Abstract
The genus of Papio (baboon) has six recognized species separated into Northern and Southern clades, each comprised of three species distributed across the African continent. Geographic origin and phenotypic variants such as coat color and body size have commonly been used to identify different species. The existence of multiple hybrid zones, both ancient and current, have complicated efforts to characterize the phylogeny of Papio baboons. More recently, mitochondrial DNA (mtDNA) and Y-chromosome genetic markers have been utilized for species identification with particular focus on the hybrid zones. Alu elements accumulate in a random manner and are a novel source of identical by descent variation with known ancestral states for inferring population genetic and phylogenetic relationships. As part of the Baboon Genome Analysis Consortium, we assembled an Alu insertion polymorphism database of nearly 500 Papio-lineage specific insertions representing all six species and performed population structure and phylogenetic analyses. In this study, we have selected a subset of 48 species indicative Alu insertions and demonstrate their utility as genetic systems for the identification of baboon species within Papio. Individual elements from the panel are easy to genotype and can be used in a hierarchical fashion based on the original level of uncertainty. This Alu-48 panel should serve as a valuable tool during the maintenance of pedigree records in captive populations and assist in the forensic identification of fossils and potential hybrids in the wild.
Keywords: retrotransposon, population genomics, evolutionary biology
Introduction
Baboons (genus Papio) cover a large geographic region on the African continent (Boissinot et al. 2014; Jolly 1993; Newman et al. 2004). There are currently six recognized species of Papio baboons: P. anubis (olive), P. hamadryas (hamadryas), P. papio (guinea), P. cynocephalus (yellow), P. kindae (kinda), and P. ursinus (chacma) (Jolly et al. 2011; Zinner et al. 2013). The six species are separated primarily into a Northern clade (olive, guinea, and hamadryas) and a Southern clade (yellow, kinda, and chacma), but the existence of natural hybrid zones, both ancient and current, located near species boundaries have been widely studied (Bergman et al. 2008; Nagel 1973; Phillips-Conroy and Jolly 1986; Szmulewicz et al. 1999). The population biology observed in the living populations shows strong evidence for active current hybrid zones between 1) P. anubis and P. hamadryas, 2) P. anubis and P. cynocephalus, 3) P. kindae and P. ursinus, 4) P. kindae and P. cynocephalus, and 5) P. anubis and P. papio. The manuscript from the Baboon Genome Analysis Consortium (Rogers et al. under revision) reported strong evidence for multiple episodes of ancient and recent admixture involving several of the recognized species, suggesting that genetic exchange and gene flow is ongoing in extant populations.
Investigators commonly use morphological characteristics, phenotypic variation and geographic locale/social groups to identify baboon species (Jolly 1993; Phillips-Conroy et al. 1991). In hybrid zones, baboon species origins are inferred using variation in coat color, body size and more recently by using genetic markers such as mitochondrial DNA (mtDNA) (Keller et al. 2010; Kopp et al. 2014; Newman et al. 2004; Zinner et al. 2013), Y-chromosome markers (Jolly et al. 2011) and microsatellites (Bergman et al. 2008; Charpentier et al. 2012; Tung et al. 2008). Most genetic variation among hybrid baboons is introduced by unidirectional introgression of males mating with native females of the other parental species, thus changing the Y-chromosome genomic component while leaving the native mitochondrial genome intact (Jolly et al. 2011; Keller et al. 2010). Alternatively, using neutral autosomal genetic markers such as retrotransposons, LINEs (long interspersed elements) and Alu, may help decipher these complex evolutionary relationships (Boissinot et al. 2014; Szmulewicz et al. 1999).
Retrotransposons have been shown to be highly valuable genetics systems to infer the evolutionary relationships between different species (Konkel et al. 2010; Murata et al. 1993; Ray et al. 2005b; Shedlock and Okada 2000). These markers, especially Alu elements and to a lesser extent L1, are now commonly used to investigate phylogenetic and population genetic relationships within the primate orders (Batzer and Deininger 1991; Konkel et al. 2010; Ray and Batzer 2005; Roos et al. 2004; Shedlock and Okada 2000; Stoneking et al. 1997; Xing et al. 2007). Alu elements are used more commonly, as they are easy to genotype with a single PCR reaction due to their relatively small size (∼300 bp). Retrotransposons such as Alu elements are identical-by-descent, have a known directionality or ancestral state and are inexpensive to genotype (Ray et al. 2006). The amplification of Alu elements has been ongoing in primate genomes for about 65 million years (Batzer and Deininger 2002; Roy-Engel et al. 2008). They mobilize via a “copy and paste” mechanism through an RNA intermediate, a process termed “target-primed reverse transcription” (TPRT) (Luan et al. 1993). Alu elements are nonautonomous and utilize the enzymatic machinery of autonomous LINE elements (L1) to mobilize (Batzer and Deininger 2002; Comeaux et al. 2009; Dewannieux et al. 2003). Due to the staggered DNA cuts of the genome by the L1-derived endonuclease during TPRT, Alu insertions are flanked by short sequences of duplicated host DNA called Target Site Duplications (TSDs) that can be used to identify the insertion event.
Because understanding the taxonomy of baboons is complicated and Alu elements are robust genetic systems for resolving primate phylogenies, we employed Papio lineage specific Alu elements in the quest to resolve the phylogenetic relationship among Papio baboons as part of the Baboon Genome Analysis Consortium. For this larger study, we identified over 500 Alu insertion polymorphisms specific to the Papio lineage with representative elements ascertained from all six Papio species (Rogers et al. under revision). A final data set of 494 insertion events were used in that study to analyze the phylogeny and population structure across 79 different baboons. Although a complete statistically robust reconstruction of Papio phylogeny remained elusive, even when using retrotransposons, the population Structure program was able to identify the existence of six distinct population clusters, one for each recognized species, and detect species admixture among some baboons, making this Alu panel a powerful tool in species identification. However, having such a large data set is cumbersome and not quite feasible for use in most field studies or captive breeding colonies. The goal of this study was to assemble a subset (≤10%) of species informative Alu insertions, and demonstrate their utility as genetic markers for the identification of Papio baboons.
Materials and Methods
DNA Samples
A complete list of all the DNA samples used in this study is shown in supplementary file S2, Supplementary Material online and is also available in the Supplementary Material, of the manuscript submitted as part of the Baboon Genome Analysis Consortium (Rogers et al. under revision). Briefly, the DNA panel included 79 individual baboon samples, 15 olive baboons (P. anubis) including DNA from the reference olive baboon individual (27861), 2 Guinea baboons (P. papio), 2 hamadryas baboons (P. hamadryas), 3 chacma baboons (P. ursinus), 15 wild caught kinda baboons (P. kindae) from Zambia, and 42 yellow baboons (P. cynocephalus) consisting of 12 captive yellow baboons from the Southwest National Primate Research Center in San Antonio, TX (SFBR-Y), which are likely descendants of baboons captured in Amboseli National Park, Kenya, and 30 wild caught yellow baboons from the Mikumi National Park in Tanzania. All the baboon DNA samples were subjected to whole genome amplification (WGA) using the illustra GenomiPhi V2 DNA amplification kit (GE Healthcare Life Sciences, Marlborough, MA, USA). This was required to obtain sufficient DNA template for PCR analysis of over 500 polymorphic Alu insertions as part of the Baboon Genome Analysis Consortium. To insure that WGA DNA was suitable for this parent study, we compared original stock DNA to WGA DNA for baboon sample 27861 (used in the reference genome assembly labelled Panu_2.0 in NCBI and papAnu2 in Ensembl) in eight PCR assays in which 27861 appeared heterozygous for the Alu insertion while being absent from Panu_2.0. In all eight cases, the genotype using the stock DNA matched the genotype using the WGA DNA. Furthermore, in a previous study comparing stock DNA and WGA DNA (Ray et al. 2005a) genotypes were 97% (473 out of 489) consistent between the original DNA and the WGA DNA. Each of the 16 (of 489) disagreements (3%) represented a single allele aberration (i.e., between heterozygous and homozygous). The ability to determine the inferred ancestry of each individual was unaffected and was 100% consistent between the original stock and WGA DNA. Therefore, WGA DNA was used throughout this study with confidence.
Alu Insertion Polymorphisms
All loci used in this study were designed as part of the Baboon Genome Analysis Consortium (Rogers et al. under revision) and ascertained from the baboon reference genome (Panu_2.0) of P. anubis, or computationally derived from sequence data from the diversity panel samples (described in detail elsewhere [Rogers et al. under revision, supplementary methods, Jordan et al. In preparation]). Briefly, whole genome sequencing data generated from diversity panel baboons (16098, 34472, 34474, 97124, 28755, 28547, and 30388) were downloaded from the Baylor College of Medicine Human Genome Sequencing Center File Transfer Database. In-house Python scripts were used to predict the insertion coordinates of taxon-specific Alu insertion candidates by aligning second generation sequencing reads both to a consensus Alu sequence (AluY) (Jurka et al. 2005) and to the reference baboon assembly (Panu_2.0) with the Burrows Wheeler Aligner (BWA mem) (Li and Durbin 2009). A confidence score was calculated for each candidate Alu locus to filter out candidates lacking sufficient computational support. The confidence score was calculated using an in-house algorithm based on several criteria, such as the number of supporting reads, number of reads that mapped both to the reference genome and the consensus Alu sequence, location within the consensus Alu to which reads mapped, local read depth and average read depth throughout the genome. To determine a reliable cutoff score, we performed PCR validation experiments on a small panel of baboon diversity samples. This was done to confirm the presence of the candidate loci in the diversity sample from which they were computationally detected, and the absence of those loci in the baboon reference genome. A complete list of the 48 Alu elements selected for this study, including the locus-specific oligonucleotide primers for PCR and genomic coordinates, is shown in supplementary file S2, Supplementary Material online.
Polymerase Chain Reaction
Oligonucleotide primers for PCR for all loci used in this study were designed as part of the Baboon Genome Analysis Consortium (Rogers et al. under revision). Briefly, the genomic coordinates of potential Alu insertions, plus 500 bp flanking both ends of the predicted insertion point, were extracted from the reference genome Panu_2.0. Orthologous human and rhesus macaque sequences were also aligned. Primer 3 software was used in all cases, either manually (Rozen and Skaletsky 2000) or using a modified version (Untergasser et al. 2012). PCR amplifications were performed in 25 μl reactions containing 25 ng of template DNA; 200 nM of each oligonucleotide primer; 1.5 mM MgCl2, 10× PCR buffer (1×:50 mM KCl; 10 mM TrisHCl, pH 8.4); 0.2 mM dNTPs; and 1–2 U Taq DNA polymerase. PCR reactions were performed under the following conditions: initial denaturation at 94 °C for 60 s, followed by 32 cycles of denaturation at 94 °C for 30 s, 30 s at optimum annealing temperature (usually 57 °C), and extension at 72 °C for 30 s. PCRs were terminated with a final extension at 72 °C for 2 min. 20 μl of each PCR product were fractionated in a horizontal gel chamber on a 2% agarose gel containing 0.2 μg/ml ethidium bromide for 60 min at 185V. UV-fluorescence was used to visualize the DNA fragments and images were saved using a BioRad ChemiDoc XRS imaging system (Hercules, CA).
Selection of the Alu-48 Panel
Structure analysis as reported for the Baboon Genome Analysis Consortium (Rogers et al. under revision) revealed that the 12 yellow baboons from the Southwest National Primate Research Center in San Antonio, TX (SFBR-Y) all exhibited evidence of mixed ancestry with olive baboons. Therefore, these 12 were removed from consideration for the initial construction of the species indicative markers. The Alu-48 panel of loci were selected from the larger data set of 494 Alu insertion polymorphisms using a combination of allele frequency data and empirical observation of agarose gels. Following gel electrophoresis, genotypic data were recorded for each allele as follows: an individual who was homozygous present for a given Alu locus was assigned the code 1, 1; homozygous absent, 0, 0; and heterozygous, 1, 0. This binomial data sheet was used to calculate the allele frequency for each Alu insertion across each population group for the final data set of 494 Alu insertion polymorphisms reported in the manuscript submitted as part of the Baboon Genome Analysis Consortium (Rogers et al. under revision). This binomial data sheet is also available on the Batzer Lab website (https://biosci-batzerlab.biology.lsu.edu/, last accessed June 6, 2017) under publications for the Baboon Genome Consortium manuscript as supplementary file S1, genotypes worksheet. In addition to using the calculated allele frequencies, gel chromatographs were also visually inspected, with selection for loci appearing to exhibit strong presence in a given species while also mostly absent from the other five species. From this filtered list of loci, seven to ten candidates representative of each of the six species were selected and the panel narrowed to 48 Alu loci.
Structure Analysis
Once the small panel of 48 species indicative markers was determined, population structure analyses were performed using Structure 2.3.4 software (Falush et al. 2003) to confirm that the Alu-48 panel properly identified six population clusters. Using genotype data from unlinked markers, this software performs a model-based clustering method to infer the population structure. For our initial analysis, the information regarding the origin of the samples was omitted. The analyses were performed under the admixture model which assumes that individuals may have mixed ancestry. To determine the value of K (where K equals the number of population clusters) with the highest likelihood, initially K was set from 1 to 8. The initial burn-in period was set at 10,000 iterations and followed by a run-length of 10,000 steps and repeated twice. These settings were based on the readme.pdf file downloaded with the software “Document for Structure Software version 2.3” listing suitable starting parameters for small data sets. The alpha statistic stabilized by about 2,000 of the 10,000 postburn-in iterations in each of the duplicate runs indicating these settings were adequate. The most likely value of K was calculated to be six based on the “estimated ln prob of data” scores generated by Structure.
The authors of Structure indicate that this method is generally accurate with small data sets, such as this one, but acknowledge it is still an estimate of K. Therefore, the discriminant analysis of principal components (DAPC) method was also used on the Alu-48 panel (Jombart et al. 2010). DAPC is a nonmodel based method to estimate the number of population clusters in a data set using the adegenet package (Jombart 2008) for the R software (R Development Core Team 2016). The DAPC method also determined K = 6.
Next, with K set to 6, another Structure analysis was performed under the admixed model using the known population information. The 12 SFBR yellow baboons (SFBR-Y) were assigned as “unknowns” or population “0” to determine if the Alu-48 panel could detect their admixed ancestry. Based on Structure’s estimate of the most likely population(s) of origin, samples were then assigned to each of the two potential source populations, Olive (population 1) and then Yellow (population 6), and admixture estimates were calculated for three parental generations. The Structure results were compared with the expected ancestry based on pedigree records for these animals obtained from the SFBR.
Results
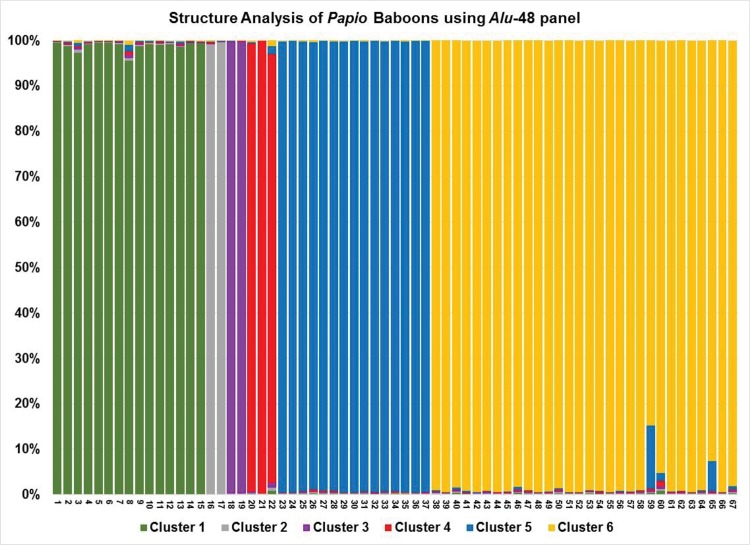
Here, we report a subset of 48 Papio lineage-specific Alu insertion polymorphisms from the Baboon Genome Analysis Consortium (Rogers et al. under revision) with species indicative distributions. A complete list of these 48 elements, including the locus specific oligonucleotide primers for PCR and genomic coordinates, is shown in supplementary file S2, Supplementary Material online. The Structure output using the Alu-48 panel across 67 Papio baboon individuals is plotted as figure 1, showing each individual assigns to their respective species cluster with near 100% probability. K = 6 clusters captures the majority of structure in the data and matches the number of recognized Papio species. The allele frequency data for each locus in the Alu-48 panel, sorted by Papio species, is shown in table 1. The allele frequency data for the 12 yellow baboon (P. cynocephalus) samples originally obtained from the Southwest National Primate Center in San Antonio, TX (SFBR-Y) are listed separately in table 1 due to their predetermined admixture with olive baboons (P. anubis) (see Methods). Colored fields in table 1 indicate the species from which the locus was ascertained. Bold font indicates an allele frequency > 0.000. The Alu-48 panel consists of 11 Alu insertions present in olive baboons, ten ascertained from the olive baboon (P. anubis) reference genome, (Panu_2.0), and one previously published Alu locus with a history of genotype data collected from a known anubis/hamadryas hybrid zone (Szmulewicz et al. 1999). The polymorphic Alu insertion used in that study, located in the baboon lipoprotein lipase (LPL) gene, was genotyped in a total of 179 baboon individuals, 58 anubis, 66 hamadryas, and 55 hybrids, providing valuable allele frequency data for these populations. The same Alu locus was also included in a later study (Boissinot et al. 2014) and was genotyped in 45 baboon individuals, reinforcing the historical precedent for including it in our analysis. We also selected seven Alu insertions ascertained from P. hamadryas diversity sample 97124 that were homozygous present in both P. hamadrayas samples we had available and homozygous absent in 77 other baboon genomes. Similarly, we selected seven Alu insertions ascertained from P. papio diversity samples (28547 or 30388) with 100% allele frequency in both Guinea baboons and absence in all 77 others. The seven Alu insertions each ascertained from P. ursinus and P. kindae baboons, and the remaining nine on the panel ascertained from P. cynocephalus, are not completely fixed present in these selected species, but do exhibit high allele frequencies and are nearly exclusive for the targeted species (table 1). Genotype data for the Alu-48 panel across all 79 Papio baboon individuals (including a T. gelada individual as the outgroup) are shown in supplementary file S3, Supplementary Material online.
Fig. 1.—
Structure analysis of 67 Papio baboon individuals using the Alu-48 panel. The y axis shows the probability of assignment of each individual to six population clusters. The x axis shows baboons numbered 1–67 listed in the same order as supplementary file S3, Supplementary Material online. Individuals 1–15 are olive baboons (P. anubis), 16–17 are hamadryas baboons (P. hamadryas), 18–19 are Guinea baboons (P. papio), 23–37 are kinda baboons (P. kindae), and 38–67 are yellow baboons (P. cynocephalus) from the Mikumi National Park, Tanzania. K = 6 population clusters matches the recognized number of Papio species and captures the majority of structure in the data.
Table 1.
Allele Frequency Distribution Data for Each Alu-48 Locus, Sorted by Papio Species
| Loci | Olive | Hamadryas | Guinea | Chacma | Kinda | Yellow | SFBR-Y | |
|---|---|---|---|---|---|---|---|---|
| 1 | Bab_LPL | 0.333 | 0.750 | 0.000 | 0.000 | 0.000 | 0.000 | 0.292 |
| 2 | TB_3063 | 1.000 | 1.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.292 |
| 3 | TB_3084 | 0.800 | 0.500 | 0.000 | 0.000 | 0.000 | 0.000 | 0.417 |
| 4 | 69388 | 0.833 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.250 |
| 5 | 46912 | 0.833 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.083 |
| 6 | 27402 | 0.667 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.292 |
| 7 | 27523 | 0.633 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.333 |
| 8 | 11507 | 0.300 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 9 | TB_3040 | 0.533 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.083 |
| 10 | TB_3023 | 0.400 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.083 |
| 11 | TB_76 | 0.433 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.125 |
| 12 | Ham-09 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 13 | Ham-16 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 14 | Ham-27 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 15 | Ham-28 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 16 | Ham-41 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 17 | Ham-43 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 18 | Ham-44 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 19 | G47-0 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 20 | G47-13 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 21 | G47-17 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 22 | G47-28 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 23 | G88-9 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 24 | G88-19 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 25 | G88-20 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 26 | C-16 | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 |
| 27 | C-38 | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 |
| 28 | C-44 | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 |
| 29 | C-05 | 0.000 | 0.000 | 0.000 | 0.667 | 0.000 | 0.000 | 0.000 |
| 30 | C-36 | 0.000 | 0.000 | 0.000 | 0.833 | 0.000 | 0.000 | 0.000 |
| 31 | C-42 | 0.000 | 0.000 | 0.000 | 0.833 | 0.000 | 0.000 | 0.000 |
| 32 | C-49 | 0.000 | 0.000 | 0.000 | 0.667 | 0.000 | 0.000 | 0.000 |
| 33 | K-20 | 0.000 | 0.000 | 0.000 | 0.000 | 0.733 | 0.000 | 0.000 |
| 34 | K-29 | 0.000 | 0.000 | 0.000 | 0.000 | 0.767 | 0.000 | 0.000 |
| 35 | K-30 | 0.000 | 0.000 | 0.000 | 0.000 | 0.500 | 0.000 | 0.000 |
| 36 | K-74-85 | 0.000 | 0.000 | 0.000 | 0.000 | 0.833 | 0.017 | 0.000 |
| 37 | K-17 | 0.000 | 0.000 | 0.000 | 0.000 | 0.600 | 0.000 | 0.000 |
| 38 | K2-10 | 0.000 | 0.000 | 0.000 | 0.000 | 0.867 | 0.017 | 0.000 |
| 39 | K-33 | 0.000 | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 | 0.083 |
| 40 | T2-103 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.600 | 0.083 |
| 41 | Y-90 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.879 | 0.125 |
| 42 | Y-65 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.414 | 0.167 |
| 43 | Y-71 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.810 | 0.375 |
| 44 | Y-141 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.733 | 0.292 |
| 45 | Y-108 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.776 | 0.292 |
| 46 | Y-119 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.914 | 0.250 |
| 47 | T2-25 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.367 | 0.042 |
| 48 | T2-29 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.567 | 0.083 |
Note.–Colored fields indicate the species from which the locus was ascertained. Bold font indicates an allele frequency >0.000.
To demonstrate the utility of our Alu-48 panel as genetic markers for the species specific identification of Papio baboons we performed a population structure analysis using Structure 2.3.4 software (Falush et al. 2003) and included 67 of our 79 total samples (the 12 SFBR-Y were omitted here). The most likely value of K was determined to be six (see Materials and Methods), matching the number of recognized species and consistent with our previous findings (Rogers et al. under revision). Next, Structure was run again, setting K = 6, and including the 12 SFBR-Y samples as population “0”, or unknowns. The results of this Structure analysis were obtained in <5 min using a 3.6 GHz processor and are shown in table 2. The 12 SFBR-Y samples as a group exhibit nearly equal membership to olive and yellow clusters with no single individual being assigned to either population with >73% probability as reflected in the data set calculations (table 3). Although not an exact match, these findings are generally consistent with the proportional admixture reported for the larger data set (Rogers et al. under revision). For example, the probability of assignment to the olive and yellow clusters, respectively, for SFBR-Y sample 6968 was 72%/28% and for sample 1X2117 was 22%/78% in that study, compared with 61%/27% and 12%/66% shown here (table 3). Pedigree records obtained from the SFBR indicated that all 12 SFBR-Y samples had at least one olive baboon ancestor within a recent generation while in captivity. In addition, these animals are likely descendants of baboons captured in the Amboseli National Park, Kenya, where a yellow/olive hybrid zone has been well documented (Alberts and Altmann 2001; Charpentier et al. 2012; Samuels and Altmann 1986). These data provided evidence to support our Alu-based findings of admixture within these individuals. This demonstrates that our Alu-48 panel can be used to identify each of the six species of Papio baboons, as well as detect the likelihood of admixture.
Table 2.
Inferred Population Structure: Probability of Assignment to Each Population Cluster
| Given Pop | Proportional Membership to Population Clusters |
Number of Individuals | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |||
| Unknown (SFBR-Y) | 0 | 0.436 | 0.030 | 0.022 | 0.033 | 0.061 | 0.419 | 12 |
| P. anubis | 1 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 15 |
| P. hamadryas | 2 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 2 |
| P. papio | 3 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 2 |
| P. ursinus | 4 | 0.000 | 0.000 | 0.000 | 0.999 | 0.000 | 0.000 | 3 |
| P. kindae | 5 | 0.000 | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 | 15 |
| P. cynocephalus | 6 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.998 | 30 |
Note.—Colored fields highlight species assignment to population clusters. Cluster 1 (olive), cluster 2 (gray), cluster 3 (lavender), cluster 4 (red), cluster 5 (blue), and cluster 6 (yellow).
Table 3.
Structure Analysis of 12 SFBR-Y Individuals as Unknowns
| Proportional Membership to Population Clusters | ||||||
|---|---|---|---|---|---|---|
| Pop 0 | 1 | 2 | 3 | 4 | 5 | 6 |
| ID | Olive | Hamadryas | Guinea | Chacma | Kinda | Yellow |
| 6968 | 0.607 | 0.032 | 0.021 | 0.030 | 0.038 | 0.273 |
| 1x1763 | 0.352 | 0.063 | 0.032 | 0.039 | 0.047 | 0.467 |
| 1x2092 | 0.725 | 0.016 | 0.014 | 0.020 | 0.025 | 0.200 |
| 9166 | 0.350 | 0.024 | 0.025 | 0.027 | 0.033 | 0.542 |
| 1x1786 | 0.447 | 0.024 | 0.021 | 0.047 | 0.060 | 0.401 |
| 1x3027 | 0.545 | 0.018 | 0.017 | 0.036 | 0.046 | 0.338 |
| 9481 | 0.286 | 0.030 | 0.036 | 0.041 | 0.045 | 0.562 |
| 8919 | 0.289 | 0.073 | 0.031 | 0.063 | 0.072 | 0.472 |
| 9656 | 0.547 | 0.017 | 0.018 | 0.024 | 0.023 | 0.371 |
| 1x2117 | 0.115 | 0.016 | 0.017 | 0.027 | 0.167 | 0.658 |
| 1x2798 | 0.529 | 0.028 | 0.019 | 0.024 | 0.156 | 0.243 |
| 8820 | 0.440 | 0.015 | 0.012 | 0.015 | 0.018 | 0.502 |
Note.—Probability of assignment to each population cluster.
However, our Alu-48 panel was unable to accurately infer the proportional ancestry, or degree of olive/yellow mixture derived from previous generations. As with field studies, most species identification within captive breeding programs has been largely based on phenotypic and behavioral observations of the animals. Therefore, the potential for multigenerational hybridization events can easily produce a phenotypically “yellow” baboon with mixed ancestry. To test our Alu-48 panel in this regard, we performed a subsequent Structure analysis, first assigning each of the SFBR-Y individuals to the Olive cluster and then each to the Yellow cluster to allow the software, with the given data set, to assign admixture going back three generations. The results of this ancestry test using the Alu-48 panel were uninformative. With the exception of sample 1x2117 the Structure analysis for ancestry indicated assignment to the Olive cluster was more likely with essentially no ability to detect which previous parental generation(s) contributed the Yellow and Olive components of the admixture (data not shown). This is not surprising given the limited number of informative loci and representative samples from each species. Estimating admixture proportions can be especially challenging if there are very few representatives of the parental populations within the data set and as such admixture estimates in these situations should be treated with caution (Falush et al. 2003; Pritchard et al. 2000). In this case, no baboons from previous generations from the Amboseli region were represented in our database.
Discussion
The availability of whole genome sequence data for all six Papio baboon species allowed Alu elements to be ascertained from individuals other than the olive baboon reference genome (Panu_2.0) and thus increased the analytical power of the data set across species. However, there are clear limitations to using a panel with a reduced number of genetic markers. A data set is only as useful as the members it contains. As shown in table 2, there were representative samples from P. anubis, P. kindae and P. cynocephalus with N = 15, 15, and 30, respectively. But, there were only two samples each from P. hamadryas and P. papio and only three from P. ursinus, one of which (18736) was likely “wild caught” originally and could have some possible admixture from neighboring population(s) of yellow or kinda baboons. Therefore, calculations of mixed ancestry are only as accurate as the depth of the parental generations represented in the data set. Our Alu-48 panel was reasonably successful at detecting the olive/yellow admixture of the SFBR-Y samples, but not at inferring the proportion of mixed ancestry because those parental generations were not available in the data set. Similarly, the two hamadryas samples in our data set are both males, originally from the Awash National Park in Ethiopia, a recognized hybrid zone between olive and hamadryas with social groups exhibiting mixed ancestry to varying degrees (Phillips-Conroy et al. 1991). These samples were not specifically labeled as hybrid individuals, but rather as “Awash hamadryas” baboons. The population Structure analysis for the Baboon Genome Analysis Consortium using the full data set of 494 Alu insertion polymorphisms detected about 25–30% admixture with olive (Rogers et al. under revision). When these two samples are subjected to a secondary Structure analysis using the population information provided, first assigning both to the hamadryas cluster and then both to the olive cluster, the admixture analysis assigns them both to the hamadryas cluster with 100% probability. This is due to the absence of individuals from nonadmixed parental generation populations in the data set. In fact, there is some evidence that some or all of the females initially classified as “pure anubis” females observed in the Awash region may have themselves been hybrids (Phillips-Conroy et al. 1991). Thus, this reinforces the need for caution with regard to sweeping conclusions regarding mixed ancestry within a limited data set.
It was not the purpose of this study to provide a comprehensive “one size fits all” Alu-based solution to solving the complex identification of Papio baboons. That is clearly unrealistic given the complex demographic and population genetic history of the species complex. Rather, the purpose of this study was to introduce a panel of Alu insertions with species indicative allele frequencies that used collectively provide an inference of Papio species identification. Individual sets of species indicative markers could be used empirically based on the initial level of ancestral uncertainty, such as in hybrid zones. The Alu-48 panel presented here was intentionally filtered from a larger database to select for high frequency alleles targeting six Papio species. The purpose of estimating the value of K clusters using both the Structure program (Falush et al. 2003) and DAPC (Jombart et al. 2010) was to insure that the true value of K could be set at six in Structure to infer species identity using the Alu-48 panel.
The Structure analysis on this data set is accomplished quickly (<5 min using a 3.6 GHz processor). Alu elements are relatively easy and inexpensive to genotype and represent a distinct advantage over large scale SNP genotyping or whole genome sequencing. They are also identical by descent, neutral autosomal markers with known directionality rather than gender derived (Y-chromosome or mtDNA). This Alu-48 panel is not intended to replace any of the existing methods for species identification, but rather it is to be used in conjunction with other widely established techniques. Baboon samples from known localities could be characterized using the most appropriate markers from the Alu-48 panel, adding to the genetic data collected for these individuals. Male introgression into hybrid zones transfers nuclear and Y-chromosome genetic data while the mitochondrial DNA of the native female parent of the other species remains unchanged. Adding more nuclear genetic markers, especially identical by descent Alu elements, collected throughout baboon populations, should provide another tool to help investigators. We are hopeful that the Alu-48 panel reported here will also assist with the maintenance of pedigrees at research centers with captive populations such as the Southwest National Primate Research Center in San Antonio, TX and the German Primate Center in Goettingen, Germany. The gradual accumulation of multigenerational genotype data from known parental matings using this genetic marker system should lead to more robust proportional ancestry estimates in the future. Such an improved data set may also be useful to augment morphological (Ackermann et al. 2014) and other methods in the forensic identification of potential hybrids from fossil records and wild populations.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Authors’ Contributions
J.A.W. and M.A.B. designed the research and wrote the paper; J.A.W., V.E.J., C.J.S., T.O.B., C.L.M., C.P.S., E.C.B., A.R., and B.N.C. conducted the experiments; M.K.K. and T.O.B. performed the Alu repeat analysis of the Panu_2.0 genome assembly; V.E.J., C.J.S., and T.O.B. performed the computational analysis of the whole genome sequencing data generated from diversity panel baboons downloaded from the Baylor College of Medicine Human Genome Sequencing Center File Transfer Database. All authors read and approved the final manuscript.
Authors’ Information
A.R. conducted experiments for this project in the Department of Biological Sciences, LSU-Baton Rouge as a participant in the Louisiana Biomedical Research Network (LBRN) while completing a degree in the Department of Biological and Physical Sciences at Northwestern State University of Louisiana, Natchitoches, LA. B.N.C. conducted experiments for this project in the Department of Biological Sciences, LSU-Baton Rouge as a member of the NIH Biomedical Research Experience for Veterinary Scholars.
Competing Interests
The authors declare that they have no competing interests.
Supplementary Material
Acknowledgments
The authors would like to thank all the members of the Batzer Lab for their helpful suggestions and the Baboon Genome Analysis Consortium. Special thanks to Dr. Laura Cox and Deborah Newman from the Southwest National Primate Research Center, Texas Biomedical Research Institute for their generous help providing SFBR-Y pedigree records. This research was supported by the National Institutes of Health R01 GM59290 (M.A.B.). A.R. was supported in part by the Louisiana Biomedical Research Network (LBRN) with funding from the National Institute of General Medical Sciences of the National Institutes of Health under Award Number P20GM103424 and by the Louisiana Board of Regents Support Fund. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or Louisiana Board of Regents. B.N.C. was funded in part by the National Institutes of Health Award Number 5T35OD011151-12 and the LSU School of Veterinary Medicine.
Literature Cited
- Ackermann RR, Schroeder L, Rogers J, Cheverud JM.. 2014. Further evidence for phenotypic signatures of hybridization in descendant baboon populations. J Hum Evol. 76:54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts SC, Altmann J.. 2001. Immigration and hybridization patterns of yellow and Anubis baboons in and around Amboseli, Kenya. Am J Primatol. 53:139–154. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL.. 2002. Alu repeats and human genomic diversity. Nat Rev Genet. 3:370–379. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL.. 1991. A human-specific subfamily of Alu sequences. Genomics 9:481–487. [DOI] [PubMed] [Google Scholar]
- Bergman TJ, Phillips-Conroy JE, Jolly CJ.. 2008. Behavioral variation and reproductive success of male baboons (Papio anubis × Papio hamadryas) in a hybrid social group. Am J Primatol. 70:136–147. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Alvarez L, Giraldo-Ramirez J, Tollis M.. 2014. Neutral nuclear variation in Baboons (genus Papio) provides insights into their evolutionary and demographic histories. Am J Phys Anthropol. 155:621–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier MJ, et al. 2012. Genetic structure in a dynamic baboon hybrid zone corroborates behavioural observations in a hybrid population. Mol Ecol. 21:715–731. [DOI] [PubMed] [Google Scholar]
- Comeaux MS, Roy-Engel AM, Hedges DJ, Deininger PL.. 2009. Diverse cis factors controlling Alu retrotransposition: what causes Alu elements to die?. Genome Res. 19:545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T.. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 35:41–48. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK.. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly CJ. 1993. Species, subspecies and baboon systematics In: Kimbel WH, Martin LB, editors. Species, species concepts and primate evolution. New York, NY: Plenum Press; p. 67–107. [Google Scholar]
- Jolly CJ, Burrell AS, Phillips-Conroy JE, Bergey C, Rogers J.. 2011. Kinda baboons (Papio kindae) and grayfoot chacma baboons (P. ursinus griseipes) hybridize in the Kafue river valley, Zambia. Am J Primatol. 73:291–303. [DOI] [PubMed] [Google Scholar]
- Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F.. 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, et al. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110:462–467. [DOI] [PubMed] [Google Scholar]
- Keller C, Roos C, Groeneveld LF, Fischer J, Zinner D.. 2010. Introgressive hybridization in southern African baboons shapes patterns of mtDNA variation. Am J Phys Anthropol. 142:125–136. [DOI] [PubMed] [Google Scholar]
- Konkel MK, Walker JA, Batzer MA.. 2010. LINEs and SINEs of primate evolution. Evol. Anthropol. 19:236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp GH, et al. 2014. The influence of social systems on patterns of mitochondrial DNA variation in baboons. Int J Primatol. 35:210–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH.. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595–605. [DOI] [PubMed] [Google Scholar]
- Murata S, Takasaki N, Saitoh M, Okada N.. 1993. Determination of the phylogenetic relationships among Pacific salmonids by using short interspersed elements (SINEs) as temporal landmarks of evolution. Proc Natl Acad Sci U S A. 90:6995–6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel U. 1973. A comparison of Anubis baboons, hamadryas baboons and their hybrids at a species border in Ethiopia. Folia Primatol. (Basel) 19:104–165. [DOI] [PubMed] [Google Scholar]
- Newman TK, Jolly CJ, Rogers J.. 2004. Mitochondrial phylogeny and systematics of baboons (Papio). Am J Phys Anthropol. 124:17–27. [DOI] [PubMed] [Google Scholar]
- Phillips-Conroy JE, Jolly CJ.. 1986. Changes in the structure of the baboon hybrid zone in the Awash National Park, Ethiopia. Am J Phys Anthropol. 71:337–350. [Google Scholar]
- Phillips-Conroy JE, Jolly CJ, Brett FL.. 1991. Characteristics of hamadryas-like male baboons living in Anubis baboon troops in the Awash hybrid zone, Ethiopia. Am J Phys Anthropol. 86:353–368. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P.. 2000. Association mapping in structured populations. Am J Hum Genet. 67:170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2016. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Ray DA, Batzer MA.. 2005. Tracking Alu evolution in New World primates. BMC Evol Biol. 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, et al. 2005a. Inference of human geographic origins using Alu insertion polymorphisms. Forensic Sci Int. 153:117–124. [DOI] [PubMed] [Google Scholar]
- Ray DA, et al. 2005b. Alu insertion loci and platyrrhine primate phylogeny. Mol Phylogenet Evol. 35:117–126. [DOI] [PubMed] [Google Scholar]
- Ray DA, Xing J, Salem AH, Batzer MA.. 2006. SINEs of a nearly perfect character. Syst Biol. 55:928–935. [DOI] [PubMed] [Google Scholar]
- Rogers J, et al. Under revision. The comparative genomics, epigenomics and complex population history of Papio baboons. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos C, Schmitz J, Zischler H.. 2004. Primate jumping genes elucidate strepsirrhine phylogeny. Proc Natl Acad Sci U S A. 101:10650–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Engel AM, Batzer MA, Deininger PL.. 2008. Evolution of human retrosequences: Alu In: Encyclopedia of life sciences. Chichester, UK: John Wiley & Sons, Ltd; p. 1–4. [Google Scholar]
- Rozen S, Skaletsky H.. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 132:365–386. [DOI] [PubMed] [Google Scholar]
- Samuels A, Altmann J.. 1986. Immigration of a Papio anubis male into a group of Papio cynocephalus baboons and evidence for an anubis-cynocephalus hybrid zone in Amboseli, Kenya. Int J Primatol. 7:131–138. [Google Scholar]
- Shedlock AM, Okada N.. 2000. SINE insertions: powerful tools for molecular systematics. BioEssays 22:148–160. [DOI] [PubMed] [Google Scholar]
- Stoneking M, et al. 1997. Alu insertion polymorphisms and human evolution: evidence for a larger population size in Africa. Genome Res. 7:1061–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szmulewicz MN, et al. 1999. An Alu insertion polymorphism in a baboon hybrid zone. Am J Phys Anthropol. 109:1–8. [DOI] [PubMed] [Google Scholar]
- Tung J, Charpentier MJ, Garfield DA, Altmann J, Alberts SC.. 2008. Genetic evidence reveals temporal change in hybridization patterns in a wild baboon population. Mol Ecol. 17:1998–2011. [DOI] [PubMed] [Google Scholar]
- Untergasser A, et al. 2012. Primer3: new capabilities and interfaces. Nucleic Acids Res. 40:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Witherspoon DJ, Ray DA, Batzer MA, Jorde LB.. 2007. Mobile elements and primate evolution. Yearbook Phys Anthropol. 50: 2–19. [DOI] [PubMed] [Google Scholar]
- Zinner D, Wertheimer J, Liedigk R, Groeneveld LF, Roos C.. 2013. Baboon phylogeny as inferred from complete mitochondrial genomes. Am J Phys Anthropol. 150:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.