Abstract
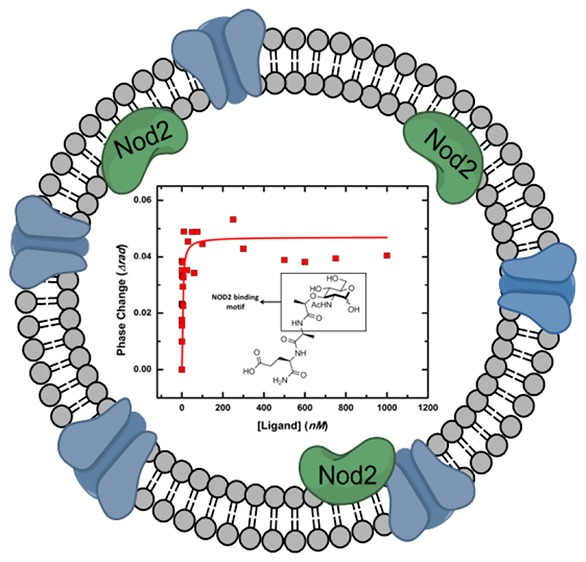
The human gut must regulate its immune response to resident and pathogenic bacteria, numbering in the trillions. The peptidoglycan component of the bacterial cell wall is a dense and rigid structure that consists of polymeric carbohydrates and highly cross-linked peptides which offers protection from the host and surrounding environment. Nucleotide-binding oligomerization domain-containing protein 2 (NOD2), a human membrane-associated innate immune receptor found in the gut epithelium and mutated in an estimated 30% of Crohn’s disease patients, binds to peptidoglycan fragments and initiates an immune response. Using a combination of chemical synthesis, advanced analytical assays, and protein biochemistry, we tested the binding of a variety of synthetic peptidoglycan fragments to wild-type (WT)-NOD2. Only when the protein was presented in the native membrane did binding measurements correlate with a NOD2-dependent nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) response, supporting the hypothesis that the native-membrane environment confers ligand specificity to the NOD2 receptor for NF-κB signaling. While N-acetyl-muramyl dipeptide (MDP) has been thought to be the minimal peptidoglycan fragment necessary to activate a NOD2-dependent immune response, we found that fragments with and without the dipeptide moiety are capable of binding and activating a NOD2-dependent NF-κB response, suggesting that the carbohydrate moiety of the peptidoglycan fragments is the minimal functional epitope. This work highlights the necessity of studying NOD2-ligand binding in systems that resemble the receptor’s natural environment, as the cellular membrane and/or NOD2 interacting partners appear to play a crucial role in ligand binding and in triggering an innate immune response.
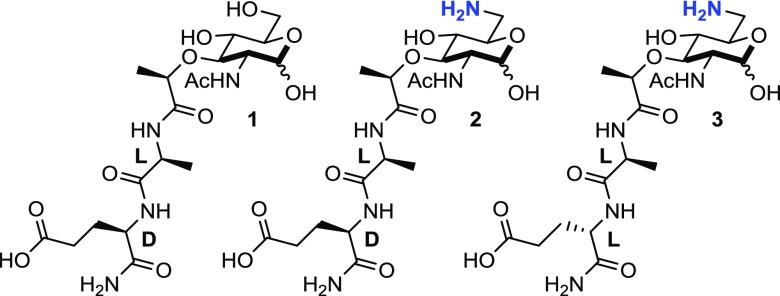
The innate immune system is the first line of defense against invading pathogens. This evolutionarily conserved system helps to maintain the proper relationship with the microbiome by detecting and responding to the molecular signatures of bacteria.1,2 To achieve this, the system uses a series of pattern recognition receptors (PRRs) that include the Nod-like receptors (NLRs).3 NOD2 (nucleotide-binding oligomerization domain-containing protein 2) is a cytosolic 110 kDa NLR protein capable of membrane association that mediates an innate immune response in the presence of bacterial cell wall fragments.4−6 It contains two N-terminal caspase recruitment domains (CARDs), a central nucleotide oligomerization domain (NOD), and a C-terminal leucine-rich repeat (LRR) domain.4 Mutations in NOD2 correlate with the onset of Crohn’s disease, an incurable, chronic inflammatory gastrointestinal disease that is proposed to be the result of an inappropriate immune response to bacteria.7−9 As an innate immune receptor, NOD2 recognizes bacterial cell wall fragments and initiates an immune response via the NF-κB pathway.10 The bacterial cell wall is made up of a strong polymer of carbohydrates and peptides known as peptidoglycan, a highly conserved component of both Gram-positive and Gram-negative bacteria. This conserved polymer serves an important role in the human innate immune system by providing a unique molecular calling card for the recognition of bacterial cells, and peptidoglycan fragments have long been known to activate an immune response.11,12 To date, N-acetyl-muramyl dipeptide (MDP-(ld); 1, Figure 1) is presented in the literature as the smallest known structural unit of peptidoglycan to bind to NOD2 and elicit a NOD2-dependent inflammatory response via the NF-κB and MAP kinase pathways.12−17
Figure 1.
Biologically relevant muramyl dipeptide fragment MDP-(ld), the 6-amino-derivatives of the naturally occurring MDP-(ld), and its unnatural diastereomer MDP-(ll). The amine functionality installed at the 6-position provides a chemical handle for surface tethering in SPR.
It has been widely debated in the field as to whether MDP binds directly to NOD2.18,19 Recently, our laboratory demonstrated that MDP and NOD2 directly interact with nanomolar affinity in an in vitro surface plasmon resonance (SPR) assay;15 the Duncan laboratory has used biotinylated MDP affinity chromatography to reach a similar conclusion.16 Elegant work from several groups has provided evidence that a series of peptide transporters20−23 exist capable of shuttling MDP across the cell membrane, thereby giving intracellular NOD2 access to this molecule. Stimulated by these new insights, we wished to investigate the assumption that MDP constitutes the minimal binding and signaling fragment via the NOD2 receptor.
Here, we test the ligand requirements for NOD2 binding and activation, following our previous report of direct binding between NOD2 and 6-amino-MDP-(ld) (2, Figure 1) as well as the unnatural diastereomer 6-amino-MDP-(ll) (3, Figure 1).15 The prior chemical synthetic route24 was modified to provide a small library of truncated peptidoglycan derivatives. Binding affinities for the peptidoglycan derivatives were established using a modified version of the previously reported SPR assay,15 as well as by backscattering interferometry (BSI), a label-free solution-based technique developed by Bornhop et al.25,26 Additionally, the compounds were profiled for their ability to activate the NOD2-dependent NF-κB driven innate immune response using a luciferase reporter assay.6 The results aid in discerning requirements for NOD2 ligand specificity and downstream signaling. Finally, the data provide evidence that NOD2 is an innate immune receptor and its interaction with the membrane is important for binding and activation.
Results and Discussion
Modular Synthesis of 6-Amino Functionalized Peptidoglycan Derivatives
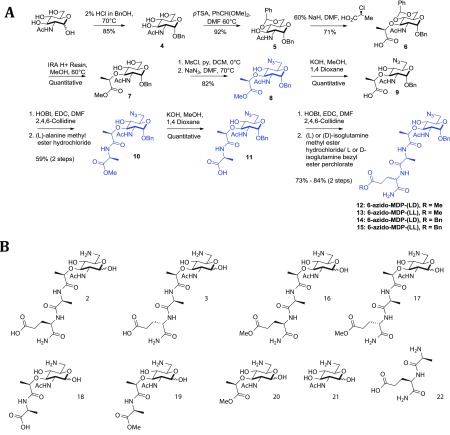
A modular synthesis (Figure 2A) was developed to produce a small peptidoglycan library with amine functionality at the 6-position of the carbohydrate.24 The amine provides a chemical handle for derivatization, allowing members of the library to be analyzed for the ability to interact with NOD2 in a SPR assay.27 We have previously demonstrated that this modification does not affect binding to NOD2 or subsequent activation of NF-κB.15,24 The synthesis began from the inexpensive and readily available carbohydrate N-acetyl-glucosamine to produce a modularly protected muramic acid fragment. Amino acid couplings allowed for the generation of peptidoglycan fragments containing 6-azido-muramic acid (9) coupled to alanine to produce the protected muramyl monopeptides (MMPs; 10 and 11) and the full dipeptide to produce the protected muramyl dipeptides (MDPs; 12–15) both as carboxylic acids or capped as methyl esters in a maximum of nine steps. Catalytic hydrogenation of the highlighted compounds (Figure 2A) yielded the final 6-amino functionalized peptidoglycan derivatives (Figure 2B). The 6-amino-N-acetyl-glucosamine fragment (21) was similarly produced by hydrogenation of protected 6-azido-N-acetyl-glucosamine (S21.1).28 Overall, the library varies in the stereochemistry of the isoglutamine residue, nature of the carboxylic acid functionality at the C-terminus of the peptide, and overall peptide chain length. It is worth noting that attempts to deprotect 9 (Figure 2A) to produce 6-amino-N-acetyl-muramic acid resulted in a product that readily degraded.
Figure 2.
An amine functionalized peptidoglycan library. (a) Synthetic route for the modular synthesis of differentially protected peptidoglycan fragments. (b) Library of amine functionalized peptidoglycan fragments.
Truncated Peptidoglycan Derivatives Bind to Purified Wild-Type (WT) NOD2
The ability of NOD2 to interact with the peptidoglycan library was assayed on a Biacore 3000 instrument (GE Healthcare) using an optimized version of the previously reported SPR binding assay.15 According to previously established methods, the amine functionalized peptidoglycan derivatives were immobilized in separate lanes to the carboxylic acid termini of thioalkanes preassembled onto a gold surface to generate peptidoglycan functionalized self-assembled monolayers.27 Purified WT-NOD2 (Figure S1) was applied to the generated surfaces at pH 6.5, the optimal pH for NOD2 and MDP interaction (Methods).15 All assayed ligands (2, 3, 16–21) bound NOD2 on the same order of magnitude, with apparent dissociation constants ranging from 4 to 36 nM (Figures S2 and S3, Table 1). Bovine serum albumin (BSA) and NOD1, an innate immune receptor homologous to NOD2, were observed not to bind to the immobilized ligands in the SPR assay, demonstrating fragment specificity for NOD2 (Figure S4). We expected some members of the synthetic peptidoglycan fragments to exhibit diminished affinity for NOD2 and that one or more structure–activity relationships could be developed. Surprisingly, however, no significant differences in binding were observed in the SPR assay.
Table 1. Dissociation Constants for Binding of Synthetic Peptidoglycan Fragments to Purified NOD2.
| NOD2 KD (nM) |
||
|---|---|---|
| ligand | SPRa | BSIb |
| 2 | 4–10 | 2–10 |
| 3 | 10–18 | 1–6 |
| 16 | 12–24 | 2–10 |
| 17 | 12–24 | 2–10 |
| 18 | 24–36 | 15–30 |
| 19 | 16–30 | 8–20 |
| 20 | 12–24 | 2–10 |
| 21 | 16–30 | 2–10 |
| 22 | 12–24 | 2–10 |
SPR measurements using surface-tethered ligands and purified protein. It should be noted that SPR, as a surface-bound technique, gives an adsorption isotherm rather than a true KD.
BSI measurements with ligands (varied concentration) and purified protein (10 nM) in solution. Affinities are reported as a range to emphasize the semiquantitative nature of these measurements as noted in the text.
To confirm NOD2-ligand binding, we employed backscattering interferometry (BSI) as a complementary label-free solution-phase method that has been used for the quantitation of a variety of protein–ligand interactions.25,29−31 BSI measures slight changes in the refractive index of a solution that occur as a result of a binding event; titration of this signal over a range of ligand concentrations allows for the determination of protein–ligand association constants.25 Using this technique, purified WT NOD2 was observed to bind the underivatized amine compounds (Figure 2B) in free solution with similar nanomolar affinities (ranging from 1 to 30 nM) to those measured by SPR (Table 1, Figure S5), confirming that all compounds in the peptidoglycan library exhibit low nanomolar affinity for purified WT NOD2 and showing that the individual components (carbohydrate and peptide) are both capable of binding NOD2 in an isolated system. The functional outcome of this interaction was assayed using the same peptidoglycan library in a NF-κB luciferase reporter assay.
Peptidoglycan Fragments Smaller than MDP Exhibit NOD2-Dependent NF-κB Activity
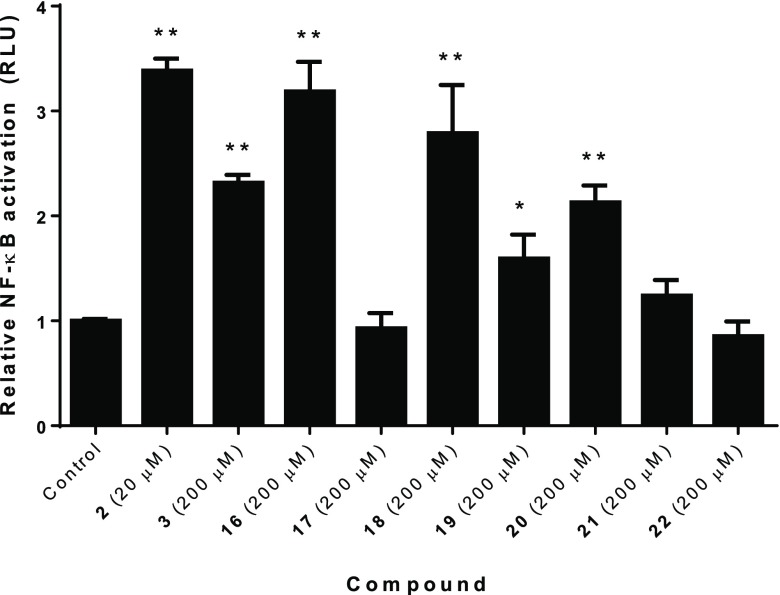
The peptidoglycan library was assayed in a standard NF-κB luciferase reporter assay to determine compounds capable of stimulating a NOD2-dependent NF-κB response. This assay employs HEK293T-NOD2myc/Tet-op cells, which demonstrate a low level of NOD2 expression in the absence of the doxycyclin promotor.32 This low level of NOD2 mirrors endogenous levels of NOD2 expression and reduces background NF-κB activity, which is commonly seen when NOD2 is highly overexpressed.18,24,33,34 These cells were stimulated by incubation with the peptidoglycan derivatives for 8 h and harvested, and measurements were taken to determine relative luciferase reporter expression (Figure 3). Minimal off-target NF-κB activation was observed in the NOD2 negative HEK293T cells used as a control (Figure S10). The NOD2 ligand, MDP, is known to enter cells via the peptide transporters hPepT1,20 SLC15A3, and SLC15A421 and by clathrin- and dynamin-dependent endocytosis.22 Preincubation of select activating and nonactivating ligands with lipofectamine LTX35 gave rise to no change in NF-κB response, indicating that the ligands are actively transported, passively permeable to some extent, or both (Figure S10).
Figure 3.
Relative NF-κB activation of synthetic peptidoglycan fragments in HEK293T-NOD2myc/Tet-op cells. Relative luciferase activity was measured after 8 h of stimulation with peptidoglycan fragments (*P < 0.03, activates in a NOD2-dependent manner; **P < 0.01). Error bars represent SEM of experiments performed at least six times. Experimental details can be found in the Supporting Information.
The data for known compounds (2–4) are consistent with those previously reported in the literature.15,36 Thus far, MDP (1) (and its amino analogue, 2) has been considered the minimal fragment necessary to activate a NOD2-dependent inflammatory response,12−17 and previous reports have shown that the N-acetyl-muramic acid (NAM) fragment does not exhibit adjuvant activity.36 We similarly showed that NAM, dipeptide (22), and a mixture of the two molecules are unable to activate an NF-κB response in this assay at high concentrations (Figure S11). However, we observed modest NOD2-dependent NF-κB responses in the presence of truncated versions of MDP (18–20, Figure 3), used at 10-fold higher concentrations than 2. In other words, smaller fragments than MDP appear to be able to activate a NOD2-dependent NF-κB response. For example, amidation of NAM at the carbohydrate 6-postion and esterification of the carboxylic acid are sufficient to turn on activatation of NOD2 in the cellular assay, albeit at a higher concentration than the binding constant of this molecule to pure NOD2 would suggest. Such differential sensitivity may relate to the cell biology of these molecules: MDP is present at very low concentrations in the human body, and therefore it is important for it be sensed at low amounts; however other ligands are known to be more prevalent.20,37 Thus, it might be expected that truncated fragments would exist at higher concentrations, making this signal relevant. Alternatively, the cell could have mechanisms to modify or selectively uptake certain PG fragments, leading to the miscorrelation between binding and activation.
These data prompted us to further investigate NOD2 binding and activation, as all compounds were observed to bind purified NOD2 (Table 1) but not all compounds activated the NF-κB pathway in a NOD2-dependent manner (Figure 3). One hypothesis is that additional interactors for NOD2 are necessary for functional ligand specificity, as these interactors would not be present in purified protein binding assays but would be present in the NF-κB luciferase reporter assay. To test binding under the latter conditions, we took advantage of NOD2 membrane association5,6 and examined ligand binding in native cell membranes using BSI.25
Native-Membrane Environment Is Crucial for NOD2 Binding Specificity
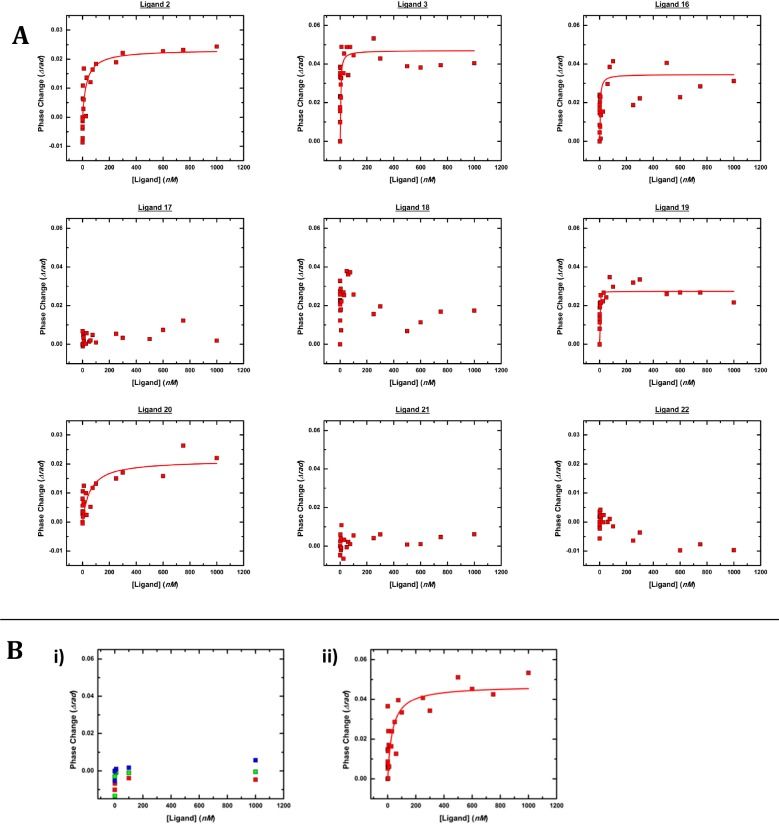
NOD2 is known to undergo a variety of interactions with membrane-bound proteins which are suspected to be necessary for a proper NOD2-dependent NF-κB response.5,6 Since BSI is compatible with complex mixtures, we re-examined NOD2-ligand binding while presenting the NOD2 receptor in a more “cell-like” environment. Therefore, we assayed the binding of our peptidoglycan library to native-membrane vesicles derived from HEK293T-NOD2myc/Tet-op cells. In contrast to the purified receptor, NOD2 is presented on the surface of these vesicles in a more natural environment where it can interact with native membrane components and other membrane-bound proteins (Figure 4, Table 2).25 The vesicles were characterized by dynamic light scattering, lipid staining (FM-1–43fx), and Western blot analysis to verify the presence of NOD2 (Supporting Information, Figure S9). Ligands 2, 3, 16, and 19 were observed in blind experiments to bind with high affinity to NOD2 in the native-membrane environment. Conversely, no binding was detected for ligands 17, 21, and 22, and 20 was observed to bind with weak affinity (Figure 4A, Table 2). Compound 18 gave indications of binding but unusual curves as it showed a strong interaction at low concentrations followed by a decrease in BSI signal with the addition of more ligand. All patterns were reproducible over several independent experiments using different cell preparations, but the apparent KD values varied somewhat, presumably due to variation in NOD2 density and orientation in the native-membrane vesicles; Table 2 therefore reports binding constant ranges of statistical confidence. Binding to the peptidoglycan library was referenced against membrane vesicles prepared from HEK293T cells not expressing NOD2 (Figure S6).
Figure 4.
Representative BSI measurements. (a) Binding data with NOD2 presented in vesicles derived from the HEK293T-NOD2myc/Tet-op cells in which the protein is expressed. Binding to the peptidoglycan library was referenced against membrane vesicles prepared from HEK293T cells not expressing NOD2 (Figure S6). Curves are fits to a single-site Langmuir binding isotherm. Experimental methods and data analysis are described in the Methods and Supporting Information. (b) Competition binding data. (i) Competition binding experiments in which membrane vesicles were preincubated with excess ligand 2 (10 μM) followed by titration of ligand 20 (red), 22 (green), or 2 (blue). (ii) Membrane vesicles preincubated with excess ligand 22 (10 μM) followed by titration of ligand 2.
Table 2. Dissociation Constants for Peptidoglycan Fragments Binding to NOD2 in Native Membrane Vesicles.
| ligand | Kd (nM)a | relative binding strength |
|---|---|---|
| 2 | 3–30 | strong |
| 3 | 3–30 | strong |
| 16 | 3–30 | strong |
| 17 | n.b. | n/a |
| 18 | bindingb | strong |
| 19 | 3–30 | strong |
| 20 | 50–200 | weak |
| 21 | n.b. | n/a |
| 22 | n.b. | n/a |
| iE-DAP | n.b. | n/a |
| N-acetyl-muramic acid | n.b. | n/a |
| 6-amino-glucose | n.b | n/a |
Values are reported as a range to reflect the scattered nature of the results in repeated experiments with different batches of cells, which could reflect differences in cell membrane composition. “n.b.” = nonbinding.
Unusual shape of binding curve did not allow the extraction of apparent Kd, perhaps suggesting an initial high-affinity interaction followed by a unique change in aggregation or other property at higher ligand concentrations. See the Supporting Information (Figure S7B and C)
Competition experiments were performed to confirm that the observed ligand binding was specific. After preincubation with ligand 2, the addition of 2 or 20 induced no BSI signal (Figure 4B), showing that these compounds access the same binding site.31 Conversely, preincubation with 22 did not block binding of 2 as detected by BSI (Figure 4B). In addition, two compounds in which the 6-amino substituent was replaced by hydroxyl [MDP-(ld) (1) and N-acetyl muramyl methyl ester (Scheme S14, 23)] were tested in these experiments to determine if the 6-amino modification affects binding to NOD2. Both compounds 1 and 23 bound with similar high affinity to that of their 6-amino analogues 2 and 20 (Figure S8), suggesting little influence by the amine group on binding. Importantly, the NOD1 ligand, iE-DAP, and the carbohydrate fragments, N-acetyl-muramic acid and 6-amino-glucose, did not bind NOD2 when presented in native membrane vesicles (Table 2, Figure S7). These results correlated very well in a qualitative sense with the NOD2-dependent NF-κB activity data (Figure 3). In quantitative terms, the correlations were also consistent with the exception that the strongly bound 19 elicited relatively weak NF-κB activation while weakly bound 20 gave moderately strong signaling. Overall, this supports the hypothesis that the cell membrane and/or presence of membrane protein NOD2 interacting partners aid in directing NOD2 substrate binding specificity.
Discussion
This work presents a detailed analytical approach to assess binding and NF-κB pathway activation requirements of the important innate immune receptor NOD2, focusing on a library of truncated versions of the natural NOD2 peptidoglycan-derived ligand, MDP. All compounds in the library exhibited nanomolar affinity for purified WT NOD2 in two independent binding assays (SPR, BSI); however only a subset of the library was capable of activating a NOD2-dependent NF-κB response. These contradictory results were resolved by measuring the ligand binding in a BSI native-membrane vesicle binding assay.
NOD2 is known to localize to the membrane, and this membrane localization is critical for activating the NF-κB response.5,6,38 Elegant work by Lipinski and co-workers showed that the membrane targeting protein, FRMPD2, is necessary for proper NOD2 localization. Moreover, they presented a model in which NOD2 mutants that do not bind to FRMPD2 are unable to localize to the membrane and signal via NF-κB.5 We therefore hypothesized that NOD2 membrane localization is critical for shaping NOD2’s bacterial peptidoglycan specificity, and further assayed NOD2 binding in native-membrane environments. The data presented in this paper, made possible by the solution based, label-free nature of the BSI binding assay and the presentation of NOD2 in native-membrane vesicles, showed that functional NOD2 exhibits a more stringent ligand binding specificity, only binding to compounds that are capable of activating a NOD2-dependent NF-κB response. This selectivity as well as the strength of the observed binding affinities suggest an important affinity of NOD2 for the 6-amino-N-acetyl-muramyl motif, as it is the only fragment common to all of the ligands that bind and activate (Figure 5). Conversely, peptidoglycan fragments consisting only of the dipeptide (22) or the structurally simpler carbohydrate 6-amino-N-acetyl glucosamine (21) were unable to bind NOD2 in the membrane environment.
Figure 5.
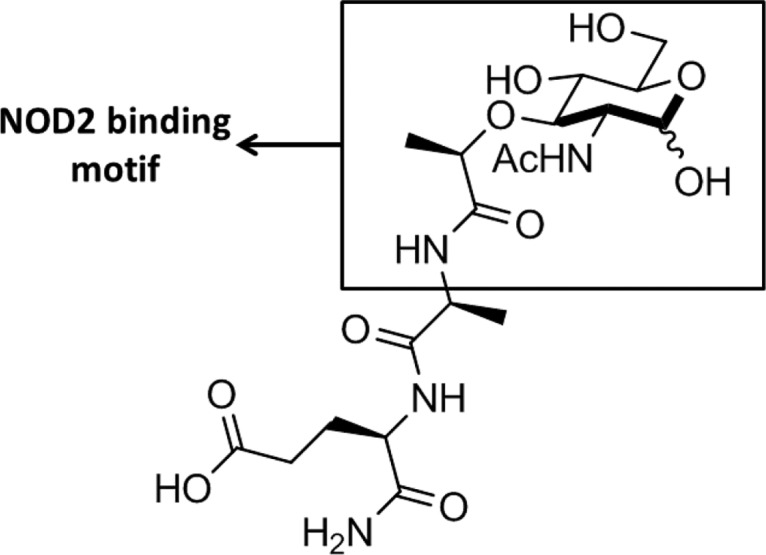
Muramyl motif of the MDP ligand proposed to be highly important for NOD2 binding.
The precise composition of the native-membrane vesicles used in the BSI assay is not defined. While we confirmed that NOD2 is present, the mechanism by which the native-membrane environment augments NOD2-ligand binding specificities is not readily apparent. It is well-known that immune receptor activity and cognate ligand binding can depend greatly on associated interacting species.39,40 We propose that the additional proteins present in the membrane environment that allow the NOD2 receptor to function in these cells may affect NOD2 interactions by (1) alteration of the NOD2 binding site through conformational changes or steric hindrance, (2) chemically modifying the ligands prior to binding, (3) assisting in the trafficking of NOD2 to the membrane, or some combination thereof. Additionally, many proteins have been found to interact with the NOD2 receptor, some of which are capable of either enhancing or inhibiting NF-κB signaling.41 The manner in which protein–protein interactions exert ligand specificities and influence NOD2 signaling is not well-known, and this work further emphasizes the importance of deciphering the roles of NOD2 accessory proteins and immune receptor interacting partners in general. It will be interesting to assess if the membrane environment also affects the substrate specificities of other NLR proteins, such as NOD1.
In addition to determining that the membrane environment is critical for shaping NOD2’s ligand specificity, this work reveals critical features of the bacterial peptidoglycan ligand. Most importantly, we observed that truncated versions of MDP containing the carbohydrate epitope are still able to bind and activate a NOD2-dependent NF-κB response, albeit at higher concentrations than MDP. Therefore, fragments smaller than MDP are capable of activating NOD2 and may be immunologically relevant in the context of Crohn’s disease. NOD2 is proposed to bind its ligands through the LRR domain, and subsequent binding triggers a conformational change and/or an oligomerization event.41−43 Recently, a crystal structure of the NOD-LRR domains of NOD2 was solved; however, this important work did not conclusively identify a ligand binding site.44 In lieu of this, the data presented here suggest that binding and activation occur through the minimal fragment, which contains the carbohydrate and an esterified lactic acid moiety (20). The data show that the electron density and/or charge of the lactic acid component of the carbohydrate influences NOD2 binding and activation (Figures 3 and 4, Table 2), as the unmodified lactic acid component (i.e., N-acetyl-muramic acid) does not bind or activate (Figures S7 and S11). Importantly, when the lactic acid moiety is esterified or extended via the amide of a peptide bond, these ligands (2, 3, 16, 18–20) are able to bind and activate a NOD2-dependent NF-κB response (Figures 3 and 4, Table 2). The very interesting exception is compound 17, which differs from 2 by having an esterified isoglutamate side chain of opposite stereochemical configuration. Either change alone (16 and 3, respectively) made little difference, but both changes (giving 17) eliminated both binding and activation of the Nod2 receptor.
While a subset of ligands that bound to purified WT NOD2 do not activate the NF-κB pathway nor bind WT NOD2 when presented in a native-membrane environment, fragments such as these could still have a NOD2-based biological function. NOD2 is currently reported to have approximately 30 cellular protein interacting partners, and the functional output of these interactions include enhancing and inhibiting NF-κB signaling, promoting MAPK signaling, increasing IL-1β secretion, and inducing autophagy.41 We note that the role of NOD2 in NF-κB signaling is the most studied functional output for the receptor to date, while the role of NOD2, its activation, and its signaling in the many other pathways is not nearly as well understood or defined. We demonstrate that NOD2 is capable of binding a range of peptidoglycan fragments and may therefore be responsible for recognizing a wide variety of peptidoglycan components, leading to a number of different outputs. Determining the potential NOD2 functional implications of both the active and inactive ligands evaluated in this study offers a direction for additional evaluation using phenotypic outputs other than NF-κB activation.
Methods
For full experimental details regarding materials, synthetic protocols, protein expression and purification, SPR/BSI procedures, and NF-κB activation assay, refer to the Supporting Information.
Synthesis of 6-amino Peptidoglycan Derivatives
All chemicals were purchased from Sigma-Aldrich, Alfa Aesar, or Invivogen and used without further purification. Solvents were reagent grade and were further dried when necessary. Analytical thin-layer chromatography was performed on glass plates precoated with silica gel (250 μm, Sorbent Technologies). Flash chromatography was carried out on silica gel (60 Å, 40–63 μm), purchased from Sorbent Technologies. Preparative HPLC was performed on an Agilent Series 1100 using a Phenomenex Luna 5 μm C18 column (250 × 10.00 mm), or using a Maxi-Clean 800 mg C18 column. NMR spectra were recorded on Bruker AV 400 MHz and AV III 600 MHz spectrometers. Mass spectra (ESI) and HRMS were obtained at the Mass Spectroscopy Facility at the Department of Chemistry, University of Delaware, (Shimadzu LCMS 2020 or Thermo Q-Exactive Orbitrap). For detailed synthetic information, see Supporting Information.
Protein Expression and Purification
Protein purity and concentration was assessed by SDS-PAGE (Supporting Information and Figure S1). All purified proteins were kept at 4 °C and was used within 3 days of purification.
Human WT Flag-Tagged NOD2 Protein
Flag-tagged human WT NOD2 expressed as previously described7 with minor adaptations to the purification.
Human GST-Tagged NOD2 Protein
GST-tagged NOD2 was generated by cloning WT NOD2 into the pFastBacM30b baculovirus expression vector (EMBL). NOD2 was amplified from the pBKCMV vector using primers (forward, 5′-GTCAATGGATCCATGGGGGAAGAGGGTGG-3′ and reverse, 5′-CAATGCGGCCGCTCAAAGCAAGAGTCTG-3′) and inserted into the pFastBacM30b vector using restriction sites BamH1 and Not1. Techniques described in the Guide to Baculovirus Expression Vector Systems and Insect Cell Culture Techniques (Invitrogen Life Technologies) were used to generate, isolate, and analyze recombinant vectors and bacmids, as well as to initially transfect insect cells. For overexpression and purification conditions, see the Supporting Information.
Human WT Flag-Tagged NOD1 Protein
NOD1 cDNA was received from the Podolsky lab. FLAG-tagged NOD1 was generated by cloning WT NOD1 into the pFastBac-CFlag baculovirus expression vector[S9] using Nde1 and Not1 restriction sites. The protein was expressed and purified as Nod2; see the Supporting Information for details.
Surface Plasmon Resonance
All SPR experiments were performed on a BIAcore 3000 instrument. Conditions for preparing the chip, mixed-SAM solutions, coupling, and binding assay methods were adapted from prior work with minor changes.7,12 Full details and raw sensograms are presented in the Supporting Information.
Backscattering Interferometry
Detection of Ligand Binding to Purified Protein
The interactions of purified NOD2 with the peptidoglycan derivatives were examined by adding increasing concentrations of each peptidoglycan ligand to a fixed concentration (10 nM) of either purified NOD2 or NOD1 (Figure S1) in PBS buffer (pH 6.5). Each solution was allowed to incubate for 40 min at RT to ensure the achievement of binding equilibrium. Samples were introduced into the BSI instrument as per standard methods, and the resulting changes in phase plotted against ligand concentration. Values observed for NOD1 under otherwise identical conditions were subtracted from the NOD2 values. The resulting curves are shown in Figure S5; fits to a single site binding model are shown. Each experiment was repeated in triplicate, giving the range of results shown in Table 1.
Native Membrane Vesicles
A cell pellet containing approximately 1 × 108 cells of either HEK293T or HEK293T-Nod2Myc/tet-op was resuspended in 10 mL of ice-cold buffer (HBSS, 1 × EDTA-free, broad-spectrum protease inhibitors). The solution was then probe-sonicated to clarity in an ice bath and transferred to a 220 nm Millipore Ultrafree-MC centrifuge tube filter. The resulting solutions were centrifuged for 2 h at 6000g and 4 °C. All solution that passed through the centrifuge tube filter was collected and stored at 4 °C for up to 4 days. Vesicle sizes were determined using a Wyatt Technologies DynaPro dynamic light scattering apparatus and lipid concentration estimated using FM1–43FX membrane stain (Invitrogen Corporation, Carlsbad CA).
Detection of Ligand Binding to Small Unilamellar Vesicles
Ligand binding to small unilamellar vesicles (SUVs) was accomplished by incubating either a fixed amount of SUV suspension or purified protein (NLRs were purified from insect cells as described above), with varying concentrations of ligands for 40 min in the dark at RT. Solutions were deposited in the channels of a microfluidic chip for analysis using a backscattering interferometer (BSI). Microfluidic chips were passivated prior to the experiment using perfluoro-octyldecyl trichlorisilatrane and rinsed extensively between each sample. The microfluidic devices were maintained at 25 °C using a feedback-controlled Peltier system. The high-contrast interference fringes produced by each sample were generated using a fiber-coupled HeNe laser as a light source and recorded on a CCD camera. Measurements were analyzed using a combination of in-house software, Microsoft Excel, and OriginPro.
Data Analysis
Spatial changes in interference fringes are measured in near real-time using high-resolution interference fringes and a fast Fourier transform, FFT. See the Supporting Information for a detailed description.
NF-κB Activation Assay
Reporter Assay with Transfected Nod2 Variants and Regulated Nod2 Cell Lines
The appropriate HEK293T cell line was transfected with Lipofectamine LTX reagent (Invitrogen) and the corresponding DNA (see Supporting Information). Activity was induced by incubating 20–200 μM of compounds with transfected cells. After 12 h of treatment, NF-κB luciferase activity was measured using the Dual-Luciferase Reporter Assay (Promega) and normalized to Renilla luciferase activity.
Acknowledgments
This work was supported by the Delaware COBRE program, with a grant from the National Institute of General Medical Sciences (NIGMS P20GM104316-01A1; C.L.G.), P50 GM103368-03 (M.G.F.), and National Science Foundation (CAREER CHE 1554967; C.L.G.). C.L.G. is a Pew Biomedical Scholar and thanks the Pew Foundation. A.K.S. thanks the National Institutes of Health for support through a CBI training grant: 5T32GM008550. Instrumentation support was provided by the Delaware COBRE and INBRE programs, supported by the National Institute of General Medical Sciences (P30 GM110758-02, P20 GM104316-01A1, and P20 GM103446).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.7b00469.
Experimental procedures and materials, synthetic methods (Schemes S1–S13), cloning details, SPR details and sensorgrams, BSI/data analysis, NF-κB assay conditions, Figures S1–S11, 1H and 13C NMR spectra, and HRMS for reported compounds (PDF)
Author Contributions
∥ J.E.M. and A.K.S. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Janeway C. A. Jr.; Medzhitov R. (2002) Innate immune recognition. Annu. Rev. Immunol. 20, 197–216. 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. (2007) Recognition of microorganisms and activation of the immune response. Nature 449, 819–826. 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- Martinon F.; Tschopp J. (2005) NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 26, 447–454. 10.1016/j.it.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Philpott D. J.; Sorbara M. T.; Robertson S. J.; Croitoru K.; Girardin S. E. (2013) NOD proteins: regulators of inflammation in health and disease. Nat. Rev. Immunol. 14, 9–23. 10.1038/nri3565. [DOI] [PubMed] [Google Scholar]
- Lipinski S.; Grabe N.; Jacobs G.; Billmann-Born S.; Till A.; Hasler R.; Aden K.; Paulsen M.; Arlt A.; Kraemer L.; Hagemann N.; Erdmann K. S.; Schreiber S.; Rosenstiel P. (2012) RNAi screening identifies mediators of NOD2 signaling: Implications for spatial specificity of MDP recognition. Proc. Natl. Acad. Sci. U. S. A. 109, 21426–21431. 10.1073/pnas.1209673109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnich N.; Aguirre J. E.; Reinecker H. C.; Xavier R.; Podolsky D. K. (2005) Membrane recruitment of NOD2 in intestinal epithelial cells is essential for nuclear factor-kB activation in muramyl dipeptide recognition. J. Cell Biol. 170, 21–26. 10.1083/jcb.200502153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott D. J.; Girardin S. E. (2009) Crohn’s disease-associated Nod2 with mutants reduce IL10 transcription. Nat. Immunol. 10, 455. 10.1038/ni0509-455. [DOI] [PubMed] [Google Scholar]
- Ogura Y.; Bonen D. K.; Inohara N.; Nicolae D. L.; Chen F. F.; Ramos R.; Britton H.; Moran T.; Karaliuskas R.; Duerr R. H.; Achkar J.-P.; Brant S. R.; Bayless T. M.; Kirschner B. S.; Hanauer S. B.; Nunez G.; Cho J. H. (2001) A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411, 603–606. 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- Hugot J.-P.; Chamaillard M.; Zouali H.; Lesage S.; Cezard J.-P.; Belaiche J.; Almer S.; Tysk C.; O’Morain C. A.; Gassull M.; Binder V.; Finkel Y.; Cortot A.; Modigliani R.; Laurent-Puig P.; Gower-Rousseau C.; Macry J.; Colombel J.-F.; Sahbatou M.; Thomas G. (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411, 599–603. 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Ting J. P.; Duncan J. A.; Lei Y. (2010) How the Noninflammasome NLRs Function in the Innate Immune System. Science 327, 286–290. 10.1126/science.1184004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindt T., Goldsby R., and Osborne B. (2007) Kuby Immunology, 6th ed., W.H. Freeman and Company, New York. [Google Scholar]
- Ellouz F.; Adam A.; Ciorbaru R.; Lederer E. (1974) Minimal Structural Requirements for Adjuvant Activity of Bacterial Peptidoglycan Derivatives. Biochem. Biophys. Res. Commun. 59, 1317. 10.1016/0006-291X(74)90458-6. [DOI] [PubMed] [Google Scholar]
- Kobayashi K. S.; Chamaillard M.; Ogura Y.; Octavian H.; Inohara N.; Nunez G.; Flavell R. A. (2005) Nod2-Dependent Regulation of Innate and Adaptive Immunity in the Intestinal Tract. Science 307, 731. 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- Wolfert M. A.; Roychowdhury A.; Boons G. J. (2007) Modification of the structure of peptidoglycan is a strategy to avoid detection by nucleotide-binding oligomerization domain protein 1. Infect. Immun. 75, 706–713. 10.1128/IAI.01597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes C. L.; Ariyananda L. D. Z.; Melnyk J. E.; O’Shea E. K. (2012) The innate immune protein Nod2 binds directly to MDP, a bacterial cell wall fragment. J. Am. Chem. Soc. 134, 13535–13537. 10.1021/ja303883c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo J.; Boyle J. P.; Howard C. B.; Monie T. P.; Davis B. K.; Duncan J. A. (2012) Pathogen sensing by nucleotide-binding oligomerization domain-containing protein 2 (NOD2) is mediated by direct binding to muramyl dipeptide and ATP. J. Biol. Chem. 287, 23057–23067. 10.1074/jbc.M112.344283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N.; Ogura Y.; Fontalba A.; Gutierrez O.; Pons F.; Crespo J.; Fukase K.; Inamura S.; Kusumoto S.; Hashimoto M.; Foster S. J.; Moran A. P.; Fernandez-Luna J. L.; Nunez G. (2003) Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J. Biol. Chem. 278, 5509–5512. 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- Murray P. J. (2005) NOD proteins: an intracellular pathogen-recognition system or signal transduction modifiers?. Curr. Opin. Immunol. 17, 352–358. 10.1016/j.coi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Meylan E.; Tschopp J.; Karin M. (2006) Intracellular pattern recognition receptors in the host response. Nature 442, 39–44. 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- Vavricka S. R.; Musch M. W.; Chang J. E.; Nakagawa Y.; Phanvijhitsiri K.; Waypa T. S.; Merlin D.; Schneewind O.; Chang E. B. (2004) hPepT1 transports muramyl dipeptide, activating NF-κB and stimulating IL-8 secretion in human colonic Caco2/bbe cells. Gastroenterology 127, 1401–1409. 10.1053/j.gastro.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Nakamura N.; Lill J. R.; Phung Q.; Jiang Z.; Bakalarski C.; de Maziere A.; Klumperman J.; Schlatter M.; Delamarre L.; Mellman I. (2014) Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature 509, 240–244. 10.1038/nature13133. [DOI] [PubMed] [Google Scholar]
- Marina-Garcia N.; Franchi L.; Kim Y. G.; Hu Y.; Smith D. E.; Boons G. J.; Nunez G. (2009) Clathrin- and dynamin-dependent endocytic pathway regulates muramyl dipeptide internalization and NOD2 activation. J. Immunol. 182, 4321–4327. 10.4049/jimmunol.0802197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charriere G. M.; Ip W. E.; Dejardin S.; Boyer L.; Sokolovska A.; Cappillino M. P.; Cherayil B. J.; Podolsky D. K.; Kobayashi K. S.; Silverman N.; Lacy-Hulbert A.; Stuart L. M. (2010) Identification of Drosophila Yin and PEPT2 as evolutionarily conserved phagosome-associated muramyl dipeptide transporters. J. Biol. Chem. 285, 20147–20154. 10.1074/jbc.M110.115584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes C. L.; Podolsky D. K.; O’Shea E. K. (2010) Synthesis of biologically active biotinylated muramyl dipeptides. Bioorg. Med. Chem. Lett. 20, 6061–6063. 10.1016/j.bmcl.2010.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baksh M. M.; Kussrow A. K.; Mileni M.; Finn M. G.; Bornhop D. J. (2011) Label-free quantification of membrane-ligand interactions using backscattering interferometry. Nat. Biotechnol. 29, 357–360. 10.1038/nbt.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornhop D. J.; Latham J. C.; Kussrow A. K.; Markov D. A.; Jones R. D.; Sorensen H. S. (2007) Free-Solution, Label-Free Molecular Interactions Studied by Back-Scattering Interferometry. Science 317, 1732–1736. 10.1126/science.1146559. [DOI] [PubMed] [Google Scholar]
- Lahiri J.; Isaacs L.; Tien J.; Whitesides G. M. (1999) A Strategy for the Generation of Surfaces Presenting Ligands for Studies of Binding Based on an Active Ester as a Common Reactive Intermediate: A Surface Plasmon Resonance Study. Anal. Chem. 71, 777–790. 10.1021/ac980959t. [DOI] [PubMed] [Google Scholar]
- Yeager A. R.; Finney N. S. (2005) Synthesis of Fluorescently Labeled UDP-GlcNac Analogues and Their Evaluation as Chitin Synthase Substrates. J. Org. Chem. 70, 1269–1275. 10.1021/jo0483670. [DOI] [PubMed] [Google Scholar]
- Kammer M. N.; Olmsted I. R.; Kussrow A. K.; Morris M. J.; Jackson G. W.; Bornhop D. J. (2014) Characterizing aptamer small molecule interactions with backscattering interferometry. Analyst 139, 5879–5884. 10.1039/C4AN01227E. [DOI] [PubMed] [Google Scholar]
- Morcos E. F.; Kussrow A.; Enders C.; Bornhop D. (2010) Free-solution interaction assay of carbonic anhydrase to its inhibitors using back-scattering interferometry. Electrophoresis 31, 3691–3695. 10.1002/elps.201000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussrow A.; Kaltgrad E.; Wolfenden M. L.; Cloninger M. J.; Finn M. G.; Bornhop D. J. (2009) Measurement of Monovalent and Polyvalent Carbohydrate-Lectin Binding by Back-Scattering Interferometry. Anal. Chem. 81, 4889–4897. 10.1021/ac900569c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanan V.; Grimes C. L. (2014) The molecular chaperone HSP70 binds to and stabilizes NOD2, an important protein involved in Crohn disease. J. Biol. Chem. 289, 18987–18998. 10.1074/jbc.M114.557686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y.; Inohara N.; Benito A.; Chen F. F.; Yamaoka S.; Nunez G. (2001) Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J. Biol. Chem. 276, 4812–4818. 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- Melnyk J. E.; Mohanan V.; Schaefer A. K.; Hou C.-W.; Grimes C. L. (2015) Peptidoglycan Modifications Tune the Stability and Function of the Innate Immune Receptor Nod2. J. Am. Chem. Soc. 137, 6987–6990. 10.1021/jacs.5b01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf A. J.; Reyes C. N.; Liang W.; Becker C.; Shimada K.; Wheeler M. L.; Cho H. C.; Popescu N. I.; Coggeshall K. M.; Arditi M.; Underhill D. M. (2016) Hexokinase Is an Innate Immune Receptor for the Detection of Bacterial Peptidoglycan. Cell 166, 624–636. 10.1016/j.cell.2016.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino S. J.; Magalhaes J. G.; Philpott D.; Bahr G. M.; Blanot D.; Girardin S. E. (2013) Identification of a synthetic muramyl peptide derivative with enhanced Nod2 stimulatory capacity. Innate Immun. 19, 493–503. 10.1177/1753425912471691. [DOI] [PubMed] [Google Scholar]
- Hasegawa M.; Yang K.; Hashimoto M.; Park J. H.; Kim Y. G.; Fujimoto Y.; Nunez G.; Fukase K.; Inohara N. (2006) Differential release and distribution of Nod1 and Nod2 immunostimulatory molecules among bacterial species and environments. J. Biol. Chem. 281, 29054–29063. 10.1074/jbc.M602638200. [DOI] [PubMed] [Google Scholar]
- Lecine P.; Esmiol S.; Metais J. Y.; Nicoletti C.; Nourry C.; McDonald C.; Nunez G.; Hugot J. P.; Borg J. P.; Ollendorff V. (2007) The NOD2-RICK complex signals from the plasma membrane. J. Biol. Chem. 282, 15197–15207. 10.1074/jbc.M606242200. [DOI] [PubMed] [Google Scholar]
- Zheng Y.; Humphry M.; Maguire J. J.; Bennett M. R.; Clarke M. C. H. (2013) Intracellular Interleukin-1 Receptor 2 Binding Prevents Cleavage and Activity of Interleukin-1a, Controlling Necrosis-Induced Sterile Inflammation. Immunity 38, 285–295. 10.1016/j.immuni.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Kim S. Y.; Pribis J. P.; Lotze M.; Mollen K. P.; Shapiro R.; Loughran P.; Scott M. J.; Billiar T. R. (2013) Signaling of High Mobility Group Box 1 (HMGB1) through Toll-like Receptor 4 in Macrophages Requires CD14. Mol. Med. 19, 88–98. 10.2119/molmed.2012.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle J. P.; Parkhouse R.; Monie T. P. (2014) Insights into the molecular basis of the NOD2 signalling pathway. Open Biol. 4, 140178. 10.1098/rsob.140178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.; Shaw M. H.; Kim Y. G.; Nunez G. (2009) NOD-like receptors: role in innate immunity and inflammatory disease. Annu. Rev. Pathol.: Mech. Dis. 4, 365–398. 10.1146/annurev.pathol.4.110807.092239. [DOI] [PubMed] [Google Scholar]
- Inohara C.; McDonald C.; Nunez G.; Chamaillard M. (2005) NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu. Rev. Biochem. 74, 355–383. 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- Maekawa S.; Ohto U.; Shibata T.; Miyake K.; Shimizu T. (2016) Crystal structure of NOD2 and its implications in human disease. Nat. Commun. 7, 11813. 10.1038/ncomms11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.