Abstract
Peptidoglycan recognition proteins (PGRPs) form a family of immune regulators that is conserved from insects to mammals. In the malaria vector mosquito Anophelescoluzzii, the peptidoglycan receptor PGRPLC activates the immune-deficiency (Imd) pathway limiting both the microbiota load and Plasmodium infection. Here, we carried out an RNA interference screen to examine the role of all 7 Anopheles PGRPs in infections with Plasmodium berghei and P. falciparum. We show that, in addition to PGRPLC, PGRPLA and PGRPS2/PGRPS3 also participate in antiparasitic defenses, and that PGRPLB promotes mosquito permissiveness to P. falciparum. We also demonstrate that following a mosquito blood feeding, which promotes growth of the gut microbiota, PGRPLA and PGRPLB positively and negatively regulate the activation of the Imd pathway, respectively. Our data demonstrate that PGRPs are important regulators of the mosquito epithelial immunity and vector competence.
Keywords: Peptidoglycan recognition protein, Anopheles, Drosophila, Immune-deficiency pathway, Antimicrobial peptides, Plasmodium, Microbiota, Bacteria
Introduction
In sub-Saharan Africa, malaria parasites are mainly transmitted by 2 sibling species of mosquitoes, Anopheles gambiae and Anopheles coluzzii. Both species mostly bite humans and are permissive to Plasmodium, two characteristics that make them very efficient malaria vectors. A better knowledge on the interactions between parasites and mosquitoes is needed to identify targets for transmission-blocking strategies. After ingestion during blood feeding, Plasmodium parasites undergo sexual reproduction inside the midgut lumen of a female Anopheles mosquito and differentiate into motile ookinetes. Upon traversing the midgut epithelium, the ookinetes transform into oocysts, where parasites multiply and differentiate into sporozoites over a period of 1–2 weeks. Oocysts rupture and release thousands of sporozoites that invade the salivary glands and are transmitted to humans during subsequent bites. The ookinete-to-oocyst transition is the main bottleneck of the Plasmodium passage through the mosquito, largely due to mosquito immune reactions that drastically reduce the parasite numbers. These reactions include the NF-κB immune-deficiency (Imd) pathway that kills parasites in the midgut lumen and epithelium through an as yet uncharacterized mechanism [1, 2] and a complement-like pathway that eliminates ookinetes soon after crossing the epithelium and before transforming into oocysts [3].
In Drosophila, the Imd pathway is elicited by bacterial peptidoglycan via the peptidoglycan recognition protein (PGRP) LC [4, 5]. This pathway is highly regulated in a tissue-dependent fashion. In the gut, it is regulated both positively by the nonpeptidoglycan-binding protein PGRPLA and negatively notably by the amidases PGRPLB and PGRPSCs that cleave peptidoglycan into nonimmunogenic fragments [6, 7, 8]. PGRPLC and PGRPLA encode transmembrane proteins while PGRPLB and PGRPSCs are secreted [7, 9, 10]. In Anopheles, the Imd pathway is induced by the microbiota growth that follows blood feeding in a PGRPLC-dependent fashion [1]. Knocking down PGRPLC by RNA interference (RNAi) reduces antimicrobial gene induction and increases mosquito permissiveness to Plasmodium [1]. Anopheles PGRPLC and PGRPLA, but also PGRPLB, are all predicted transmembrane proteins.
The Anopheles genome encompasses 6 additional PGRP genes [11], encoding the predicted amidases PGRPLB, PGRPS2, and PGRPS3, and the noncatalytic PGRPLA, PGRPS1, and PGRPLD. The PGRPLA gene is reported to encode 2 isoforms referred to as PGRPLA1 and PGRPLA2 [11], which share an intracellular N terminus but differ in their C terminus that encompasses the PGRP domain [11]. Here, we investigated the role of PGRPs in regulating malaria infection in A.coluzzii mosquitoes (formerly known as A. gambiae M form). We identify PGRPLA and PGRPS2/S3 as new Plasmodium antagonists and reveal that PGRPLB enhances mosquito tolerance to Plasmodium infection. We also show that PGRPLA1 and PGRPLB positively and negatively regulate the Anopheles Imd pathway, respectively, similarly to their Drosophila orthologs.
Materials and Methods
Ethics Statement
Experiments performed in the UK were carried out in accordance with the UK Animals (Scientific Procedures) Act 1986. Mosquito infections with Plasmodium berghei by blood feeding on parasitized mice were approved by the Imperial College Ethical Review Committee and carried out under the UK Home Office License PPL70/6347. All the procedures were of mild-to-moderate severity, and protocols were designed to minimize the numbers of animals used. Opportunities for reduction, refinement, and replacement of animal procedures were constantly reviewed and considered.
Experiments performed in Cameroon, including volunteer recruitment and mosquito maintenance, were carried out following protocols approved by the WHO and the Cameroon National Ethics Committee (agreement 039/CNE/MP/06). Screening of human volunteers was carried out on a voluntary basis after all participants and families were given extensive information on the program. Written informed consent was taken from each volunteer and/or their legal guardian prior to enrolment.
Mosquito Rearing
The Ngousso colony of A. coluzzii (formerly referred to as A. gambiae M form) was reared at 27°C and 80% humidity in a 12-h/12-h dark/light cycle. Plasmodium falciparum experiments in Cameroon were performed on a colony maintained on rabbit blood and 10% glucose solution. All other experiments were performed on a colony maintained on mouse and human blood and 10% fructose solution.
Double-Stranded RNA Preparation and Gene Silencing
PCR amplification of 120- to 600-bp-long regions of the PGRP genes was performed using T7-tagged primers (online suppl. Table S1; for all online suppl. material, see www.karger.com/doi/10.1159/000452797) and Ngousso mosquito cDNA or the control bacterial gene encoding β-galactosidase (LacZ) as templates. After purification with the QIAquick PCR Purification Kit (Qiagen), PCR products were used as templates for double-stranded RNA (dsRNA) synthesis using the T7 MEGAscript kit (Ambion). Remaining DNA was degraded with 1 μl TURBO DNase (Ambion) for 15 min at 37°C. dsRNAs were purified using the RNeasy Mini Kit (Qiagen). One- to 2-day-old mosquitoes were injected with 69 nL of 3 mg·mL−1 (Fig. 1, 2, 3, online suppl. Fig. S1, S2, S3) or 6 mg·mL−1 (Fig. 5, online suppl. Fig. S4) dsRNA solution in the thorax using a Nano-injector (Drummond). For PGRPS2/S3 RNAi, dsRNA targeting each gene was designed separately, and both dsRNA solutions were mixed prior to injection. For regular Plasmodium infections, an infectious blood meal was offered to mosquitoes 4 days after injection. For experiments involving antibiotic treatment, dsRNAs were injected 24 h after the start of antibiotic feeding, and Plasmodium infections were performed 48 h after injection. For experiments involving Enterobacter cloacae oral infections, dsRNAs were injected into freshly emerged mosquitoes 48 h before bacterial oral infections, and Plasmodium infections were carried out 48 h after bacterial infections. Quantification of the efficiency of PGRPLA dsRNA targeting was performed on midguts 24 h after injection and 24 h after a blood feed offered to the mosquitoes 30 h after injection.
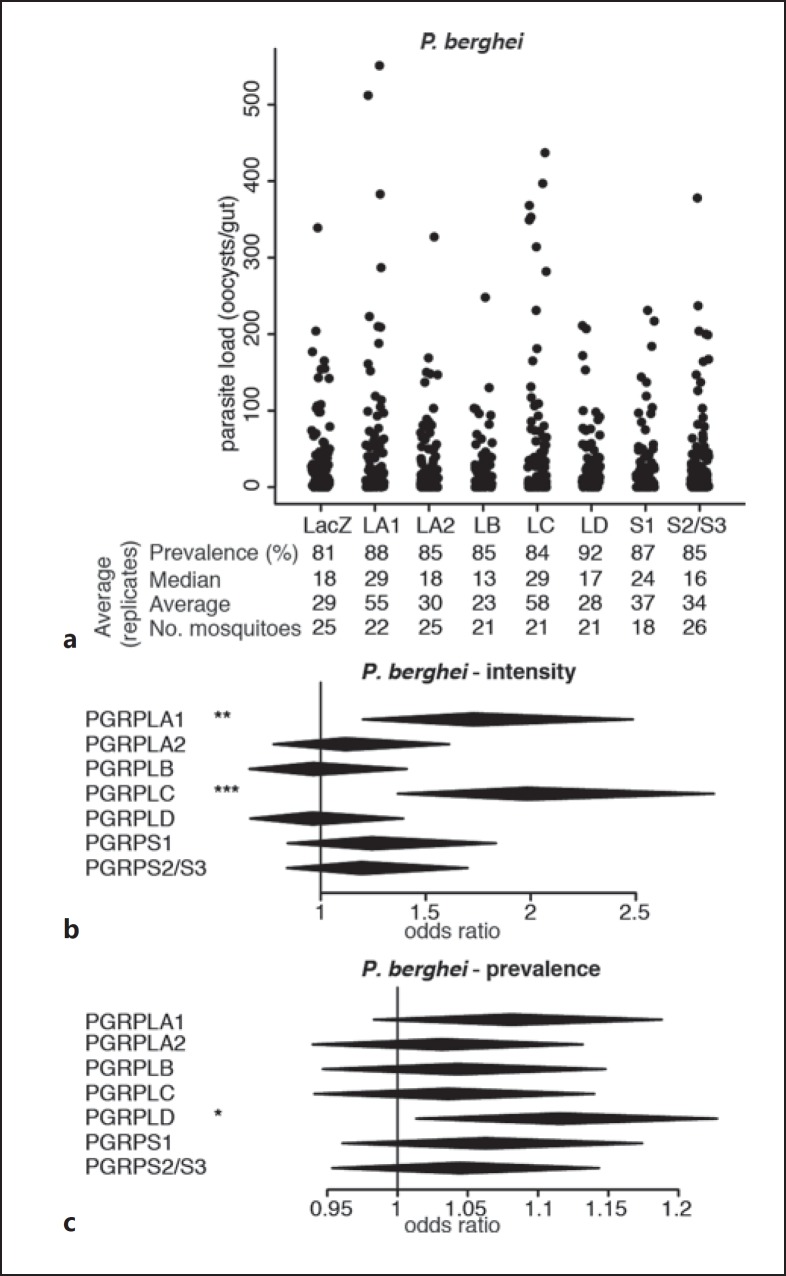
Fig. 1.
Effect of RNAi on PGRP genes on P. berghei infections. a Strip-chart representation of the oocyst load of P. berghei upon RNAi on the genes encoding PGRPs. RNAi targeting the bacterial gene LacZ was used as a control. Each dot shows the oocyst load in an individual gut, and data show pools of 4 independent replicates. b, c Forest plots showing the effect of knocking down Anopheles PGRP genes on the intensity (b) and prevalence (c) of P. berghei infection, resulting from the same experiments as data shown in a. Statistical data show the result of a Wald Z test following zero-inflated negative binomial regression model fitting. The center and horizontal diagonal of each diamond show the odds ratio of the fixed effect and its confidence interval, respectively. * p < 0.05; ** p < 0.01; *** p < 0.001.
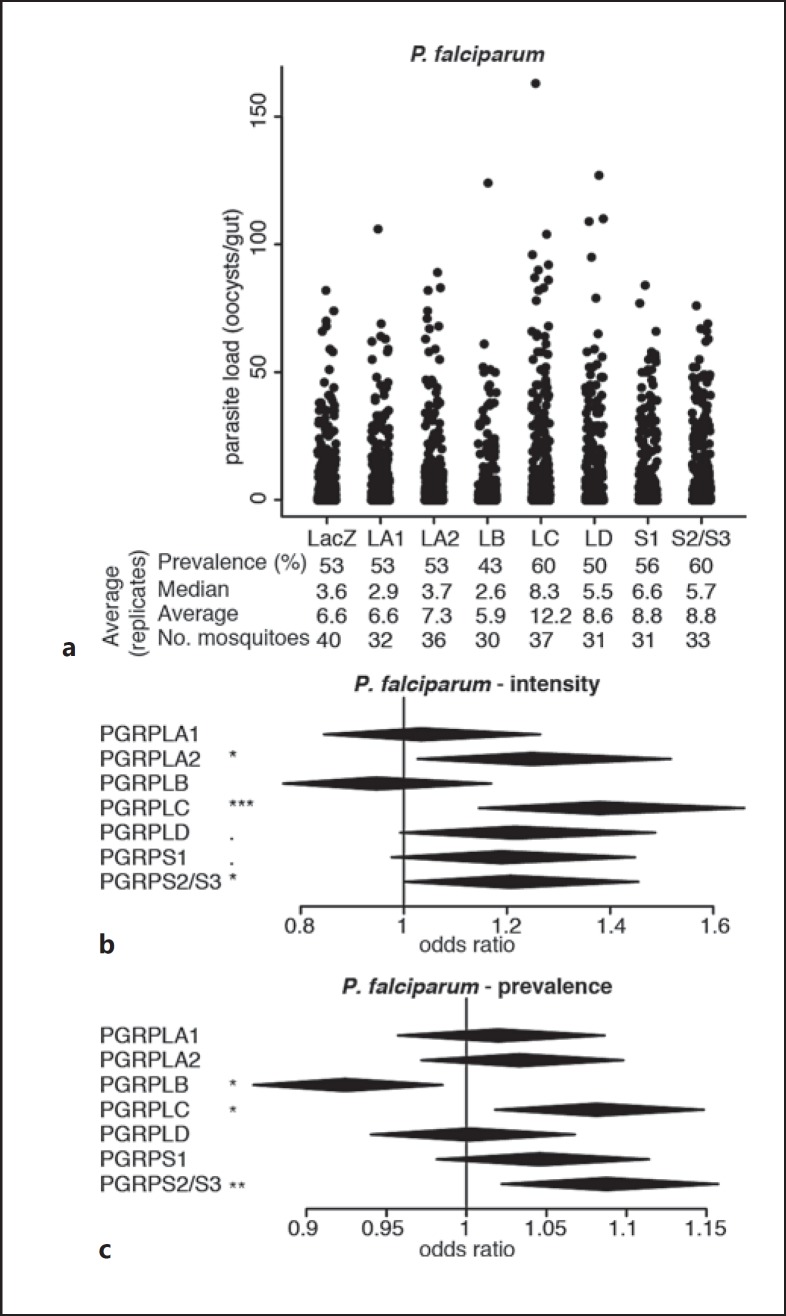
Fig. 2.
Effect of RNAi on PGRP genes on P. falciparum infections. a Strip-chart representation of the oocyst load of P. falciparum upon RNAi on the genes encoding PGRPs. RNAi targeting the bacterial gene LacZ was used as a control. Each dot shows the oocyst load in an individual gut, and data show pools of 11 independent replicates. b, c Forest plots showing the effect of knocking down Anopheles PGRP genes on the intensity (b) and prevalence (c) of P. falciparum infection, resulting from the same experiments as data shown in a. Statistical data show the result of a Wald Z test following zero-inflated negative binomial regression model fitting (b) and of an ANOVA following a logistic regression model fitting (c). The center and horizontal diagonal of each diamond show the odds ratio of the fixed effect and its confidence interval, respectively. ·p < 0.1; * p < 0.05; ** p < 0.01; *** p < 0.001.
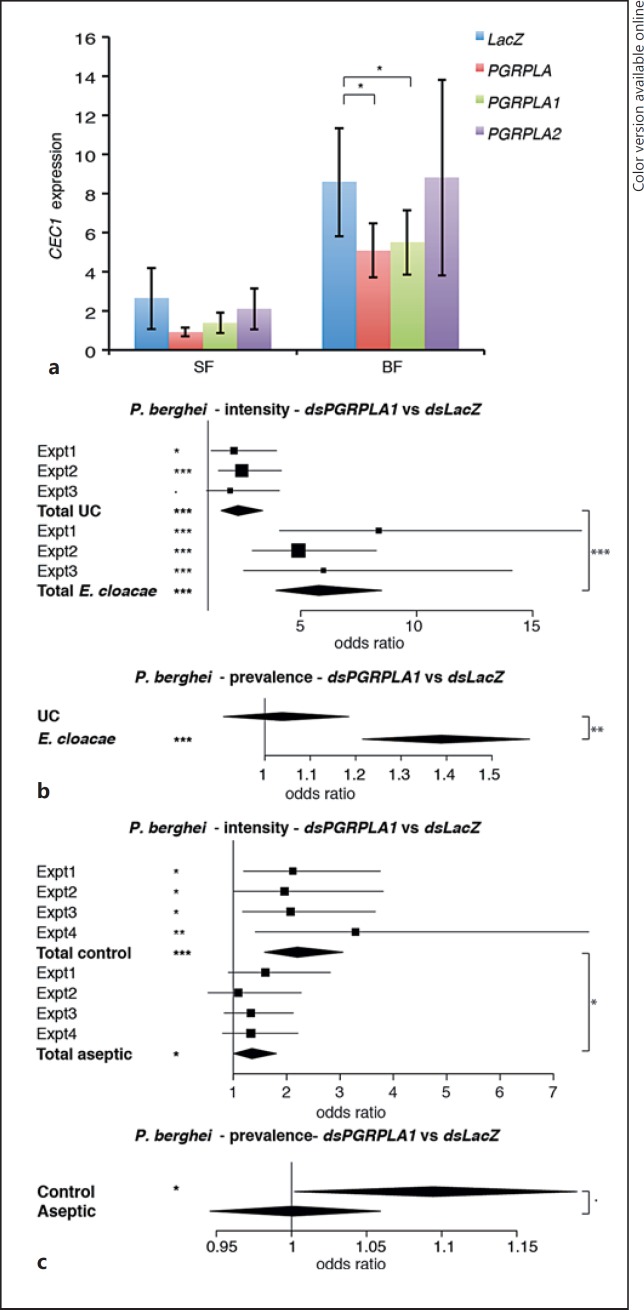
Fig. 3.
Effect of PGRPLA RNAi on Imd pathway induction and on P. berghei infection. aCEC1 expression in midguts of sugar-fed mosquitoes (SF) or 24 h after blood feeding (BF) when targeting the whole PGRPLA gene or only one of its isoforms (PGRPLA1 or PGRPLA2). RNAi targeting the bacterial gene LacZ was used as a control. b Effect of PGRPLA1 RNAi on P. berghei live oocyst intensity (upper panel) and prevalence (lower panel) in unchallenged (UC) or E. cloacae orally infected mosquitoes. c Effect of PGRPLA1 RNAi on P. berghei live oocyst intensity (upper panel) and prevalence (lower panel) in control or gentamycin-treated (aseptic) mosquitoes. Squares represent the odds ratio of the fixed effect for each gene in each independent replicate, and error bars represent 95% confidence intervals. In each chart, square sizes are proportional to mosquito numbers. Diamonds represent the summary of independent replicates. Statistical data show the result of ANOVA following a common linear regression model fitting (a), a Wald Z test following zero-inflated negative binomial regression model fitting (b, c; intensity data), and ANOVA following a logistic regression model fitting (b, c; prevalence data). ·p < 0.1; * p < 0.05; ** p < 0.01; *** p < 0.001.
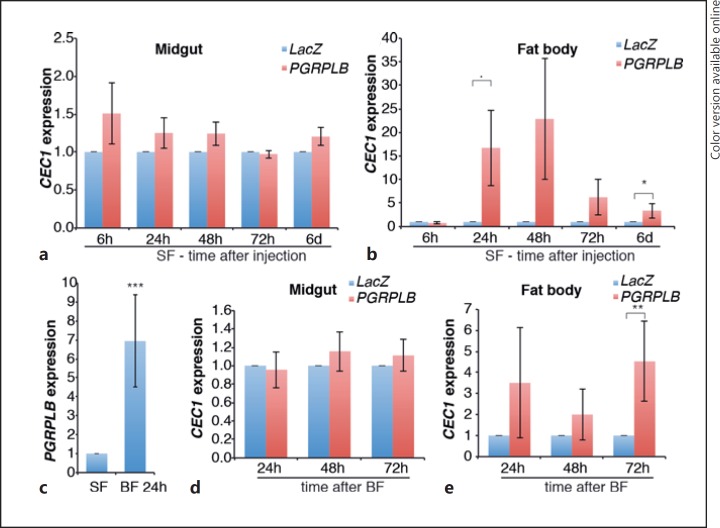
Fig. 5.
Effect of PGRPLB RNAi on the local and systemic activation of the Imd pathway. a, b, d, eCEC1 expression in the midguts (a, d) and fat bodies (b, e) of mosquitoes after injection of PGRPLB or LacZ control dsRNA in sugar-fed (SF; a, b) or blood-fed (BF; d, e) conditions. cPGRPLB expression in sugar-fed mosquitoes or 24 h after blood feeding. Statistical data show the results of ANOVA following a common linear regression model fitting. ·p < 0.1; * p < 0.05; ** p < 0.01; *** p < 0.001.
Antibiotic Treatment
Freshly emerged adult mosquitoes were fed on sterile water supplemented with 25 μg·mL−1 gentamycin (Sigma), offered on a cotton pad that was changed every 12 h for 3 days [12]. Gentamycin was diluted in water rather than in sucrose to prevent its retention in the crop, the mosquito sugar storage organ. Sugar cubes were provided as an alternative source of sugar. Treatment efficacy was checked by plating homogenized mosquitoes on LB-agar plates. No colony grew from the gentamycin-treated mosquitoes while many colonies were observed in the untreated control sample.
E. cloacae Oral Infections
E. cloacae cultures were grown overnight at 37°C in LB medium, washed twice in sterile PBS, and resuspended in sterile sucrose. Bacterium-containing sucrose solutions were offered to recently emerged mosquitoes on cotton pads for 48 h.
P. berghei Infections
GFP-expressing P. berghei parasites [13] were injected intramuscularly into Balb/C or CD1 mice. Two days later, parasitemia was assessed by Giemsa staining (typically 4–6%), and the presence of gametocytes was verified. Mosquitoes were allowed to feed for 10–15 min on anesthetized infected mice and maintained at 21°C. Unfed mosquitoes were discarded 48 h after the blood meal. Midguts were dissected 7–8 days after the blood meal, fixed in 4% paraformaldehyde for 45 min, washed 3 times in PBS for 20 min, and mounted on microscope slides using Vectashield medium (Vector Laboratories). Fluorescent parasites were counted using a fluorescence microscope. Experiments were included in the analysis if at least 1 mosquito was alive in each RNAi condition at the time of dissection (n = 9–36 per condition, average 22).
P. falciparum Infections
Infections with P. falciparum field isolates were carried out during the rainy seasons from May to June in 2007 and 2008. School children volunteers were enrolled at the village of Mfou, near Yaoundé, Cameroon. Gametocyte carriers were identified by Giemsa staining of thick blood smears. Venous blood was drawn from antecubital fossa by venipuncture, and the serum was replaced by malaria-naïve AB serum. Blood was maintained at 37°C in a membrane-feeding device covered with Parafilm M (Bemis) as a skin substitute, and mosquitoes were allowed to feed on infected blood for 30 min. Blood-fed mosquitoes were separated from unfed mosquitoes 5–6 h later and subsequently kept at 27 ± 2°C. Mosquitoes were dissected 8 days later and their midguts were stained with a solution of 0.4% Mercurochrome. Red-stained oocysts were counted by light microscopy. Experiments were included in the analysis if at least 15 mosquitoes were alive in each RNAi condition at the time of dissection (n = 17–60 per condition, average 34).
Gene Expression
Gene expression was examined in the midgut, the region of the gut located between the proventriculus (included) and the malpighian tubules (excluded), and the fat body, the external part of the abdomen including the cuticle and the underlying tissue. Tissues were homogenized in TRIzol (Invitrogen) using a Precellys 24 homogenizer (Bertin) with 0.5-mm-wide glass beads (Bertin). RNA was extracted in chloroform and precipitated with isopropanol, washed with 70% ethanol, and resuspended in molecular-biology grade water. cDNA was synthesized using PrimeScript Reverse Transcriptase (Takara) and 700 ng RNA as a template. qPCR was carried out on an ABI 7700 real-time PCR machine (Applied Biosystems), using SYBR Premix Ex-Taq-Tli RNase H Plus (Takara). Primer sequences are indicated in online supplementary Table S2. The ribosomal transcript S7 was used as normalization control.
Statistical Analyses
Statistical analyses based on generalized linear mixed models (GLMM) were performed in R (version 3.2.3). Prevalence data were examined by ANOVA χ2 test on a logistic regression (glmer). Intensity data are based on oocyst counts of all mosquitoes, including noninfected ones; they were analyzed by Wald Z test on a zero-inflated negative binomial regression (glmmADMB). qPCR data were analyzed by ANOVA χ2 test on a common linear regression (lmer). GLMM analyses extract from the data the contribution of a fixed component (the covariates in question, here RNAi challenge) and a random component (here experiments). The odds ratios shown in the figures were calculated as the exponential of the fixed effect estimates. Sample size was determined according to standard protocols (oocyst counts and gene expression). Mosquitoes were excluded if they died during the experiment or did not blood feed (oocyst counts and expression data on blood-fed samples).
Accession Numbers
The accession numbers of the genes and transcript mentioned in this paper are reported in Table 1.
Table 1.
VectorBase accession numbers of the genes and transcripts mentioned in the paper
| Gene or transcript | Accession No. |
|---|---|
| PGRPLA | AGAP005205 |
| PGRPLA1 | AGAP005205-RC = RF |
| PGRPLA2 | AGAP005205-RA and RD |
| PGRPLB | AGAP001212 |
| PGRPLC | AGAP005203 |
| PGRPLD | AGAP005552 |
| PGRPS1 | AGAP000536 |
| PGRPS2 | AGAP006343 |
| PGRPS3 | AGAP006342 |
| S7 | AGAP010592 |
| CEC1 | AGAP000693 |
Results
PGRPs Influence Mosquito Permissiveness to Plasmodium
We investigated the roles of PGRPs in mosquito permissiveness to the rodent parasite P.berghei through an RNAi screen. We injected mosquitoes with dsRNA targeting each PGRP gene and challenged them with P. berghei 4 days later via blood feeding on infected mice. One week later, we monitored infection by counting the number of oocysts in each individual midgut and assessing the proportion of infected mosquitoes, referred to as prevalence, and the number of oocysts per midgut, referred to as intensity. We confirmed the previously reported increase in infection intensity upon PGRPLC knockdown [1] and observed that PGRPLA1 silencing had a similar effect on P. berghei infection (Fig. 1a, b; online suppl. Table S1). The prevalence of infection was not affected when silencing PGRPLC or PGRPLA1 but increased when targeting PGRPLD (Fig. 1a, c).
Next, we screened for the effects of PGRPs on human malaria parasites P. falciparum. We chose to infect mosquitoes with the blood of gametocyte carriers in Cameroon as this experimental setup allows us to assess infections from a naturally diverse set of parasites having developed in vivo. We injected mosquitoes with dsRNA and offered them an infectious blood sample 4 days later. In addition to the strong negative impact of PGRPLC on infection intensity reported previously [1], we observed that silencing PGRPLA2, but not PGRPLA1, also increased mosquito permissiveness to P. falciparum (Fig. 2a, b; online suppl. Table S2). Silencing PGRPS2 and PGRPS3, which we could only target together due to their high sequence similarity, increased P. falciparum infection intensity and prevalence, while silencing PGRPLB significantly decreased infection prevalence (Fig. 2; online suppl. Table S2).
Together, these data indicate that beside PGRPLC, PGRPLA, PGRPLD, and PGRPS2/PGRPS3 also participate in the resistance to Plasmodium, while PGRPLB may be implicated in the tolerance to parasites. The antiparasitic function of PGRPLC is linked to the induction of the Imd pathway due to the microbial growth following a blood meal [1]. We then focused on PGRPLA and PGRPLB as their Drosophila orthologs are known to positively and negatively regulate the Imd pathway, respectively [7, 8]. We investigated whether their effect on parasite infections was mediated by similar regulatory roles on the Anopheles Imd pathway.
PGRPLA Is a Positive Regulator of the Imd Pathway in the Midgut
First, we investigated the role of PGRPLA in the induction of antimicrobial peptides in the midgut, where its Drosophila ortholog positively affects immune responses. We followed the expression of Cecropin1 (CEC1), an antibacterial gene induced by the Imd pathway, in the midguts of sugar- and blood-fed mosquitoes 3 and 4 days after dsRNA injection, respectively, in order to follow the blood meal-induced Imd response [1]. We found that CEC1 was induced 4.9-fold in the midgut of blood-fed versus sugar-fed mosquitoes. Simultaneous knockdown of both PGRPLA1 and PGRPLA2 reduced the blood meal-induced CEC1 expression (Fig. 3a). The same effect was observed upon specific PGRPLA1 knockdown, while knockdown of PGRPLA2 had no effect on CEC1 expression (Fig. 3a). These results suggested that PGRPLA1 is a positive regulator of the Imd pathway. When testing the efficiency of knockdown of PGRPLA1 and PGRPLA2, we found that PGRPLA1 was reduced by 44 and 37% in sugar- and blood-fed mosquitoes, respectively, while PGRPLA2 was only reduced by 18 and 24%, respectively. In terms of isoform specificity, PGRPLA2 dsRNA did not consistently affect PGRPLA1, while PGRPLA1 dsRNA slightly decreased PGRPLA2 expression by 7 and 15% prior to and after blood feeding, respectively. Thus, we cannot rule out that the specific effect found on PGRPLA1 and not PGRPLA2 is not due to the difference in efficiency and specificity of these dsRNAs.
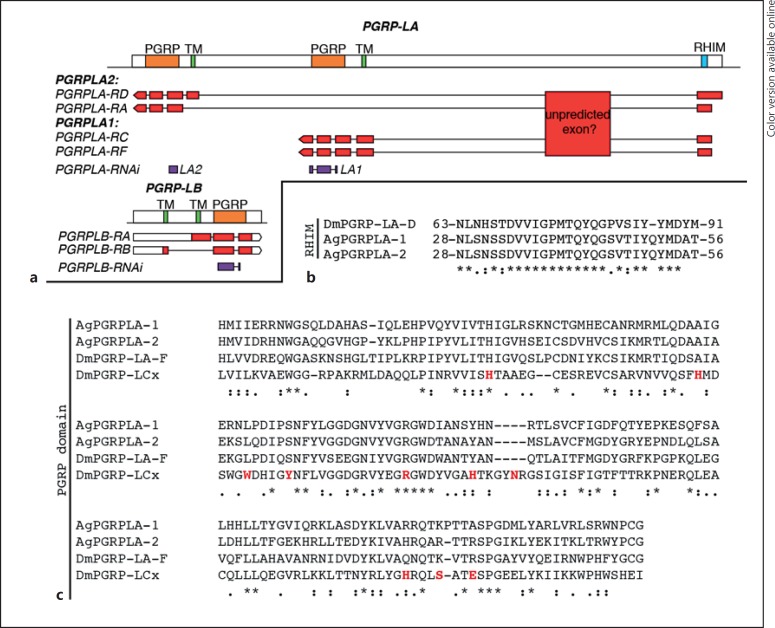
VectorBase annotation of PGRPLA suggests that this gene is transcribed as 4 transcripts, 2 of them sharing the exact same sequence encoding for PGRPLA1, while PGRPLA2 would actually correspond to 2 isoforms sharing the same PGRP domain, 1 of which lacks the transmembrane and intracellular domains (Fig. 4a). According to RNA-sequencing data added to VectorBase, a whole additional region is also transcribed in PGRPLA [14, 15, 16]. As this region is ortholog to a PGRPLA region in Anophelesmerus and Anopheles quadriannulatus[16], it might be an additional unpredicted exon of the gene. This suggests that the exact sequences of PGRPLA transcripts may need further investigation.
Fig. 4.
Map of PGRPLA and PGRPLB and sequence features of PGRPLA. a Map of the PGRPLA and PGRPLB gene loci. Predicted PGRP domains are shown in orange, predicted transmembrane domains (TM) in green and the predicted RHIM in turquoise (see online version for colors). The map of each VectorBase-predicted transcript is shown below each gene, including untranslated regions in white and coding DNA sequences in red. According to RNA-Seq data [14, 15, 16], a transcribed region that may participate in the sequence of PGRPLA transcripts is also indicated as “unpredicted exon.” dsRNA-targeted regions are shown in purple. b, c Sequence alignments of PGRPLA RHIM (b) and PGRP domain (c) with their homologous sequences in Drosophila PGRPLA (b, c) and PGRPLCx (c). In Drosophila, there is no single isoform encompassing both an RHIM and a PGRP domain, hence the sequence of PGRPLAD isoform is shown for the RHIM and the sequence of PGRPLAF isoform for the PGRP domain. c Amino acids shown in red are in direct contact with the peptidoglycan monomer “TCT” (tracheal cytotoxin) [18]. Sequence alignments were performed with ClustalW [19] and manually curated. *, positions with fully conserved residues;:, positions with conservation of “strong” groups (STA, NEQK, NHQK, NDEQ, QHRK, MILV, MILF, HY, FYW); ·, positions with conservation of “weaker” groups (CSA, ATV, SAG, STNK, STPA, SGND, SNDEQK, NDEQHK, NEQHRK, FVLIM, HFY).
In Drosophila, the receptors PGRPLC and PGRLE induce the Imd pathway through a RIP Homotypic Interaction Motif (RHIM), which is also present in Drosophila PGRPLA [17]. Alignment of this RHIM to Anopheles PGRPLA revealed a high similarity in a sequence of 27 amino acids, in a region shared by both PGRPLA1 and PGRPLA2 (Fig. 4b). These data suggest that both isoforms of Anopheles PGRPLA carry a RHIM domain.
We then investigated whether PGRPLA1 promotes resistance to P. berghei in a bacterium-dependent fashion. Two days after dsRNA injection, mosquitoes were orally infected with E. cloacae, a Gram-negative bacterium previously found in the mosquito microbiota, and infected with P. berghei 48 h later. We observed that silencing PGRPLA1 had a stronger effect on intensity and prevalence of P. berghei infection in E. cloacae-infected than noninfected mosquitoes, as shown by a significant increase in the PGRPLA1-to-LacZ odds ratios (Fig. 3b; online suppl. Fig. S3A). To examine whether the PGRPLA1-mediated response was induced by the midgut microbiota, we orally treated mosquitoes with the antibiotic gentamycin and infected them with P. berghei 3 days later. We observed that the antibiotic treatment significantly reduced but did not eliminate the effect of PGRPLA1 on P. berghei infection intensity, while the effect on prevalence was lost (Fig. 3c; online suppl. Fig. S3B). These results suggested that PGRPLA1 protection is partly microbiota independent. This prompted us to investigate whether the PGRP domain of PGRPLA1, as previously reported in Drosophila PGRPLA and Anopheles PGRPLA2, lacks essential features for peptidoglycan binding [8]. Indeed, sequence alignment revealed that 8/10 amino acids involved in binding peptidoglycan in PGRPLCx are absent from PGRPLA1 (Fig. 4c).
PGRPLB Downregulates Systemic Induction of the Imd Pathway
We also investigated whether PGRPLB is a negative regulator of the Imd pathway, similarly to its Drosophila ortholog [6, 7]. According to VectorBase annotations, PGRPLB encodes 2 isoforms, which share the same PGRP domain and differ in their transmembrane domains (Fig. 4a). We silenced PGRPLB by targeting a region shared by both predicted transcripts and monitored the expression of CEC1 6 h to 6 days after dsRNA injection in the mosquito midgut and fat body to examine local and systemic Imd responses, respectively. We found that PGRPLB silencing had no effect on CEC1 expression in the midgut, but it significantly increased CEC1 expression in the fat body 6 days later (Fig. 5a, b; online suppl. Fig. S4A). As PGRPLB is highly induced in the gut after blood feeding (Fig. 5c), we studied its role in the induction of the immune response after a blood meal provided 30 h after dsRNA injection. Our results indicate that CEC1 is induced in the midgut, but not in the fat body, after blood feeding (online suppl. Fig. S4B). PGRPLB silencing did not affect CEC1 expression in the gut but significantly induced CEC1 in the fat body 72 h after the blood meal (Fig. 5d, e). These data suggested that, similarly to Drosophila, PGRPLB is involved in preventing activation of a systemic immune response. Interestingly, PGRPLB expression was efficiently knocked down in the midgut but not in the fat body (online suppl. Fig. S4C, S4D). This is probably due to its very low expression in the fat body, which is 41- and 15-fold lower than in the midgut of sugar- and blood-fed mosquitoes, respectively (online suppl. Fig. S4C, S4D). These data suggested that PGRPLB is required in the gut and/or potentially other tissues, but not in the fat body, to prevent the activation of a late systemic response.
Discussion
In this study, we identify 2 new A. coluzzii PGRPs involved in the vector-parasite interactions. We reveal that PGRPLA has an antiparasitic effect and positively regulates the Imd pathway, similar to what was shown previously for PGRPLC [1]. Although our data suggest that PGRPLA1 and PGRPLA2 affect negatively P. berghei and P. falciparum, respectively, previous experiments performed in our laboratory using the G3 mosquito strain infected with P. berghei revealed an antiparasitic function for PGRPLA2 [20]. As both isoforms share the exons encoding the intracellular domain, including the RHIM motif, and are predicted not to bind peptidoglycan, we hypothesize that their differential effects on Imd pathway activation and anti-Plasmodium responses depend on their expression levels rather than functional differences. Indeed, P. falciparum and P. berghei infection of the mosquito midgut occur with different timing and at different temperatures. In that respect, published data suggest that PGRPLA1 and PGRPLA2 expression differs between mosquito strains and Plasmodium infection stages [21, 22, 23, 24], and we found that PGRPLA2 expression is 14 and 10 times lower than that of PGRPLA1 in the midguts of sugar- and blood-fed mosquitoes of our colony, respectively. Moreover, as mentioned in the results, the observed phenotypic difference may arise from PGRPLA1 dsRNA having a higher silencing efficiency and being less isoform specific than PGRPLA2 dsRNA. However, we cannot rule out that the differences found between PGRPLA1 and PGRPLA2 are not biologically relevant, they may notably be linked to the absence of a transmembrane domain in one of the predicted PGRPLA2 isoforms.
Oral bacterial challenge and antibiotic treatments revealed that the effect of PGRPLA1 is only partly bacteria dependent. Therefore, the effect of PGRPLA1 on Plasmodium in aseptic mosquitoes may be due to basal induction of the Imd pathway by PGRPLA1 dimerization. The RHIM domain of PGRPLA1 is highly conserved with that of Drosophila PGRPLA, which is shown to induce the Imd pathway [8]. The observed increase in the antiparasitic effect of PGRPLA1 silencing upon bacterial challenge suggests that PGRPLA1 also participates in a bacterium-mediated induction of the Imd pathway, perhaps through heterodimerization with PGRPLC. Induction of the Imd pathway by heterodimerization has been reported in Drosophila between PGRPLCx, which binds peptidoglycan, and PGRPLCa, which does not [25].
Our results also identify PGRPLB as a negative regulator of the Imd pathway, promoting tolerance to P. falciparum. More specifically, our data suggest that PGRPLB is needed in the gut and perhaps other tissues, but not in the fat body, to prevent induction of a systemic immune response. This is in contrast to what is known in Drosophila, where PGRPLB is required both in the gut and in the fat body to control systemic responses [7]. This difference between Anopheles and Drosophila PGRPLB may relate to the fact that the former is a predicted transmembrane protein, potentially scavenging peptidoglycan fragments as they traverse the gut epithelium, while the latter is a secreted protein [7], likely scavenging peptidoglycan monomers in the gut lumen and potentially in the hemolymph. Indeed, peptidoglycan fragments are known to diffuse through the gut and other epithelia into the hemolymph, where they remotely induce a systemic response [26]. Anopheles PGRPLB expression is enriched in the midgut and the salivary glands [27], 2 organs harboring microbial communities, further implicating this protein in epithelial tolerance to microbiota. Alternatively, the effect that we report on the systemic response may also happen via an antibacterial effect of PGRPLB, which may prevent bacteria from entering the mosquito hemolymph. In the tsetse fly, PGRPLB is shown to be a negative regulator of the Imd pathway, participating in tolerance to symbionts [28], and also to have a direct antiparasitic effect against trypanosomes [29]. Our results do not point to such an antiparasitic effect of Anopheles PGRPLB against Plasmodium. In our experimental setting, the role of PGRPLB on Plasmodium infection is probably reduced due to the fact that PGRPLB is itself a target of the Imd pathway [30]; hence, its silencing may induce Imd pathway activation leading to its overexpression. A deletion of the gene is required to determine the full effect of PGRPLB in Plasmodium infections.
Taken together, our data demonstrate that PGRPs are key regulators of the mosquito immune system and permissiveness to Plasmodium. We reveal that PGRPLA is a positive regulator of the Imd pathway and that its antiparasitic activity is partly bacteria dependent, while PGRPLB negatively regulates the same pathway, participating in the tolerance to the gut microbiota.
Author Contributions
M.G., F.T., F.H.R., and G.K.C. designed the experiments. M.G., F.T. and F.H.R. performed the experiments. A.C. and I.M. organized gametocyte carrier recruitment in Cameroon and provided infrastructure and intellectual input for work on P. falciparum. M.G., F.T., and G.K.C wrote this paper. All the authors commented on the paper and approved the final version.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
We are grateful to Mfou primary school pupils and their parents or guardians for volunteering to participate in this study and for blood donations. We also thank Jean-Pierre Agbor, Rose Nyambam, Etienne Onana, Isaac Tchikangwa, Katarzyna Sala, and Tibebu Habtewold for technical assistance, and Engelbert Manga, Emmanuel Bodzwan, and Constance Efemba for medical assistance. We thank Antonio Mendes and Sandrine Nsango for helping with P. falciparum experiments. We also thank João Trindade Marques for insights while writing this paper.
The work was supported by the Biotechnology and Biological Sciences Research Council Project BB/K009338/1, the European Commission FP7 Collaborative Project grant “TransMalariaBloc” (HEALTH-F3-2008-223736), the NIH Program Project grant 2PO1AI044220-06A1, and the ANR-06-MIME-001-01 grant.
References
- 1.Meister S, Agianian B, Turlure F, Relogio A, Morlais I, Kafatos FC, Christophides GK. Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog. 2009;5:e1000542. doi: 10.1371/journal.ppat.1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meister S, Kanzok SM, Zheng XL, Luna C, Li TR, Hoa NT, Clayton JR, White KP, Kafatos FC, Christophides GK, Zheng L. Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proc Natl Acad Sci USA. 2005;102:11420–11425. doi: 10.1073/pnas.0504950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Povelones M, Bhagavatula L, Yassine H, Tan LA, Upton LM, Osta MA, Christophides GK. The CLIP-domain serine protease homolog SPCLIP1 regulates complement recruitment to microbial surfaces in the malaria mosquito Anopheles gambiae. PLoS Pathog. 2013;9:e1003623. doi: 10.1371/journal.ppat.1003623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- 5.Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- 6.Paredes JC, Welchman DP, Poidevin M, Lemaitre B. Negative regulation by amidase PGRPS shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity. 2011;35:770–779. doi: 10.1016/j.immuni.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 7.Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, Oh BH, Ueda R, Mengin-Lecreulx D, Lemaitre B. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 2006;24:463–473. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 8.Gendrin M, Zaidman-Remy A, Broderick NA, Paredes J, Poidevin M, Roussel A, Lemaitre B. Functional analysis of PGRP-LA in Drosophila immunity. PLoS One. 2013;8:e69742. doi: 10.1371/journal.pone.0069742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurata S. Peptidoglycan recognition proteins in Drosophila immunity. Dev Comp Immunol. 2014;42:36–41. doi: 10.1016/j.dci.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neyen C, Poidevin M, Roussel A, Lemaitre B. Tissue- and ligand-specific sensing of Gram-negative infection in Drosophila by PGRP-LC isoforms and PGRP-LE. J Immunol. 2012;189:1886–1897. doi: 10.4049/jimmunol.1201022. [DOI] [PubMed] [Google Scholar]
- 11.Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa NT, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Moita LF, Muller HM, Osta MA, Paskewitz SM, Reichhart JM, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J, von Mering C, Xu J, Zheng L, Bork P, Kafatos FC. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 12.Wotton RS, Chaloner DT, Yardley CA, Merritt RW. Growth of Anopheles mosquito larvae on dietary microbiota in aquatic surface microlayers. Med Vet Entomol. 1997;11:65–70. doi: 10.1111/j.1365-2915.1997.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 13.Franke-Fayard B, Trueman H, Ramesar J, Mendoza J, van der Keur M, van der Linden R, Sinden RE, Waters AP, Janse CJ. A Plasmodium berghei reference line that constitutively expresses GFP at a high level throughout the complete life cycle. Mol Biochem Parasitol. 2004;137:23–33. doi: 10.1016/j.molbiopara.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Gibbons JG, Janson EM, Hittinger CT, Johnston M, Abbot P, Rokas A. Benchmarking next-generation transcriptome sequencing for functional and evolutionary genomics. Mol Biol Evol. 2009;26:2731–2744. doi: 10.1093/molbev/msp188. [DOI] [PubMed] [Google Scholar]
- 15.Pitts RJ, Rinker DC, Jones PL, Rokas A, Zwiebel LJ. Transcriptome profiling of chemosensory appendages in the malaria vector Anopheles gambiae reveals tissue- and sex-specific signatures of odor coding. BMC Genomics. 2011;12:271. doi: 10.1186/1471-2164-12-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.VectorBase: Chromosome 2L: 12,211,358-12,220,758 https://www.vectorbase.org/Anopheles_gambiae/Location/View?db=core;g=AGAP005205;r=2L:12211358-12220758 (accessed January 6, 2017)
- 17.Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, Oshima Y, Peach C, Erturk-Hasdemir D, Goldman WE, Oh BH, Kurata S, Silverman N. PGRP-LC and PGRP-LE have essential yet distinct functions in the drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol. 2006;7:715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- 18.Chang CI, Chelliah Y, Borek D, Mengin-Lecreulx D, Deisenhofer J. Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science. 2006;311:1761–1764. doi: 10.1126/science.1123056. [DOI] [PubMed] [Google Scholar]
- 19.ClustalW multiple sequence alignment program, version 1.83, 2003.
- 20.Meister S. The Role of PGRP Proteins in Innate Immunity Pathways in the Malaria Vector Anopheles gambiae; thesis. Ruperto Carola University of Heidelberg. 2006 [Google Scholar]
- 21.Fossog Tene B, Poupardin R, Costantini C, Awono-Ambene P, Wondji CS, Ranson H, Antonio-Nkondjio C. Resistance to DDT in an urban setting: common mechanisms implicated in both M and S forms of Anopheles gambiae in the city of Yaoundé Cameroon. PLoS One. 2013;8:e61408. doi: 10.1371/journal.pone.0061408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fossog Tene B, Poupardin R, Costantini C, Awono-Ambene P, Wondji CS, Ranson H, Antonio-Nkondjio C. VectorBase: AGAP005205 expression in experiment ddt resistant field isolates. https://www.vectorbase.org/expression-browser/gene/AGAP005205/experiment/E-MTAB-1382 (accessed January 6, 2017)
- 23.Mendes AM, Awono-Ambene PH, Nsango SE, Cohuet A, Fontenille D, Kafatos FC, Christophides GK, Morlais I, Vlachou D. VectorBase: AGAP005205 expression in experiment Plasmodium falciparum infections. https://www.vectorbase.org/expression-browser/gene/AGAP005205/experiment/Plasmodium%20falciparum%20infections%20(Mendes%20et%20al.,%202011) (accessed January 6, 2017)
- 24.Mendes AM, Awono-Ambene PH, Nsango SE, Cohuet A, Fontenille D, Kafatos FC, Christophides GK, Morlais I, Vlachou D. Infection intensity-dependent responses of Anopheles gambiae to the African malaria parasite Plasmodium falciparum. Infect Immun. 2011;79:4708–4715. doi: 10.1128/IAI.05647-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mellroth P, Karlsson J, Hakansson J, Schultz N, Goldman WE, Steiner H. Ligand-induced dimerization of Drosophila peptidoglycan recognition proteins in vitro. Proc Natl Acad Sci USA. 2005;102:6455–6460. doi: 10.1073/pnas.0407559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gendrin M, Welchman DP, Poidevin M, Herve M, Lemaitre B. Long-range activation of systemic immunity through peptidoglycan diffusion in Drosophila. PLoS Pathog. 2009;5:e1000694. doi: 10.1371/journal.ppat.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker DA, Nolan T, Fischer B, Pinder A, Crisanti A, Russell S. A comprehensive gene expression atlas of sex- and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genomics. 2011;12:296. doi: 10.1186/1471-2164-12-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Wu Y, Yang G, Aksoy S. Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc Natl Acad Sci USA. 2009;106:12133–12138. doi: 10.1073/pnas.0901226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Aksoy S. PGRP-LB is a maternally transmitted immune milk protein that influences symbiosis and parasitism in tsetse's offspring. Proc Natl Acad Sci USA. 2012;109:10552–10557. doi: 10.1073/pnas.1116431109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maccallum RM, Redmond SN, Christophides GK. An expression map for Anopheles gambiae. BMC Genomics. 2011;12:620. doi: 10.1186/1471-2164-12-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data