sizes are indicated to the left of the gel. PTGS-specific RNA is indicated by an arrow to the right of the gel.
Figure 2.
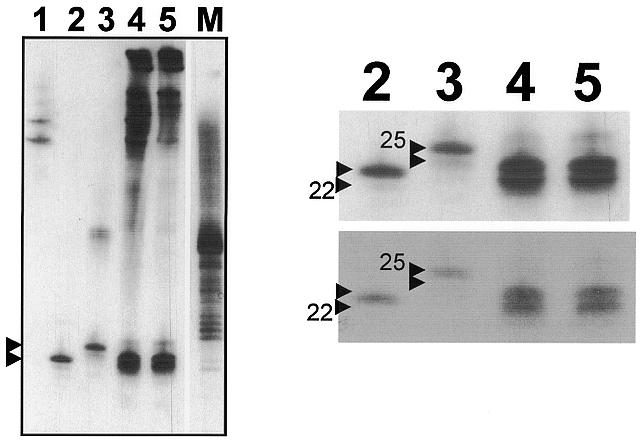
Northern analysis for detection of PTGS-typical small RNAs. Samples were hybridised with a full length PSTVd(–) RNA transcript. (A) Lane 1, healthy control, contains an unspecific cross-hybridisation signal detected under these low stringency conditions (compare with Fig. 1); lane 2, 23mer DNA marker (lower arrowhead); lane 3, 25mer DNA marker (upper arrowhead); lane 4, crude extract of tomato plants inoculated with PSTVd isolate KF440-2; lane 5, LiCl-soluble fraction of the extract shown in lane 4; lane M, mixture of synthetic RNA transcripts, consisting prominently of lengths between 27 and 39 nt. (B) A close-up of the relevant section of lanes 2–5 in different exposure times, detailing the signals originating from the small RNAs. The positions corresponding to 22–25 nt are indicated by arrowheads.
Figure 4.

Northern analysis for detection of PTGS-typical small RNAs from LiCl-fractionated extracts from leaves of tomato plants infected with three different PSTVd isolates. The hybridisation probe was the sense probe (Fig. 3, right panel). The positions of the two relevant signals are indicated with arrowheads.
One millilitre of 8 M LiCl was added to the dried pellet of the crude extract and shaken at 4°C overnight. Following centrifugation at 4°C, 4000 g for 30 min, the supernatant was collected and the extraction procedure repeated for 6 h. The combined supernatants were precipitated with 2.5 vol ethanol.
The samples were again collected by centrifugation, followed by washing several times in 70% ethanol and drying. The LiCl-soluble fraction was dissolved by adding 100 µl water.
Hybridisation probes
Hybridisation was performed using in vitro synthesised 32P-labelled RNA transcripts generated as previously described [(17) (Fig. 1) or (35) (Figs 2–4)]. Templates used were (i) pHa106 (33) linearised with HindIII which was transcribed with T7 RNA polymerase for synthesis of a full-length PSTVd(–) RNA probe and (ii) plasmids containing either of the two AvaI fragments of PSTVd cDNA (36) to synthesise sense or antisense probes of the ‘left’ and ‘right’ halves of PSTVd.
Markers
Markers shown in Figure 1 were 32P-labelled mGFP5 RNA, transcribed in vitro and digested with RNAase T1. In Figures 2–4 the following DNA oligonucleotides were end-labelled with [γ-32P]ATP: 23mer 5′-GGGGTAGGTGCGGGTTTCTTCAC; 25mer 5′-AGGTTTGGGTTTCGCCACTCTCTCG. For the transcription marker, we used a mixture of plasmid pT3T7lac (Boehringer Mannheim) linearised with either AvaI or SalI. Transcription with T7 RNA polymerase resulted in RNA transcripts of ∼27 and 39 nt.
Northern analysis
The analysis shown in Figure 1 was carried out according to Hamilton and Baulcombe (17). In those shown in Figures 2–4, samples equivalent to ∼100 mg of plant tissue were heat-treated in formamide buffer and loaded onto a 12% polyacrylamide slab gel (12% acrylamide, 0.6% bisacrylamide) containing 7 M urea and 50 mM TBE buffer (50 mM Tris, 41.5 mM boric acid, 0.5 mM EDTA) and separated by electrophoresis. The samples were electroblotted to Nytran®N membrane (Schleicher & Schuell) and fixed by UV cross-linking. Pre-hybridisation and hybridisation, which included the RNA probe at ∼106 c.p.m./ml, was carried out in 5× SSC, 1× Denhardt solution (37), 1% SDS, 0.25 mg/ml tRNA carrier at 58°C for 2 and 16 h, respectively. After hybridisation, the following washing steps were used: two washes with 2× SSC for 5 min at room temperature followed by two washes with 2× SSC, 0.5% SDS for 30 min at 58°C. Signals were visualised by autoradiography.
RESULTS
In vitro synthesised RNA transcripts were used to inoculate tomato plants at the four-leaf stage with the severe PSTVd isolate KF440-2. After the plants developed PSTVd symptoms, leaves were harvested at day 30 post-inoculation, which is an early stage of viroid symptom development at the low temperature conditions applied. Low molecular weight RNA was detected in PSTVd infected tomato which was apparently identical in size to the PTGS-specific, low molecular weight GFP RNA from GFP-silenced transgenic Nicotiana benthamiana (Fig. 1). This size class of RNA has been found in all examples of PTGS tested in this way but in none of the corresponding controls that did not exhibit PTGS (16–25).
As an alternative to the previously described procedure for extraction of these RNAs (17), we tested whether they could be enriched by LiCl solubilisation from total RNA and DNA extracts of infected plants as used for the enrichment of viroid RNA (38), except that the LiCl concentration was increased to 8 M. Under these conditions tRNAs, 5S rRNA, DNA and traces of rRNA were dissolved in the LiCl extraction solution and the longer RNAs remained in the pellet. Since the LiCl-soluble fraction includes most of the low molecular weight RNAs (and chromosomal DNA) this extraction resulted in enrichment for small RNAs.
The LiCl extracted material was desalted by ethanol precipitation and extensive washing and subjected to northern blot analysis by hybridisation with a full length PSTVd antisense RNA probe (Fig. 2A). Two prominent low molecular weight RNA fragments could be detected in extracts originating from infected plants. Based on the migration of oligonucleotide markers (Fig. 2B), we concluded that these RNAs were ∼22 and 23 nt long. However, neither of these RNAs migrated exactly in parallel with the markers, and precise size determination was not possible. Nevertheless, the size of small RNAs in viroid-infected plants appeared to be identical to that in plants exhibiting PTGS of transgenes (Fig. 1).
Next we assayed for representation of the sense and antisense strands of viroid RNA in the population of the small RNAs in viroid-infected plants. We also determined whether the small RNAs could hybridise to different regions of the PSTVd genome and whether the production of these RNAs varied at different stages of the infection or with mild and aggressive strains.
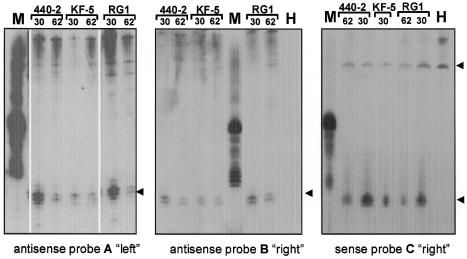
To carry out these tests we inoculated tomato plants with three PSTVd strains of different virulence. Of these, KF5 was the least aggressive, inducing only mild symptoms. Strain KF440-2 induced severe symptoms but isolate RG1, which arose spontaneously during an infection experiment (34), was the most aggressive. Plant tissues were harvested at 30 days post-inoculation and a second sample of the same plants was collected 62 days after inoculation. LiCl-fractionated RNA extracts of these samples were analysed by northern blotting using three different probes. Probe A was an antisense RNA that covered the left half of the PSTVd, ranging from nucleotides 284 to 359/1–97 (Fig. 3, left). Probe B was also of antisense polarity and detected the right half of the PSTVd molecule, ranging from nucleotides 92 to 289 (Fig. 3, middle). Probe C was the sense equivalent to probe B (Fig. 3, right).
Figure 3.
Northern analysis for detection of PTGS-typical small RNAs. Tomato plants were inoculated with PSTVd isolates KF440-2 (440-2), KF5 and RG1, and leaves were harvested at days 30 and 62 post-inoculation to prepare extracts that were fractionated by LiCl. Hybridisation probes were as described under each panel. H, healthy control; M, transcription marker. The position of the small RNAs is indicated by an arrowhead. The cross hybridisation signal obtained with the sense probe is also indicated (upper arrow).
The characteristic small RNAs were detected in all PSTVd-infected samples. The occurrence of the small RNAs in the left and the right domain of PSTVd indicated that different domains of the PSTVd RNA sequence are represented in the population and that RNAs of both polarities are produced.
The antisense probes showed additional signals originating from larger PSTVd(+) RNAs that were absent in the healthy control. These larger PSTVd(+) RNAs are most probably unrelated to PTGS. In accordance with the lower concentration of PSTVd replication intermediates of (–) polarity (39) there was a lower background hybridisation detected with the sense probe C. However, there was an additional signal in the healthy control that is due to cross hybridisation of the chloroplast large 5S rRNA of tomato (40). This 121 nt RNA is 100% identical to its tobacco homologue (accession number M10360; 41) and can cross hybridise due to a match of 15/16 nt between positions 215 and 230 of PSTVd(+) RNA.
This cross hybridisation to 5S RNA was used as an internal standard in our analysis for PSTVd-specific small RNAs in plants infected with PSTVd strains of different virulence. The hybridisation signals of 5S rRNA and small PTGS-induced PSTVd(–) RNAs were quantified with a phosphorimager. Using the 5S rRNA as a reference, we found that the samples taken at day 30 contained ∼2–4-fold more small RNAs than the samples taken at day 62. Re-probing of the blot shown in Figure 3, right, with a U1 antisense RNA confirmed this slight difference (data not shown). Although we cannot exclude some variation in the concentration of short RNAs in individual plants, these data indicate that PTGS is induced at the early stage of PSTVd replication, when plants start to develop symptoms. Comparing independent experiments we could not, however, detect any significant difference in the PTGS response between PSTVd isolates of different virulence (Fig. 4).
DISCUSSION
Small 21–25 nt RNAs have been associated with several examples of PTGS. These include different examples of PTGS in plants (16,17,23–25). Similar RNA species have been detected in Drosophila in response to treatment with interfering dsRNAs (RNAi) (18–22). Recently, these RNAs have been shown to be the specificity determinants of a sequence-specific ribonuclease which carries out the degradation of mRNA that is characteristic of PTGS (18–22). Nevertheless, it is possible to use the presence of these RNAs as an indicator of PTGS or PTGS-like mechanisms in plants. Here we show the presence of small PSTVd homologous RNAs of both polarities covering different domains of the PSTVd genome in infected plants. This indicates for the first time that plants respond to viroid infection by initiating PTGS directed against the viroid RNA.
Originally the short PTGS-associated RNAs were described as 25 nt in length. However, similar RNAs in extracts of Drosophila were assigned lengths of 21 and 23 nt (19) and here, using DNA markers, we determined a length of 22 and 23 nt. Whatever the true length of these molecules, it does seem likely that they exist in at least two major size classes. In addition, two less abundant RNAs were observed in the PSTVd-infected plants. One of these was 21 nt, and the other was ∼26 nt. One way in which these classes of molecules could differ in a manner that affects migration on gels is in the chemical nature of the terminal residue. Recently, Hutvágner et al. (25) showed that the PTGS-typical short RNAs can be phosphorylated, indicating that they have a 5′-hydroxyl group. However, that analysis did not distinguish between the two classes of molecule and it remains possible that they differ in the nature of the terminal residues. The chemical nature of the terminal residue could explain the anomalous migration of the short RNAs relative to the marker oligonucleotides that had 5′-terminal phosphate groups.
PTGS in plants can be initiated by cytoplasmically replicating viruses and is usually targeted against mature rather than precursor mRNAs. However, it is not yet clear whether the process is exclusively cytoplasmic and the results reported here add to this ambiguity because PSTVd replicates in the nucleus. PTGS can also be induced by transgenes (reviewed in 42), and in some instances causes an increase in the level of precursor mRNAs corresponding to the target species (43). In virus-infected cells there are several lines of evidence to indicate that PTGS is part of a defence system that prevents unrestricted accumulation of the viral RNA. Initially, as the virus enters the cell, viral RNA accumulates rapidly. However, at later times, after PTGS has been targeted against the viral RNA, its rate of accumulation slows down and may even stop. It is tempting to speculate that similar processes are active in PSTVd-infected cells. Thus, at 30 days post-inoculation when viroid replication is most rapid, the small PSTVd-specific RNAs are more abundant than at later stages when the viroid RNA is present at a lower level. According to the idea that PTGS protects against viroids, the reduced level of viroid RNA in the later stages of infection would be due to PTGS induced by the rapidly replicating viroid RNA in the early stages of the infection process. To test this idea it would be necessary to assess viroid accumulation in mutant plants that are defective for PTGS or in the presence of virus-encoded suppressors of PTGS. If viroids are susceptible to restriction by PTGS then this also provides a likely molecular basis for the observed cross protection between related viroid strains.
The finding that PTGS restricts viroid RNA accumulation implies that the execution of PTGS could take place in the nucleus where PSTVd replicates and accumulates. Attempts at detecting short RNAs in nuclear RNA preparations have failed so far. Perhaps the PTGS machinery has both cytoplasmic and nuclear phases. However, viroid RNA would have to be present in the cytoplasm as it moves from cell to cell and spreads systemically in the entire plant and it cannot be ruled out that the results reported here reflect a response to the cytoplasmic phase of the viroid RNA. On the other hand, it has already been shown that agents known to induce PTGS (44–47) also induce homology dependent transcriptional gene silencing (TGS; a nuclear process by definition) and implies that the signaling molecules conveying sequence specificity may be active in both nucleus and cytoplasm. Notably, TGS was also accompanied by the accumulation of the same sized short RNAs characteristic of PTGS. Our works extends this interrelationship of TGS and PTGS to viroids as PSTVd was previously shown to induce methylation of homologous cDNA sequences (48–50).
It was notable that we could not correlate aggressiveness of the viroid isolates with the PTGS response as determined by the concentration of the PTGS-specific small RNAs. This is reminiscent of the observation that virulence of different viroid isolates is not correlated with the titer of accumulating viroid RNA (34). It is unlikely, therefore, that the variation in aggressiveness is related to the differential effects of PTGS on the overall accumulation of the viroid RNA. A second untested possibility is that PTGS is directly involved with symptom development in viroid-infected plants. Since the specificity determinants of PTGS are short RNAs, similarly short lengths of chance homology between parts of the viroid genome and cellular mRNAs may be sufficient to render those host mRNAs targets of the PTGS response induced by the viroid. In this hypothesis, the differential virulence of viroids might be explained not by their overall level of accumulation in the plant, but by subtle sequence variation in the region sharing homology with the targeted host genes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Gerhard Steger, University of Düsseldorf for providing cDNA clones of PSTVd isolate RG1 and KF5, and A. Emilio Martinez de Alba (IMBB) for subcloning the AvaI fragments of PSTVd cDNA. This work was in part supported by the Greek Ministry of Development, (PENED 99ED376) and the European Union (Biotech BIO4 CT97-2300). A.J.H. and D.C.B. are grateful to the Gatsby Charitable Foundation for financial support.
References
- 1.Jorgensen R. (1990) Altered gene expression in plants due to trans interactions between homologous genes. Trends Biotechnol., 8, 340–344. [DOI] [PubMed] [Google Scholar]
- 2.van der Krol A.R., Mur,L.A., Beld,M., Mol,J.N. and Stuitje,A.R. (1990) Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell, 2, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baulcombe D.C. (1999) Viruses and gene silencing in plants. Arch. Virol., 15, S189–S201. [DOI] [PubMed] [Google Scholar]
- 4.Baulcombe D.C. (1999) Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol., 2, 109–113. [DOI] [PubMed] [Google Scholar]
- 5.Kooter J.M., Matzke,M.A. and Meyer,P. (1999) Listening to the silent genes: transgene silencing, gene regulation and pathogen control. Trends Plant Sci., 9, 340–347. [DOI] [PubMed] [Google Scholar]
- 6.Meins F. (2000) RNA degradation and models for post-transcriptional gene silencing. Plant Mol. Biol. 43, 261–273. [DOI] [PubMed] [Google Scholar]
- 7.Voinnet O., Vain,P., Angell,S. and Baulcombe,D.C. (1998) Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell, 95, 177–187. [DOI] [PubMed] [Google Scholar]
- 8.Ratcliff F.G., MacFarlane,S.A. and Baulcombe,D.C. (1999) Gene silencing without DNA. RNA-mediated cross-protection between viruses. Plant Cell, 11, 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ketting R.F., Haverkamp,T.H., van Luenen,H.G. and Plasterk,R.H. (1999) Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell, 99, 133–141. [DOI] [PubMed] [Google Scholar]
- 10.Cogoni C. and Macino,G. (1997) Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc. Natl Acad. Sci. USA, 94, 10233–10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 12.Kennerdell J.R. and Carthew,R.W. (1998) Use of double-stranded RNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell, 95, 1017–1026. [DOI] [PubMed] [Google Scholar]
- 13.Wargelius A., Ellingsen,S. and Fjose,A. (1999) Double-stranded RNA induces specific developmental defects in zebrafish embryos. Biochem. Biophys. Res. Commun., 263, 156–161. [DOI] [PubMed] [Google Scholar]
- 14.Li Y.X., Farrell,M.J., Liu,R., Mohanty,N. and Kirby,M.L. (2000) Double-stranded RNA injection produces null phenotypes in zebrafish. Dev. Biol., 217, 394–405. [DOI] [PubMed] [Google Scholar]
- 15.Wianny F. and Zernicka-Goetz,M. (2000) Specific interference with gene function by double-stranded RNA in early mouse development. Nat. Cell Biol., 2, 70–75. [DOI] [PubMed] [Google Scholar]
- 16.Dalmay T., Hamilton,A., Rudd,S., Angell,S. and Baulcombe,D.C. (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell, 101, 543–553. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton A.J. and Baulcombe,D.C. (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- 18.Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- 19.Zamore P.D., Tuschl,T., Sharp,P.A. and Bartel,D.P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]
- 20.Yang D., Lu,H. and Erickson,J.W. (2000) Evidence that processed small dsRNAs may mediate sequence-specific mRNA degradation during RNAi in Drosophila embryos. Curr. Biol., 10, 1191–1200. [DOI] [PubMed] [Google Scholar]
- 21.Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 22.Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001) RNA interference is mediated by 21-and 22-nucleotide RNAs. Genes Dev., 18, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dalmay T., Hamilton,A., Mueller,E. and Baulcombe,D.C. (2000) Potato virus X amplicons in arabidopsis mediate genetic and epigenetic gene silencing. Plant Cell, 12, 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Llave C., Kasschau,K.D. and Carrington,J.C. (2000) Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl Acad. Sci. USA, 97, 13401–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutvágner G., Mlynárová,L. and Nap,J.P. (2000) Detailed characterization of the posttranscriptional gene-silencing-related small RNA in a GUS gene-silenced tobacco. RNA, 6, 1445–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flores R., de la Peña,M., Navarro,J.A., Ambrós,S. and Navarro,B. (1999) Molecular biology of viroids. In Mandahar,C.L. (ed.) Molecular Biology of Plant Viruses. Kluwer Academic Press, Dordrecht, The Netherlands, pp. 225–239.
- 27.Mühlbach H.-P. and Sänger,H.L. (1979) Viroid replication is inhibited by α-amanitin. Nature, 278, 185–188. [DOI] [PubMed] [Google Scholar]
- 28.Schindler I.M. and Mühlbach,H.P. (1992) Involvement of the nuclear DNA-dependent RNA polymerases in potato spindle tuber viroid replication: a re-evaluation. Plant Sci., 84, 221–229. [Google Scholar]
- 29.Harders J., Lukacs,N., Robert-Nicoud,M., Jovin,T.M. and Riesner,D. (1989) Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridisation and confocal laser scanning microscopy. EMBO J., 8, 3941–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabler M. and Sänger,H.L. (1984) Cloned single- and double-stranded DNA copies of potato spindle tuber viroid (PSTV) and co-inoculated subgenomic DNA fragments are infectious. EMBO J., 3, 3055–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lakshman D.K. and Tavantzis,S.M. (1993) Primary and secondary structure of a 360-nucleotide isolate of potato spindle tuber viroid. Arch. Virol., 128, 319–331. [DOI] [PubMed] [Google Scholar]
- 32.Schnölzer M., Haas,B., Ramm,K., Wang,Z.F. and Sänger,H.L. (1985) Correlation between structure and pathogenicity of potato spindle tuber viroid (PSTV). EMBO J., 4, 2181–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsagris M., Tabler,M. and Sänger,H.L. (1991) Ribonuclease T1 generates circular RNA molecules from viroid-specific RNA transcripts by cleavage and intramolecular ligation. Nucleic Acids Res., 19, 1605–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gruner R., Fels,A., Qu,F., Zimmat,R., Steger,G. and Riesner,D. (1995) Interdependence of pathogenicity and replicability with potato spindle tuber viroid. Virology, 209, 60–69. [DOI] [PubMed] [Google Scholar]
- 35.Hammann C., Hormes,R., Sczakiel,G. and Tabler,M. (1997). A spermidine-induced conformational change of long-armed hammerhead ribozymes: ionic requirements for fast cleavage kinetics. Nucleic Acids Res., 25, 4715–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez de Alba A.E. (2000) Isolation and Characterisation of Viroid-Binding Proteins. PhD thesis, Universidad del Pais Vasco, Bilbao, Spain.
- 37.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Tabler M., Günther,I., Kern,R. and Sänger,H.L. (1989) A microscale procedure for isolating and sequencing the viroid RNA present in one gram of infected leaf tissue. J. Virol. Methods, 23, 111–126. [DOI] [PubMed] [Google Scholar]
- 39.Spiesmacher E., Mühlbach,H.P., Tabler,M. and Sänger,H.L. (1985) Synthesis of (+) and (–) RNA molecules of potato spindle tuber viroid (PSTV) in isolated nuclei and its impairment by transcription inhibitors. Biosci. Rep., 5, 251–265. [DOI] [PubMed] [Google Scholar]
- 40.Herold T. (1990) Zusammenhänge zwischen der Struktur und Pathogenität der Viroide am Beispiel des Potato Spindle Tuber Viroids, sowie die sequenzierung und molekulare klonierung einer zelleigenen RNA, die mit PSTV (+) RNA hybridisiert. PhD thesis, University of Giessen, Germany.
- 41.Takaiwa F. and Sugiura,M. (1981) Heterogeneity of 5S RNA species in tobacco chloroplasts. Mol. Gen. Genet., 182, 385–389. [Google Scholar]
- 42.Sijen T. and Kooter,J.M. (2000) Post-transcriptional gene-silencing: RNAs on the attack or on the defense? Bioessays, 6, 520–531. [DOI] [PubMed] [Google Scholar]
- 43.Mishra K.K. and Handa,A.K. (1998) Post-transcriptional silencing of pectin methylesterase gene in transgenic tomato fruits results from impaired pre-mRNA processing. Plant J., 14, 583–592. [Google Scholar]
- 44.Jones L., Hamilton,A.J., Voinnet,O., Thomas,C.L., Maule,A.J. and Baulcombe,D.C. (1999) RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell, 11, 2291–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mette M.F., Aufsatz,W., van der Winden,J., Matzke,M.A. and Matzke,A.J. (2000) Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J., 19, 5194–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang M.B., Wesley,S.V., Finnegan,E.J., Smith,N.A. and Waterhouse,P.M. (2001) Replicating satellite RNA induces sequence-specific DNA methylation and truncated transcripts in plants. RNA, 7, 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas C.L., Jones,L., Baulcombe,D.C. and Maule,A.J. (2001) Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using Potato virus X vector. Plant J., in press. [DOI] [PubMed] [Google Scholar]
- 48.Wassenegger M., Heimes,S., Riedel,L. and Sänger,H.L. (1994) RNA-directed de novo methylation of genomic sequences in plants. Cell, 76, 567–576. [DOI] [PubMed] [Google Scholar]
- 49.Pelissier T. and Wassenegger,M. (2000) A DNA target of 30 bp is sufficient for RNA-directed DNA methylation. RNA, 6, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wassenegger M. (2000) RNA-directed DNA methylation. Plant Mol. Biol., 43, 203–220. [DOI] [PubMed] [Google Scholar]