Abstract
Inflammation in the perinatal brain caused by maternal or intrauterine fetal infection is now well established as an important contributor to the development of perinatal brain injury. Exposure to inflammatory products can impair perinatal brain development and act as a risk factor for neurological dysfunction, cognitive disorders, cerebral palsy, or preterm birth. Pre-exposure to inflammation significantly exacerbates brain injury caused by hypoxic/ischaemic insult. Tumour necrosis factor (TNF) is a family of cytokines largely involved in inflammation signalling. In our previous study, we identified the importance of TNF-related apoptosis-inducing ligand (TRAIL) signalling in the development of perinatal brain injury. We observed a significant increase in the expression levels of a soluble decoy receptor for TRAIL, osteoprotegerin (OPG). Besides TRAIL, OPG is able to bind the receptor activator of the NF-κB (RANK) ligand (RANKL) and inhibit its signalling. The function of the RANK/RANKL/OPG system in the brain has not come under much scrutiny. The aim of this research study was to elucidate the role of RANK, RANKL, and OPG in microglial responses to the proinflammatory stimuli lipopolysaccharide (LPS) and polyinosinic-polycytidylic acid (Poly I:C). Here, we show that RANK signalling is important for regulating the activation of the BV2 microglial cell line. We found that LPS treatment causes a significant decrease in the expression of RANK in the BV2 cell line while significantly increasing the expression of OPG, Toll-like receptor (TLR)3, and the adaptor proteins MyD88 and TRIF. We found that pretreatment of BV2 cells with RANKL for 24 h before the LPS or Poly I:C exposure decreases the expression of inflammatory markers such as inducible nitric oxide synthase and cyclooxygenase. This is accompanied by a decreased expression of the TLR adaptor proteins MyD88 and TRIF, which we observed after RANKL treatment. Similar results were obtained in our experiments with primary mouse microglia. Using recently developed CRISPR/Cas9 technology, we generated a BV2 cell line lacking RANK (RANK-/- BV2). We showed that most effects of RANKL pretreatment were abolished, thereby proving the specificity of this effect. Taken together, these findings suggest that RANK signalling is important for modulating the inflammatory activation of microglial cells to a moderate level, and that RANK attenuates TLR3/TLR4 signalling.
Keywords: Hypoxia-ischaemia, Inflammation, Microglia, RANK, RANKL, Osteoprotegerin, Toll-like receptors, MyD88, TRIF
Introduction
Microglial cells are the most abundant resident macrophages in the central nervous system (CNS) and are the first line of defence against injury and infection. As a consequence of brain injury or inflammatory stimuli, microglial cells respond by changing their phenotype from ramified morphology to an activated form. These activating stimuli can range from pathogens and pathogenic products to cytokines, or the efflux of intracellular contents following injury [1]. In the healthy brain, microglial cells are continually surveying their local micro-environment with their highly dynamic processes and are responsible for the elimination and refinement of synaptic connections. This microglial function is especially important and pronounced during the development of the brain [2]. The main function of the microglial cell in pathological situations is the clearance of the damaged cells after injury and phagocytosing and neutralizing infectious agents [3]. Moreover, the activated microglia cells secrete a range of signalling cytokines into their surroundings in order to communicate with the rest of the cells in the brain. Cytokine secretion is aimed at guiding other immune-competent cells to the site of injury/infection to destroy infected or damaged neurons and oligodendrocytes, but uncontrolled phagocytosis can cause a large amount of bystander damage to the healthy cells. Among the most important stimuli leading to microglial activation are lipopolysaccharides (LPS) from Gram-negative bacteria and double-stranded (ds)RNA from viruses. These are detected by Toll-like receptor (TLR)4 localized on the surface of the microglial cell and TLR3 localized in the endosomes [4,5]. The inflammatory signalling of activated TLR3 and TLR4 is mediated by adaptor proteins such as myeloid-differentiation primary-response gene 88 (MyD88) and TIR-domain-containing adaptor-inducing interferon (IFN)-β (TRIF). MyD88 is involved in the signalling of all TLRs except TLR3, where TRIF is the main adaptor. The precise regulation of microglial activation is not fully understood and is believed to be dependent on many different external and internal factors. The disruption of this regulation can contribute to the development or aggravation of existing brain disorders like neurodegeneration [6], traumatic injury [7], and perinatal brain injury [8]. Inflammation has been recognized as one of the key contributing factors for the development of cerebral palsy in term-born infants [9]; this association is supported by experimental data from animal models of foetal infection showing that LPS-exposed ovine foetuses develop white-matter damage similar to that seen in human infants [10]. Both clinical and experimental evidence also suggest that systemic infection/inflammation can affect cerebral vulnerability to a later insult such as hypoxia-ischaemia (HI) in term infants [11].
In our previous work investigating the involvement of tumour necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) signalling in the development of perinatal brain injury [12], we found a significant increase in osteoprotegerin (OPG) expression in vivo in an animal model of neonatal brain injury as well as in vitro in neurons and oligodendrocyte precursor cells (OPCs) subjected to TNF-α/IFN-γ treatment. OPG is a soluble decoy receptor member of the TNF family and is able to bind and inhibit both TRAIL and RANKL signalling [13]. In TRAIL signalling, the inhibition caused by the increased levels of OPG may be protective, as the pro-apoptotic role of TRAIL through binding with death receptor (DR)4 or DR5 in the CNS is well established [14] including in the context of neonatal brain injury [12]. Despite the fact that RANKL, its corresponding receptor RANK, and OPG are expressed in the brain tissue, little is known about the action of OPG in the inhibition of RANKL signalling in the CNS. The RANK/RANKL interaction has been involved in regulating fever responses in females following inflammation [15]. More importantly, the OPG/RANKL/RANK axis has been described as critical for the inflammatory response in ischaemic brains in mice. It was shown that enhanced RANKL/RANK signalling in OPG−/− mice or treatment with recombinant RANKL in wild-type mice leads to the reduction of infarct volume and brain oedema via reduced post-ischaemic inflammation [16].
Most of the studies on the mechanism of action of the RANK/RANKL/OPG interaction have been performed in the context of inflammation-dependent bone resorption [17,18] and, unlike TRAIL signalling, RANK activation from RANKL promotes the differentiation of osteoclast cells [19,20]. As microglia and osteoclasts have a shared embryological origin (they both differentiate from common premonocyte lineage [21]) and display many characteristics typical of macrophages, this makes microglia obvious candidates for the action of RANK/RANKL/OPG signalling in the brain.
The aim of this study was to elucidate the mechanism of RANK, RANKL, and OPG in the regulation of microglial responses to LPS and polyinosinic:polycytidylic acid (Poly I:C). We hypothesized that RANK/RANKL signalling influences microglial activation in the brain by attenuating the expression levels of the key components of TLR signalling.
Materials and Methods
Primary Microglia
Primary microglia were prepared from newborn Sprague-Dawley rats as described previously [22,23]. In brief, brain cortices were dissected from the rat pups on postnatal day (P)0–2, roughly chopped, and enzymatically digested with DNase I and 0.25% trypsin-EDTA. Cells were resuspended in minimum essential Eagle's medium supplemented with 10% FBS, 2 mML-glutamine, 100 µg/mL penicillin-streptomycin solution (all from Sigma), and 0.6% glucose, and plated in T75 flasks at a density of 2 × 105/cm2. Mixed glial cell cultures were grown for 11 days, and the microglia were separated from the OPCs and astrocytes through shaking. The mixed glial cell culture was shaken for 1.5 h at 260 rpm to detach the microglia from the layer of attached OPCs and astrocytes. Purified microglia were plated in DMEM supplemented with 10% FBS and 100 µg/mL penicillin-streptomycin solution (all from Sigma).
BV2 Cell Culture
The immortalized microglia cell line, BV2, was donated by Professor R. Donato (University of Perugia, Perugia, Italy); it was generated by infecting primary mouse microglia cultures with a v-raf/v-myc oncogene-carrying retrovirus (J2) [24]. BV2 cells were cultured in RPMI medium containing 100 µg/mL penicillin-streptomycin solution (both from Sigma) and supplemented with 10% FBS. Pilot experiments to determine the correct cell density to prevent overcrowding and standardize treatments across plate types were performed. All experiments were repeated on at least 3 separate occasions, representing separate experimental days at least 1 passage apart. Activation of BV2 cells was achieved by treatment with LPS or Poly I:C (Sigma) at final concentrations of 10 ng/mL and 10 µg/mL, respectively. RANKL treatment was performed using recombinant murine RANKL (200 ng/mL; Peprotech Inc., USA), with the concentration chosen after previous optimization experiments (data not shown).
RT-qPCR
For analysis of the expression levels of target genes, the cells were solubilized in DireCtQuant 100 (Spain) and processed according to the manufacturer's instructions. Before the real-time quantitative PCR (RT-qPCR) reactions, the samples were treated with dsDNase (ThermoFisher Scientific, UK) according to the manufacturer's instructions to remove the double-genomic DNA. The reverse-transcription reaction was performed using an Amethyst One-Step RT-kit with ROX and SYBR (Cambridge Bioscience) or TaqMan® RNA-to-Ct™ 1-Step kit according to the manufacturer's instructions. RT-qPCR experiments were performed using the StepOnePlus™ Real-Time PCR Systems (ThermoFisher Scientific). The TaqMan probes for TLR3, RANK, and GAPDH were purchased from ThermoFisher Scientific and the primers from Integrated DNA Technologies (Table 1). All reactions were conducted in duplicate and corrected to the expression of the internal control genes GAPDH, actin-β, and RNA polymerase II polypeptide A. Data was analyzed using the delta threshold cycle (CT) method [25].
Table 1.
Primer sequence and Taqman probe reference numbers used in the study
| SYBR primers | |
| Mouse TRIF_F | GCC AGC CAC CTA GAG ATC AG |
| Mouse TRIF_R | TGT CCA GCG GTG TGT TAC AT |
| Mouse TLR4_F | GCT CCT GGC TAG GAC TCT GA |
| Mouse TLR4_R | TGT CAT CAG GGA CTT TGC TG |
| Mouse MyD88_F | CAC CTG TGT CTG GTC CAT TG |
| Mouse MyD88_R | AGG CTG AGT GCA AAC TTG GT |
| Mouse GAPDH_F | GGT GCT GAG TAT GTC GTG GA |
| Mouse GAPDH_R | CAC ACC CAT CAC AAA CAT GG |
| Rat RANK_F | GTGCTGGAGTCTGTGGACCT |
| Rat RANK_R | CCGGTCCGTGTACTCATCTT |
| Mouse RANK_Genot_F | CCT CCG ACA GTG TGT GTC TG |
| Mouse RANK_Gento_R | CAG ACT TTA TGC AGC AAG CA |
| Puromycin_Seq_F | CTT CAC CGT CAC CGC C |
| 18S_RNA_F | CGC GGT TCT ATT TTG TTG GT |
| 18S_RNA_R | AGT CGG CAT CGT TTA TGG TC |
| Taqman probes | |
| Mouse OPG | Mm01205928_m1 |
| Mouse RANKL | Mm00441906_m1 |
| Mouse RNA polymerase II polypeptide A | Mm00839502_m1 |
| Mouse iNOS | Mm00440502_m1 |
| Mouse TLR3 | Mm01207404_m1 |
| Mouse RANK | Mm00437132_m1 |
| Rat RANKL | Rn00589289_m1 |
| Rat OPG | Rn00563499_m1 |
| Rat β-actin | Rn00667869_m1 |
RANK Gene Knockout
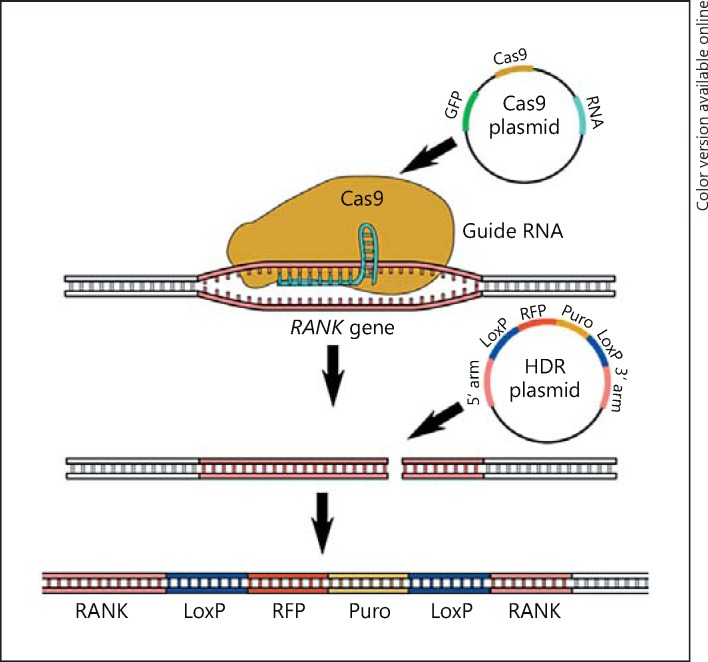
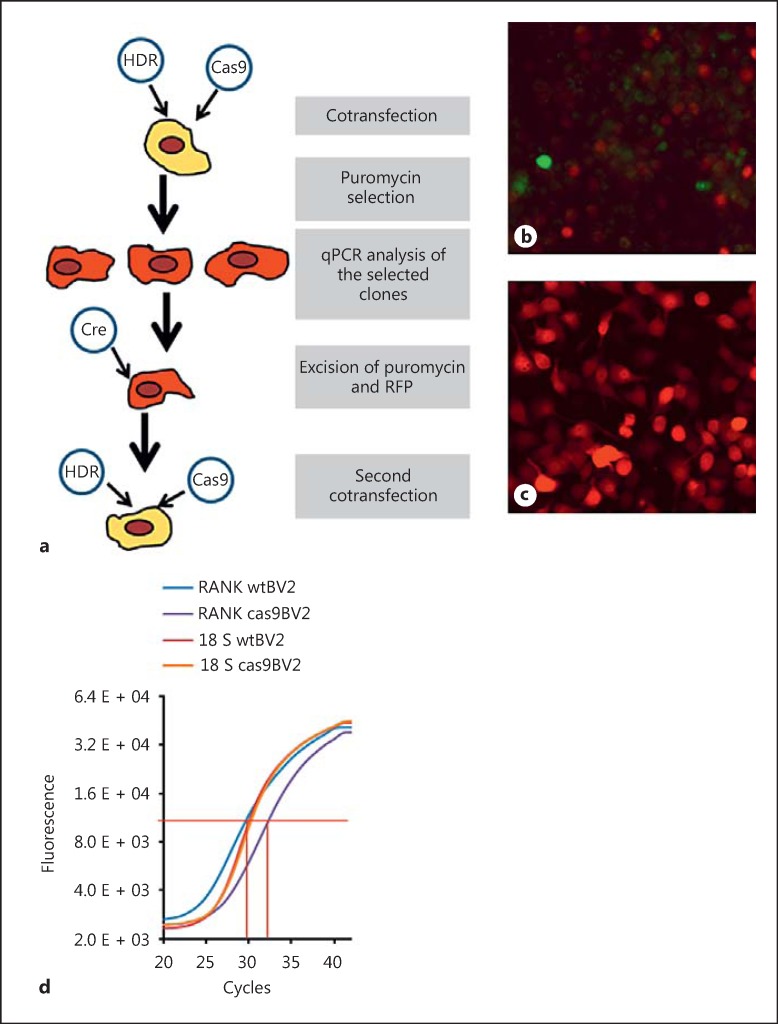
The RANK receptor gene was excised from the BV2 genome using CRISPR-Cas9 genome editing (Fig. 1). BV2 cells were co-transfected with Cas9 and homologous directed repair (HDR) plasmids using Ultracruz transfection reagent and plasmid transfection medium (all from Santa Cruz Biotechnology). Cas9 plasmid carries the gene for Cas9 enzyme synthesis, an RNA guide sequence, and for green fluorescent protein (GFP) expression. The RNA guide sequence consists of a 20-nucleotide sequence complementary to the mouse RANK gene (guide sequence) and a loop for the recognition and binding of the Cas9 enzyme. When in complex, the RNA guide and the Cas9 enzyme are capable of performing dsDNA cleavage at a precise place in the target gene, in this case, the first exon of RANK. GFP encoded by the Cas9 plasmid is used as positive control for the transfection, allowing transfected cells to be visualized by fluorescence microscopy 24 h after the transfection. The HDR plasmid carries the sequence used as a template for HDR of the dsDNA cleavage generated by Cas9. This template sequence is flanked by the 3′ and 5′ arms complementary to the RANK gene; these direct the sequence to the precise place of the DNA break. The template sequence between the 2 arms is inserted into the RANK gene, breaking the first exon and rendering the gene inactive. The inserted sequence consists of gene coding for red fluorescent protein (RFP) and the puromycin resistance gene flanked by LoxP sites. RFP expression allows the visualization of the transfected cells by fluorescence microscopy and the puromycin resistance gene is used to select the positive cells by plating them on media supplemented with 5 µg/mL puromycin. The LoxP sites allow the removal of RFP and puromycin genes from the cell DNA while preserving the target gene breakage. The cells were visualized using EVOS FL cell imaging system (ThermoFisher Scientific).
Fig. 1.
Generating RANK knockout using the CRISPR/Cas9 system. Schematic representation of the double-transfection with Cas9 and HDR plasmids, the binding of the Cas9 enzyme to guide RNA with consecutive double-strand cleavage in the RANK gene, and the introduction of the LoxP/RFP/Puro/LoxP sequence from the HDR plasmid through homologous directed repair.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism v5.0 software. Data is expressed as mean ± standard error of the mean (SEM). Comparisons between the experimental groups were made using one-way analysis of variance (ANOVA) followed by the Dunnett test (treatment conditions vs. control) or the Tukey test (treatment vs. control and vs. another treatment).
Results
TNF-α/INF-γ Treatment and Oxygen-Glucose Deprivation Inhibit RANK Signalling by Decreasing RANK/RANKL Expression in the Primary Microglia
Previously, we showed that the expression levels of OPG mRNA increased significantly after HI injury in P9 mice [12]. We also found a significant upregulation of OPG mRNA expression in the primary neurons after oxygen-glucose deprivation (OGD) and/or TNF-α/INF-γ treatment and also in the OPCs after TNF-α/INF-γ treatment [12]. The fact that OPG expression is increased suggests that RANK signalling is inhibited; increased levels of OPG will outcompete RANK for RANKL binding.
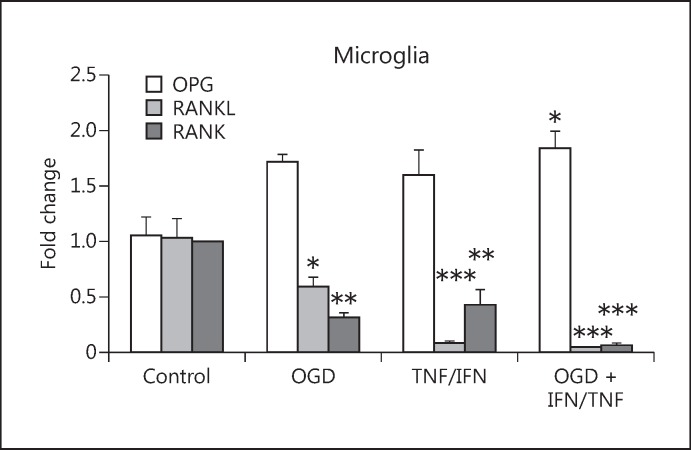
In this study, we initially determined whether the expression levels of OPG, RANK, and RANKL were changed in microglial cells after the HI and/or inflammatory insult. We performed in vitro experiments with primary microglia subjected to OGD to mimic HI and with TNF- α and INF-γ to mimic inflammatory insult. The mRNA expression of OPG, RANK, and RANKL was measured 48 h after the insult, and we found that the expression increased significantly only when the OGD and TNF-α/INF-γ treatment were combined (Fig. 2). However, the change in the expression levels of RANK and RANKL was much more dramatic. OGD and TNF-α/INF-γ treatments both reduced the expression of RANK and RANKL, and, in combination, they reduced the expression to 6% (for RANK) and 3% (for RANKL) of their normal expression levels. We speculated that this reduction, together with the general increase of OPG expression after insult, shuts down RANKL signalling in the microglial cells, and that this happens when the cells shift to a pro-inflammatory phenotype.
Fig. 2.
The effect of OGD and/or TNF-α/INF-γ treatment on the expression levels of OPG, RANKL, and RANK in primary rat microglia, measured 24 h after the treatment. Bars, mean; error bars, SEM. Results are expressed as fold change compared with non-treated control cells; * p < 0.05; ** p < 0.01; *** p < 0.001. n = 5.
RANKL Pretreatment Prevents an Increase in Inflammatory Markers in BV2 Cells Caused by LPS and Poly I:C Treatment
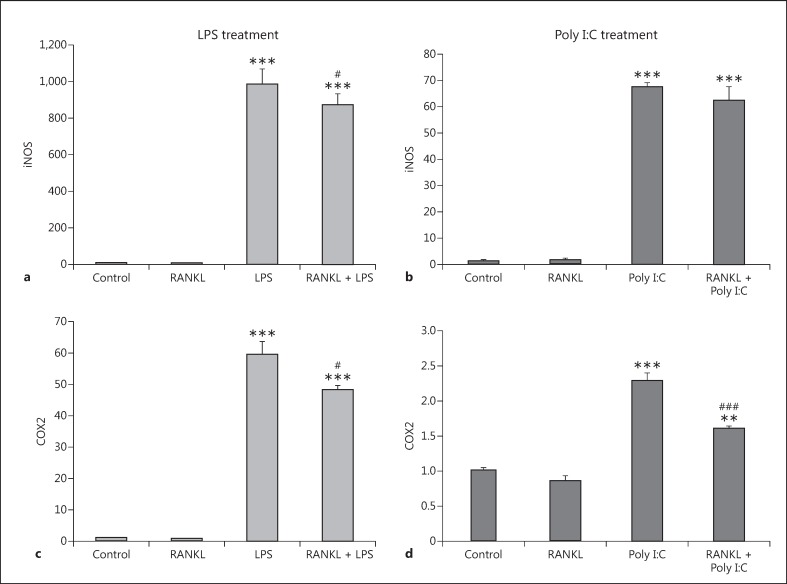
As the transfection of the primary microglia usually produces a very low number of transfected cells and the generation of knockout primary cell culture is not feasible, we decided to use the mouse microglial cell line BV2 in order to further clarify the role of RANK signalling in the development of an inflammatory phenotype of microglial cells. The BV2 cell line has been reported as a valid substitute for primary microglia in many experimental settings [26] and is widely used in in vitro experiments. These cells are highly responsive to inflammatory stimuli like LPS and Poly I:C, which cause a significant increase in the expression of inflammatory markers such as inducible nitric oxide synthase (iNOS, Fig. 3a, 3b) and cyclooxygenase (COX)2, Fig. 3c, 3d). We pretreated BV2 cells with recombinant RANKL for 24 h before exposure to LPS and Poly I:C and measured the mRNA expression levels 6 h later. RANKL pretreatment was able to significantly decrease the expression level of COX2 after LPS and Poly I:C insult compared with cells treated with vehicle only (Fig. 3c, 3d). The pretreatment was able to significantly reduce the expression of iNOS with LPS treatment (Fig. 3a) but not when the cells were treated with Poly I:C (Fig. 3b). The results showed that the activation of RANK by RANKL renders the BV2 cells less reactive to LPS and Poly I:C.
Fig. 3.
Expression levels of the inflammatory markers iNOS and COX2 in the BV2 cell line after RANKL pretreatment and LPS or Poly I:C treatment. BV2 cells were pretreated with 200 ng/mL recombinant RANKL for 24 h before the treatment with 10 ng/mL LPS (a, c) or 10 µg/mL Poly I:C (b, d). mRNA levels of iNOS (a, b) and COX2 (c, d) were measured 6 h later by RT-qPCR and normalized with respect to the reference gene GAPDH. Bars, mean; error bars, SEM. Results are expressed as fold change compared with non-treated control cells. Statistical significance was calculated with respect to non-treated controls (** p < 0.01; *** p < 0.001) or LPS/Poly I:C non-RANKL-treated cells (#p < 0.05; ###p < 0.001). n = 4.
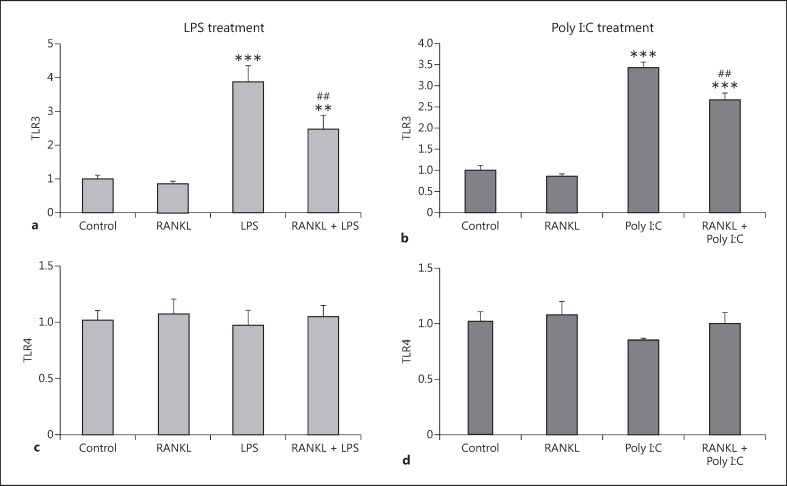
RANKL Pretreatment Restricts the Increase in TLR3 Expression Caused by LPS and Poly I:C Exposure
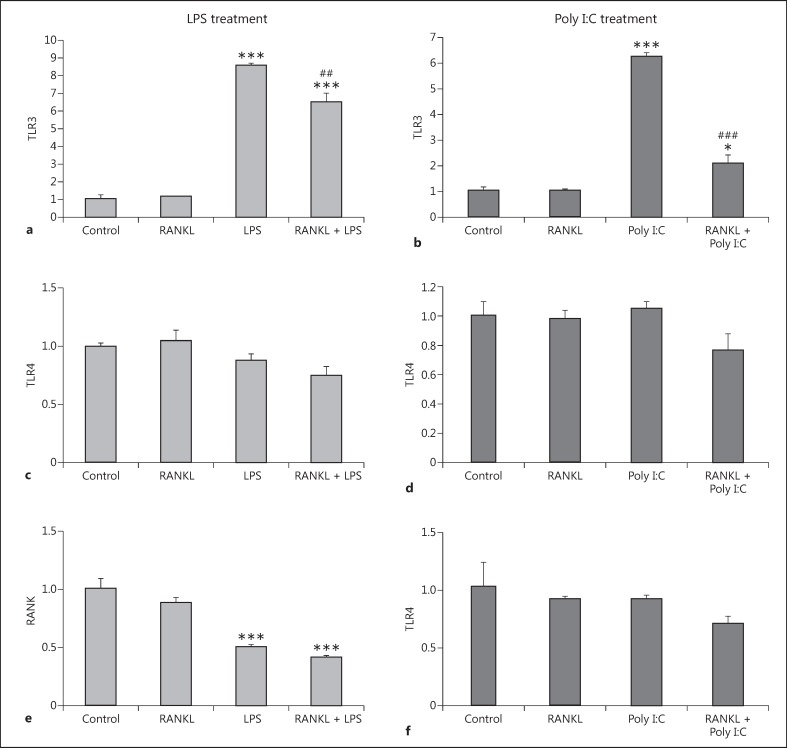
Given our previous results, we investigated the possible mechanism of the inhibition of inflammatory phenotype caused by RANKL pretreatment. We hypothesized that RANKL pretreatment would reduce the expression levels of the key proteins responsible for the recognition of LPS and Poly I:C. We therefore assessed the mRNA expression levels of TLR3 (recognizing Poly I:C) and TLR4 (recognizing LPS), in response to the inflammatory stimuli after the RANKL pretreatment. We found that both the LPS and the Poly I:C treatment resulted in a significant increase in TLR3 expression (Fig. 4a, 4b), while TLR4 expression remained unaffected (Fig. 4c, 4d). RANKL pretreatment significantly restricted the upregulation of TLR3 caused by both the LPS and Poly I:C treatments (Fig. 4a, 4b). Interestingly, the RANKL pretreatment did not affect the basal levels of TLR3 expressed when the BV2 cells were not challenged by an inflammatory stimulus. In addition, we assessed the RANK receptor to determine whether expression can be reduced by either LPS or Poly I:C, the same way as they are reduced in the microglia by TNF-α/INF-γ. We found that LPS (Fig. 4e), but not Poly I:C (Fig. 4f), reduced the mRNA level of RANK. RANKL pretreatment did not influence the reduction in RANK expression caused by LPS (Fig. 4e).
Fig. 4.
Expression levels of TLR3, TLR4, and RANK receptor in the BV2 cell line after RANKL pretreatment and LPS or Poly I:C treatment. BV2 cells were pretreated with 200 ng/mL recombinant RANKL for 24 h before the treatment with 10 ng/mL LPS (a, c, e), or 10 µg/mL Poly I:C (b, d, f). mRNA levels of TLR3 (a, b), TLR4 (c, d), and RANKL (e, f) were measured 6 h later by RT-qPCR and normalized with respect to the reference gene GAPDH. Bars, mean; error bars, SEM. Results are expressed as fold change compared with non-treated control cells. Statistical significance was calculated with respect to non-treated controls (* p < 0.05; *** p < 0.001) or LPS/ Poly I:C non-RANKL-treated cells (##p < 0.01; ###p < 0.001). n = 4.
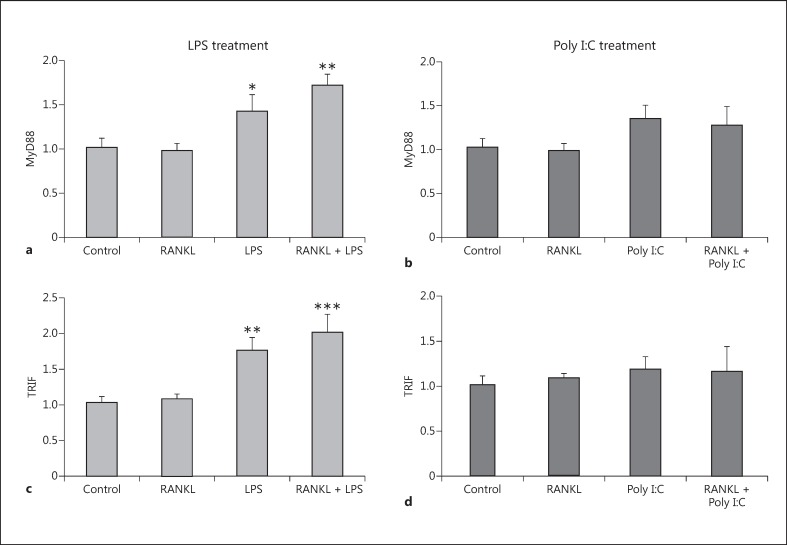
RANKL Pretreatment Prevents the Increased Expression of the Adaptor Proteins MyD88 and TRIF Caused by LPS and Poly I:C Treatment
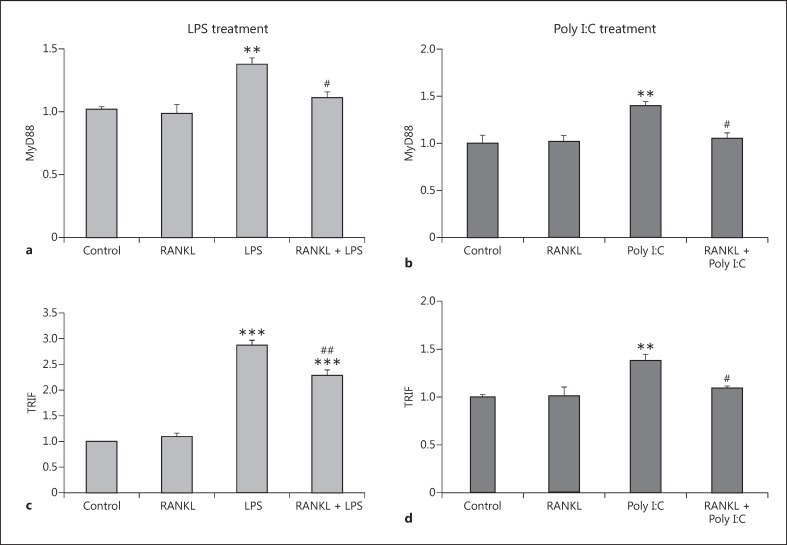
Following the binding of the ligand to TLR3 and TLR4, propagation of the inflammatory signal requires the recruitment of the adaptor proteins MyD88 and TRIF. We assessed changes in expression after an inflammatory challenge, and found that LPS and Poly I:C increased the expression levels of both MyD88 (Fig. 5a, 5b) and TRIF (Fig. 5c, 5d). Pretreatment with RANKL attenuated the increases in MyD88 and TRIF expression after both LPS and Poly I:C treatment.
Fig. 5.
Expression levels of the adaptor proteins MyD88 and TRIF in the BV2 cell line after RANKL pretreatment and LPS or Poly I:C treatment. BV2 cells were pretreated with 200 ng/mL recombinant RANKL for 24 h before the treatment with 10 ng/mL LPS (a, b) or 10 µg/mL Poly I:C (c, d). mRNA levels of MyD88 (a, b) and TRIF (c, d) were measured 6 h later by RT-qPCR and normalized with respect to the reference gene GAPDH. Bars, mean; error bars, SEM. Results are expressed as fold change compared with non-treated control cells. Statistical significance was calculated with respect to non-treated controls (** p < 0.01; *** p < 0.001) or LPS/ Poly I:C non-RANKL-treated cells (#p < 0.05; ##p < 0.01). n = 4.
RANKL Pretreatment Modulates MyD88 Expression in the Primary Microglia
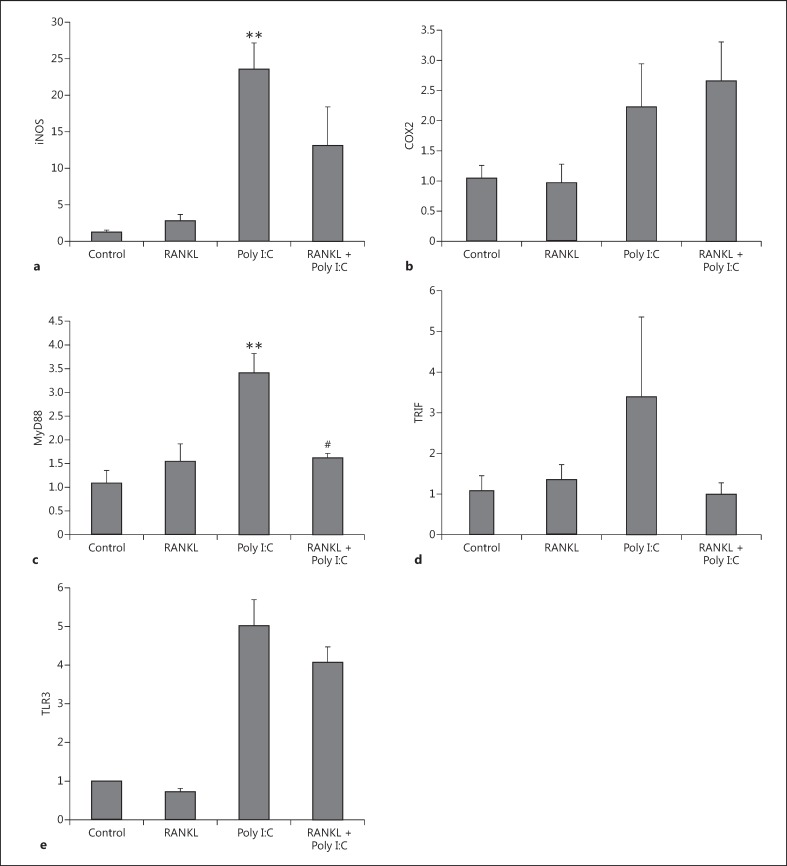
Although the BV2 cell line mimics many of the properties of microglia, we confirmed the effects of RANKL pretreatment in primary mouse microglia cultures exposed to Poly I:C (Fig. 6). We observed a significant inhibition of Poly I:C-induced MyD88 overexpression if the culture was pretreated with RANKL (Fig. 6c). We also observed a trend towards the inhibition of iNOS (Fig. 6a), TRIF (Fig. 6d), and TLR3 (Fig. 6e) overexpression in the same experiment.
Fig. 6.
Expression levels of inflammatory markers iNOS, COX2 of adaptor proteins MyD88 and TRIF, and TLR3 in primary mouse microglia after RANKL pretreatment and Poly I:C treatment. Primary mouse microglia cells were pretreated with 200 ng/mL recombinant RANKL for 24 h before the treatment with 10 µg/mL Poly I:C. mRNA levels of iNOS (a), COX2 (b), MyD88 (c), TRIF (d), and TLR3 (e) were measured 6 h later by RT-qPCR and normalized with respect to the reference gene GAPDH. Bars, mean; error bars, SEM. Results are expressed as fold change compared with non-treated control cells. Statistical significance was calculated with respect to non-treated controls (** p < 0.01) or Poly I:C-treated non-RANKL-treated cells (#p < 0.05). n = 3.
Development of a Stable BV2 RANK-/- Cell Line Using CRISPR/Cas9 Technology
In order to verify that the effects observed after RANKL pretreatment were caused by the activation of the RANK signalling pathway, we generated a BV2 cell line lacking the RANK receptor (BV2 RANK-/-) using CRISPR/Cas9 technology. This RANK knockout BV2 cell line was generated by co-transfecting the cells with HDR and Cas9 plasmids (Fig. 7a). In the first 24–48 h after co-transfection, the cells successfully transfect with Cas9 plasmid display green fluorescence, and in the next 24–48 h, the green fluorescence is gradually lost and substituted by red fluorescence in the cells where the double-transfection with Cas9 and HDR plasmids has been successful (Fig. 7b). Cells with genomic (permanent) insertion of the puromycin resistance gene were positively selected for growth in puromycin-supplemented medium. The selected cells display intensive red fluorescence (Fig. 7c).
Fig. 7.
Generation of RANK knockout BV2 cell line. a Schematic representation of the process of generation of the homo-knockout RANK BV2 cell line. b Fluorescent live imaging of BV2 cells transfected with Cas9 and HDR plasmids; 48 h after the transfection, green cells (expressing GFP from the Cas9 plasmid) and red cells (expressing RFP from the HDR plasmid) can be seen. c Fluorescent live imaging of puromycin-selected clones of BV2 cells; 1 week after the transfection, all cells are expressing RFP from the HDR plasmid construct introduced into the cell genomic DNA. d qPCR verification of the deletion in the RANK gene after the first transfection; normal BV2 cells (RANK wtBV2; blue line) with CT = 29.00, Cas9/HDR-transfected BV2 (RANK cas9BV2; purple line) CT = 30.60, and the control reactions with the 18S RNA gene normal BV2 cells (18S wtBV2; magenta line) CT = 28.77, Cas9/HDR-transfected BV2 (18S cas9BV2; orange line) CT = 29.05.
We picked 4 different BV2 colonies and analyzed them by direct DNA sequencing (Beckman Coulter Genomics, UK) with sequencing primers generated from the puromycin gene (Puromycin_Seq_F; Table 1). Based on the information from the sequencing, we designed a set of primers (Mouse RANK_Genot_F and Mouse RANK_Genot_F; Table 1) with the reverse primer spanning the site of the HDR insert. Using this set of genotyping primers, we verified the RANK gene knockout by qPCR. The CT values calculated for the cells with both RANK genes intact was around 30, while the cells carrying only 1 intact copy of the RANK gene showed CT values of approximately 31 (Fig. 7d). The gene for 18S ribosomal RNA was used as a control for the total amount of genomic DNA in the reaction [27]. Unfortunately, all 4 colonies selected were heterozygous; only 1 of the RANK genes cleaved while preserving the intact gene in the other homologous chromosome. For this reason, we transduced one of the clones with the Cre recombinase gene-carrying adenovirus VQAd Cre (ViraQuest, USA). The Cre recombinase excised the RFP and puromycin genes from the cell genome, making the cells sensitive to puromycin and non-fluorescent again. After selecting non-fluorescent cells, we performed a second double-transfection with Cas9 and HDR plasmids (Fig. 7a). The colonies selected after the second transformation were double knockouts for the RANK gene, verified by a non-detectable signal using our qPCR assay.
Knocking out the RANK Receptor from BV2 Cells Abolishes the Anti-Inflammatory Effect of Recombinant RANKL Pretreatment
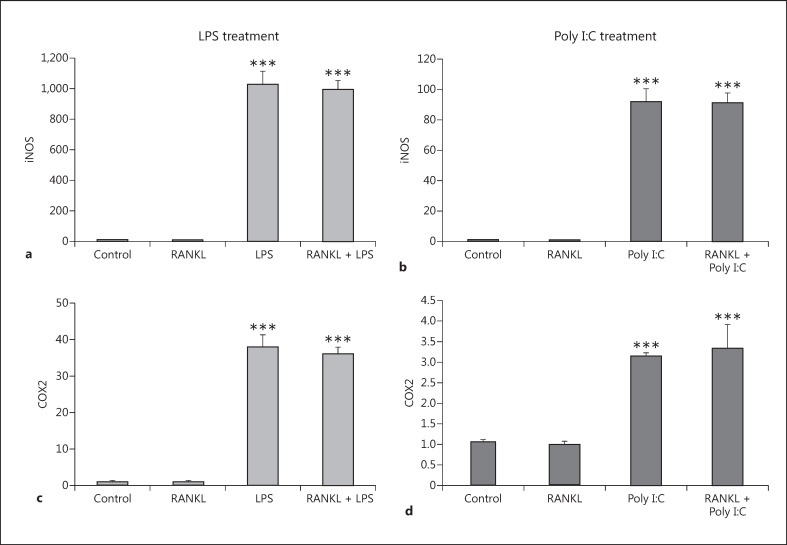
To characterize our new BV2 RANK-/- cell line, we first explored the response of the inflammatory markers iNOS and COX2 to inflammatory LPS or Poly I:C in the presence and absence of RANKL pretreatment. Unlike the previous BV2 results (Fig. 3), the RANKL pretreatment was unable to prevent the significant upregulation of iNOS and COX2 caused by both LPS (Fig. 8a, 8c) and Poly I:C (Fig. 8b, 8d). This finding confirms that RANK is essential for the reduction in the expression of inflammatory markers observed in the BV2 wild-type mice cells due to RANKL pretreatment.
Fig. 8.
Expression levels of inflammatory markers iNOS and COX2 in the RANK-/- BV2 cell line after RANKL pretreatment and LPS or Poly I:C treatment. RANK-/- BV2 cells were pretreated with 200 ng/mL recombinant RANKL for 24 h before the treatment with 10 ng/mL LPS (a, c) or 10 µg/mL Poly I:C (b, d). mRNA levels of iNOS (a, b) and COX2 (c, d) were measured 6 h later by RT-qPCR and normalized with respect to the reference gene GAPDH. Bars, mean; error bars, SEM. Results are expressed as fold change compared with non-treated control cells. Statistical significance was calculated with respect to non-treated controls (*** p < 0.001) or LPS/ Poly I:C-treated cells. n = 6 for control and RANK; n = 3 for LPS, Poly I:C, RANKL + LPS, and RANKL + Poly I:C.
Knocking out the RANK Receptor from BV2 Cells Does Not Alter the Inhibition of TLR3 Expression Caused by RANKL Pretreatment
As previously, we assessed the expression levels of TLR3 and TLR4 in the BV2 RANK-/- cell line after the RANKL pretreatment followed by LPS and Poly I:C. Interestingly, unlike with the inflammatory markers iNOS and COX2, here the significant decrease in the expression of TLR3 after RANKL pretreatment was not abolished by the removal of the RANK receptor (Fig. 9a, 9b). This may suggest an alternative unreported receptor with a RANKL-specific affinity for TLR3 signalling. TLR4 expression remained unaffected (Fig. 9c, 9d).
Fig. 9.
Expression levels of TLR3 and TLR4 in the RANK-/- BV2 cell line after RANKL pretreatment and LPS or Poly I:C treatment. RANK-/- BV2 cells were pretreated with 200 ng/mL recombinant RANKL for 24 h before the treatment with 10 ng/mL LPS (a, c) or 10 µg/mL Poly I:C (b, d). mRNA levels of TLR3 (a, b) and TLR4 (c, d) were measured 6 h later by RT-qPCR and normalized with respect to the reference gene GAPDH. Bars, mean; error bars, SEM. Results are expressed as fold change compared with non-treated control cells. Statistical significance was calculated with respect to non-treated controls (** p < 0.01; *** p < 0.001) or LPS/Poly I:C non-RANKL-treated cells (##p < 0.01). n = 6 for control and RANK; n = 3 for LPS, Poly I:C, RANKL + LPS, and RANKL + Poly I:C.
BV2 RANK-/- Cells Are Resistant to the Reduction in Expression of the Adaptor Proteins MyD88 and TRIF Caused by RANKL Pretreatment
The expression levels of the adaptor proteins MyD88 and TRIF were studied in experiments performed with BV2 RANK-/- cells. Here again, as with the inflammatory markers iNOS and COX2, the absence of the RANK receptor completely abolished the effect of RANKL pretreatment over the expression levels of MyD88 and TRIF (Fig. 10).
Fig. 10.
Expression levels of adaptor proteins MyD88 and TRIF in the RANK-/- BV2 cell line after RANKL pretreatment and LPS or Poly I:C treatment. RANK-/- BV2 cells were pretreated with 200 ng/mL recombinant RANKL for 24 h before the treatment with 10 ng/mL LPS (a, c) or 10 µg/mL Poly I:C (b, d). mRNA levels of MyD88 (a, b) and TRIF (c, d) were measured 6 h later by RT-qPCR and normalized with respect to the reference gene GAPDH. Bars, mean; error bars, SEM. Results are expressed as fold change compared with non-treated control cells. Statistical significance was calculated with respect to non-treated controls (* p < 0.05; ** p < 0.01; *** p < 0.001). n = 6 for control and RANK; n = 3 for LPS, Poly I:C, RANKL + LPS, and RANKL + Poly I:C.
Discussion
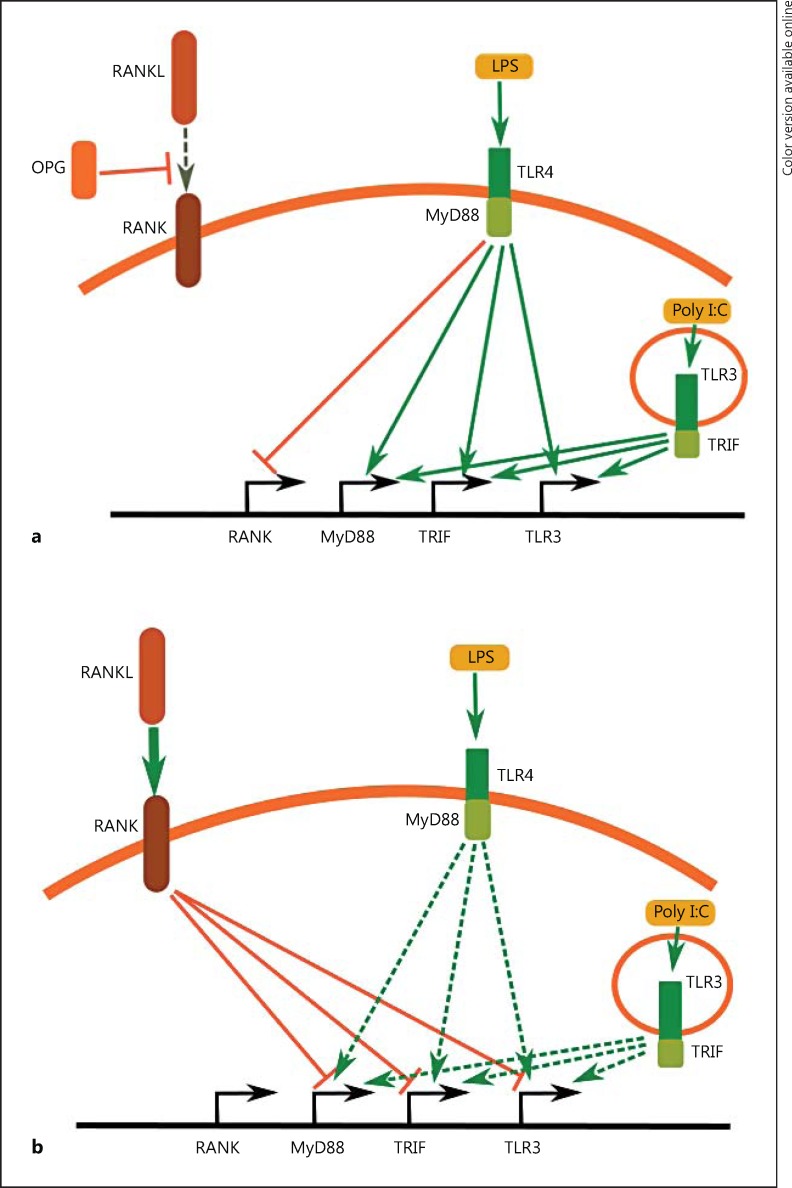
In this study, we showed a possible mechanism for the regulation of the microglial responses to LPS and Poly I:C by the RANKL/RANK pathway. We propose that there is an antagonistic relationship between TLR3/TLR4 signalling and RANK signalling (Fig. 11). The activation of TLR4 by LPS suppresses the expression of the RANK receptor in a similar way to TNF-α/INF-γ treatment. LPS treatment concomitantly increases the expression of TLR3 and the adaptor proteins MyD88 and TRIF. A similar activation of TLR3 by Poly I:C was observed, with the exception of the suppression of RANK expression. Increased levels of TLR3, MyD88 and TRIF suggest that the cells are becoming more susceptible to consecutive inflammatory stimuli, and this may result in a self-perpetuating increase of the inflammation response (Fig. 11a).
Fig. 11.
Antagonistic action of TLR3/TLR4 signalling and RANK signalling. a Activation of TLR4 suppresses the expression of the RANK receptor and increases the expression of TLR3 and the adaptor proteins MyD88 and TRIF. A similar action of TLR3 activation was observed, with the exception of the suppression of RANK. This mechanism results in a self-perpetuating increase of the inflammation. b Pretreatment of BV2 cells with recombinant RANKL decreases the ability of TLR3/TLR4 signalling to activate the expression of TLR3, MyD88, and TRIF. This suppresses BV2 microglia activation, confirmed by the reduction of the inflammatory markers iNOS and COX2.
At the same time, the inhibition of RANK signalling after HI insult in the neonatal mouse brain caused by overexpression of the decoy receptor OPG [12] is not restricted only to the microglial cells, as OPG is markedly overexpressed in primary neurons. In primary microglia treated with the inflammatory cytokines TNF-α/INF-γ or subjected to OGD, the levels of RANK and RANKL were significantly decreased (Fig. 2). A similar decrease in the expression of RANK in the BV2 cells was observed after the treatment with LPS (Fig. 4e). All these changes suggest that, during inflammation, the OPG/RANKL ratio shifts in favour of OPG and, as a consequence, the level of free RANKL able to bind the RANK receptor will decrease significantly. This, combined with the significant decrease in the RANK receptor in microglial cells, suggests a significant decrease in RANK signalling in the microglial cells. Previous studies have identified the importance of the RANK signalling pathway in inflammation in the ischaemic brain. In an adult mouse model of HI, animals lacking OPG (OPG-/-) experienced a reduction in infarct volume accompanied by reduced post-ischaemic inflammation [16]. In the same study, the authors showed that enhancing RANKL/RANK signalling in wild-type animals by recombinant RANKL is able to significantly reduce the infarct volume.
In our experiments, pretreatment of BV2 cells with recombinant RANKL decreased the ability of TLR3/TLR4 signalling to activate the expression of TLR3, MyD88, and TRIF. This suppressed BV2 microglia activation, confirmed by the reduction of the inflammatory markers iNOS and COX2. In addition, our previous work suggested that MyD88 plays a pivotal role in LPS-induced sensitization to HI, resulting in the potentiation of neonatal brain injury [28]. Therefore, keeping the expression of adaptor proteins at physiological levels by RANK activation is important to prevent the overactivation of TLR signalling in microglial cells. All the effects of recombinant RANKL treatment are present only if cells are treated prior to the inflammatory insult. In our experiments we failed to observe any effect if the cells were treated with RANKL after/at the same time as LPS and Poly I:C (data not shown). This may be due to a much faster metabolic reaction to the inflammatory stimuli shutting down the RANK signalling (including the downregulation of RANK expression; Fig. 4e), thus preventing the action of RANKL.
Another interesting finding was the ability of LPS treatment to significantly increase the expression of TLR3 and the TRIF adaptor, which suggests that the activation of TLR4 can render the microglial cells more sensitive to dsRNA/Poly I:C. On the other hand, the activation of TLR3 was able to increase the expression of the MyD88 adaptor but not of TLR4. As MyD88 is the adaptor protein for all TLRs (except for TLR3) this may suggest that the activation of either TLR3 or TLR4 will increase the sensitivity of the microglial cells to inflammatory stimuli in general.
The removal of the RANK receptor from BV2 cells abolished the effect of RANKL pretreatment, except for the increase in TLR3 expression caused by the LPS and Poly I:C treatment. This unexpected finding can be attributed to an undescribed alternative receptor of RANKL. As most of the TNF receptor family members are similar in structure, it is not unlikely that RANKL binds and activates another receptor of the same family with a lower affinity, in the same way that OPG is able to bind both RANKL and TRAIL, or TNF-α and lymphotoxin-α both bind TNF receptor-1 and TNF receptor-2 [29].
The method of generating the RANK knockout BV2 cell line described in this paper using commercial CRISPR/Cas9 technology is a relatively cheap and quick way of generating genetically modified cells. Compared with traditional ways of decreasing the expression levels of certain proteins using siRNA, here we had 100% removal of the target protein in all cells in our culture which was maintained through subsequent passages in the cell line. The precision of the CRISPR/Cas9 in targeting the desired gene substantially decreases the chances of off-site effects. Because of all these features, the results we obtained were very reliable and reproducible. This technology can be used as a valuable tool for proof-of-concept as well as for studying the effect of single or multiple gene knockouts in a cell line. The convenient production of both homo- and hetero-knockout clones using this technique and the use of the same cell line for multiple-gene manipulation makes this system a valuable tool for in vitro research. Special attention should be focussed on the fact that many of the cell lines available are polypoid and >2 rounds of transfection may be needed in order to achieve the full removal of the target gene from the genome.
To summarize our findings, we conclude that RANK signalling plays an important role in the regulation of microglial responses to inflammatory stimuli. The blockage of this signalling pathway may lead to the overstimulation of microglial cells, and, as a consequence, exacerbate damage after brain injury. In contrast, the initiation of RANK signalling decreases the inflammatory response and has the potential to be used as a treatment after brain injury. This study is the first to implicate RANK signalling in the regulation of perinatal brain injury. More studies are therefore needed to fully understand the impact of RANK signalling.
Disclosure Statement
No conflict of interest exists.
Acknowledgments
We gratefully acknowledge the support of the Department of Perinatal Imaging and Health and financial support from the Wellcome Trust (WT094823), the Medical Research Council, the Swedish Medical Research Council (VR 2015–02493), the Brain Foundation (HH), the Ahlen Foundation (HH), ALF-GBG (426401), ERA-net (EU;VR 529-2014-7551), and the Leducq Foundation (DSRRP34404), Action Medical Research and The Henry Smith Charity to enable the completion of this study. In addition, the authors acknowledge financial support from the Department of Health via the National Institute for Health Research (NIHR) Comprehensive Biomedical Research Centre Award to Guy's & St Thomas' NHS Foundation Trust in partnership with King's College London and King's College Hospital NHS Foundation Trust.
References
- 1.Nakajima K, Kohsaka S. Microglia: activation and their significance in the central nervous system. J Biochem. 2001;130:169–175. doi: 10.1093/oxfordjournals.jbchem.a002969. [DOI] [PubMed] [Google Scholar]
- 2.Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 3.Hanisch U-KK, Kettenmann H. Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat Neurosci. 2007;10:1387–1394. doi: 10.1038/nn1997. [DOI] [PubMed] [Google Scholar]
- 4.Botos I, Segal DM, Davies DR. The structural biology of Toll-like receptors. Structure. 2011;19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee BL, Barton GM. Trafficking of endosomal Toll-like receptors. Trends Cell Biol. 2014;24:360–369. doi: 10.1016/j.tcb.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perry VH, Perry VH, Nicoll JAR, Holmes C. Microglia in neurodegenerative disease. Nat Publ Gr. 2010;6:193–201. doi: 10.1038/nrneurol.2010.17. [DOI] [PubMed] [Google Scholar]
- 7.Loane DJ, Kumar A. Microglia in the TBI brain: the good, the bad, and the dysregulated. Exp Neurol. 2016;275:316–327. doi: 10.1016/j.expneurol.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagberg H, Mallard C, Ferriero DM, Vannucci SJ, Levison SW, Vexler ZS, et al. The role of inflammation in perinatal brain injury. Nat Rev Neurol. 2015;11:192–208. doi: 10.1038/nrneurol.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson KB. Infection in pregnancy and cerebral palsy. Dev Med Child Neurol. 2009;51:253–254. doi: 10.1111/j.1469-8749.2008.03256.x. [DOI] [PubMed] [Google Scholar]
- 10.Svedin P, Kjellmer I, Welin A-K, Blad S, Mallard C. Maturational effects of lipopolysaccharide on white-matter injury in fetal sheep. J Child Neurol. 2005;20:960–964. doi: 10.1177/08830738050200120501. [DOI] [PubMed] [Google Scholar]
- 11.Hagberg H, Mallard C. Effect of inflammation on central nervous system development and vulnerability. Curr Opin Neurol. 2005;18:117–123. doi: 10.1097/01.wco.0000162851.44897.8f. [DOI] [PubMed] [Google Scholar]
- 12.Kichev A, Rousset CI, Baburamani A a, Levison SW, Wood TL, Gressens P, et al. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) signaling and cell death in the immature central nervous system after hypoxia-ischemia and inflammation. J Biol Chem. 2014;289:9430–9439. doi: 10.1074/jbc.M113.512350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vitovski S, Phillips JS, Sayers J, Croucher PI. Investigating the interaction between osteoprotegerin and receptor activator of NF-κB or tumor necrosis factor-related apoptosis-inducing ligand: evidence for a pivotal role for osteoprotegerin in regulating two distinct pathways. J Biol Chem. 2007;282:31601–31609. doi: 10.1074/jbc.M706078200. [DOI] [PubMed] [Google Scholar]
- 14.Mc Guire C, Beyaert R, van Loo G. Death receptor signalling in central nervous system inflammation and demyelination. Trends Neurosci. 2011;34:619–628. doi: 10.1016/j.tins.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Hanada R, Leibbrandt A, Hanada T, Kitaoka S, Furuyashiki T, Fujihara H, et al. Central control of fever and female body temperature by RANKL/RANK. Nature. 2009;462:505–509. doi: 10.1038/nature08596. [DOI] [PubMed] [Google Scholar]
- 16.Shimamura M, Nakagami H, Osako MK, Kurinami H, Koriyama H, Zhengda P, et al. OPG/RANKL/RANK axis is a critical inflammatory signaling system in ischemic brain in mice. Proc Natl Acad Sci USA. 2014;111:8191–8196. doi: 10.1073/pnas.1400544111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson DM, Maraskovsky E, Billingsley WL, Dougall WC, Tometsko ME, Roux ER, et al. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature. 1997;390:175–179. doi: 10.1038/36593. [DOI] [PubMed] [Google Scholar]
- 18.Kong YY, Feige U, Sarosi I, Bolon B, Tafuri A, Morony S, et al. Activated T cells regulate bone loss and joint destruction in adjuvant arthritis through osteoprotegerin ligand. Nature. 1999;402:304–309. doi: 10.1038/46303. [DOI] [PubMed] [Google Scholar]
- 19.Simonet WS, Lacey DL, Dunstan CR, Kelley M, Chang MS, Lüthy R, et al. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell. 1997;89:309–319. doi: 10.1016/s0092-8674(00)80209-3. [DOI] [PubMed] [Google Scholar]
- 20.Hsu H, Lacey DL, Dunstan CR, Solovyev I, Colombero A, Timms E, et al. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc Natl Acad Sci USA. 1999;96:3540–3545. doi: 10.1073/pnas.96.7.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollard JW. Trophic macrophages in development and disease. Nat Rev Immunol. 2009;9:259–270. doi: 10.1038/nri2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levison SW, McCarthy KD. Characterization and partial purification of AIM: a plasma protein that induces rat cerebral type 2 astroglia from bipotential glial progenitors. J Neurochem. 1991;57:782–794. doi: 10.1111/j.1471-4159.1991.tb08220.x. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell RHB, Goldstein BI. Inflammation in children and adolescents with neuropsychiatric disorders: a systematic review. J Am Acad Child Adolesc Psychiatry. 2014;53:274–296. doi: 10.1016/j.jaac.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Henn A, Lund S, Hedtjärn M, Schrattenholz A, Pörzgen P, Leist M. The suitability of BV2 cells as alternative model system for primary microglia cultures or for animal experiments examining brain inflammation. ALTEX. 2009;26:83–94. doi: 10.14573/altex.2009.2.83. [DOI] [PubMed] [Google Scholar]
- 27.Podlesniy P, Figueiro-Silva J, Llado A, Antonell A, Sanchez-Valle R, Alcolea D, et al. Low CSF concentration of mitochondrial DNA in preclinical Alzheimer's disease. Ann Neurol. 2013;74:655–668. doi: 10.1002/ana.23955. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Stridh L, Li W, Dean J, Elmgren A, Gan L, et al. Lipopolysaccharide sensitizes neonatal hypoxic-ischemic brain injury in a MyD88-dependent manner. J Immunol. 2009;183:7471–7477. doi: 10.4049/jimmunol.0900762. [DOI] [PubMed] [Google Scholar]
- 29.Bodmer JL, Schneider P, Tschopp J. The molecular architecture of the TNF superfamily. Trends Biochem Sci. 2002;27:19–26. doi: 10.1016/s0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]