Abstract
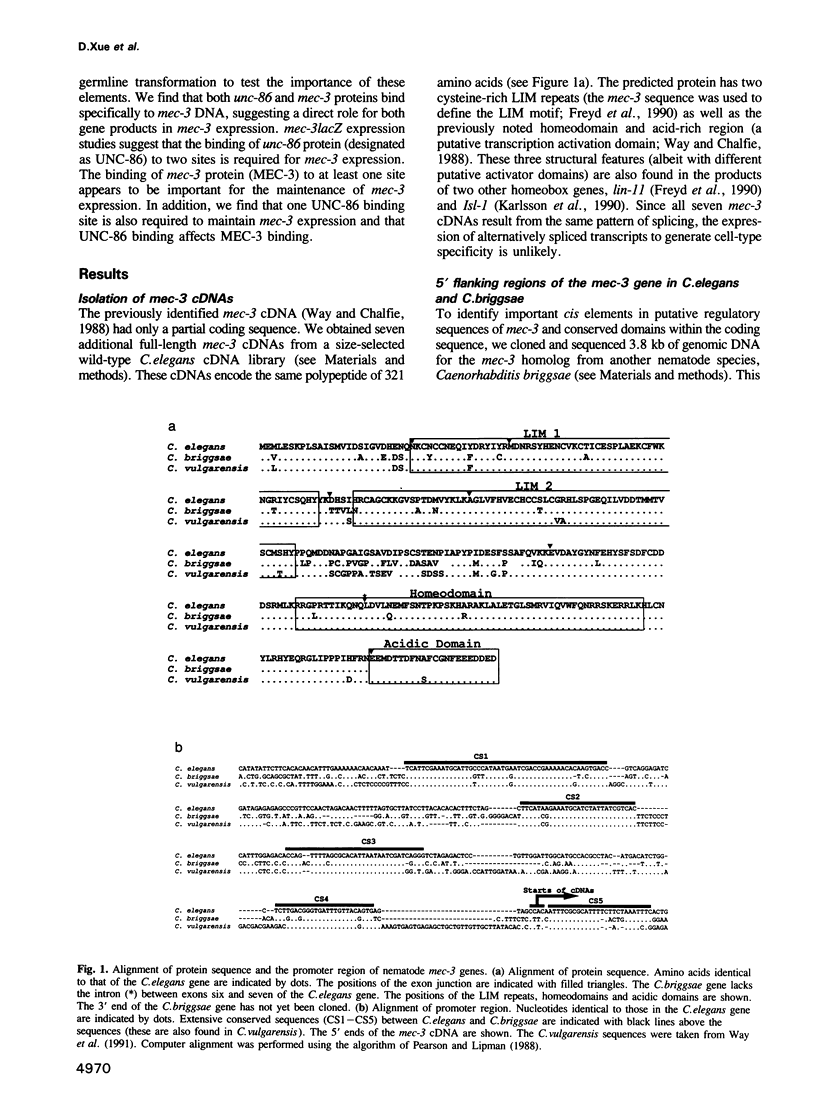
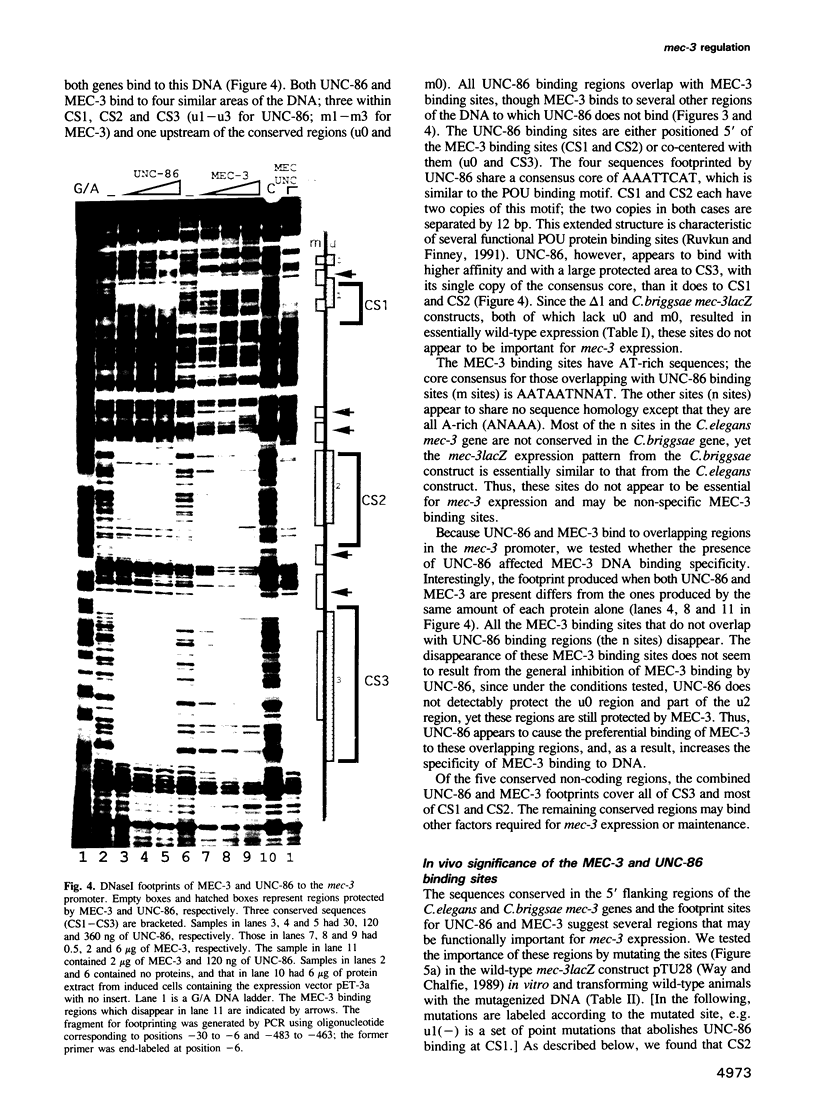
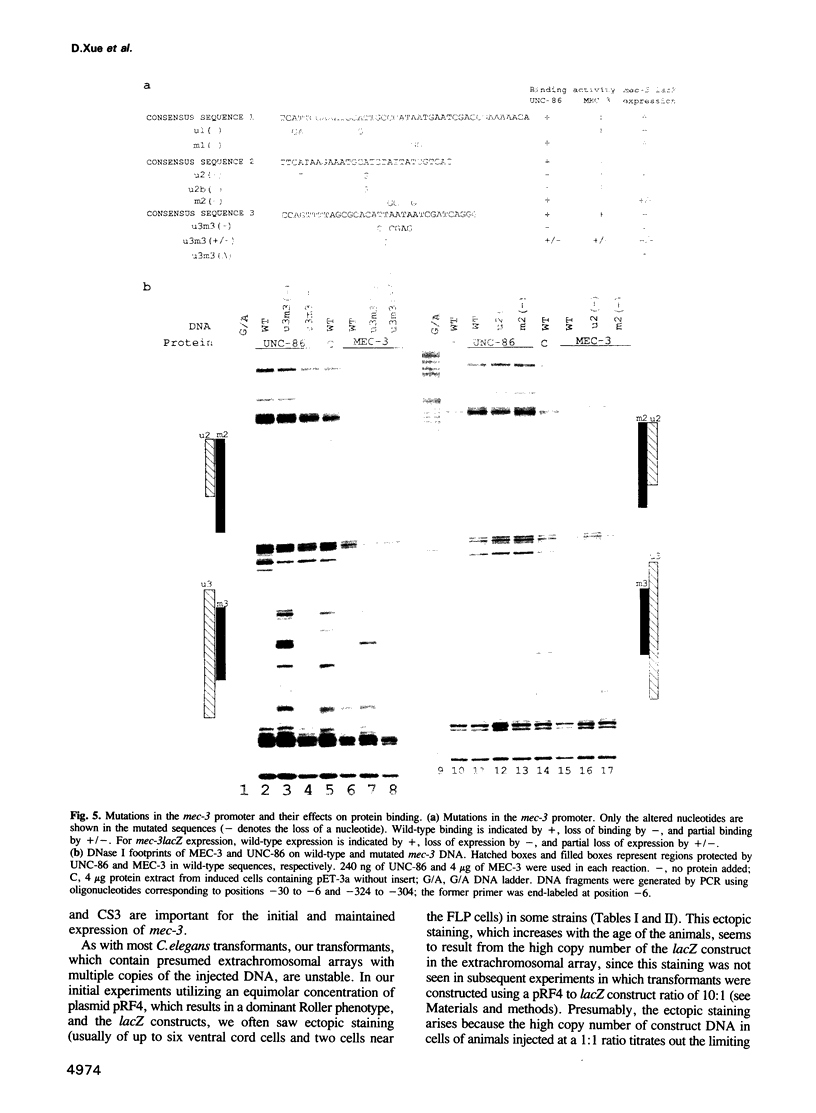
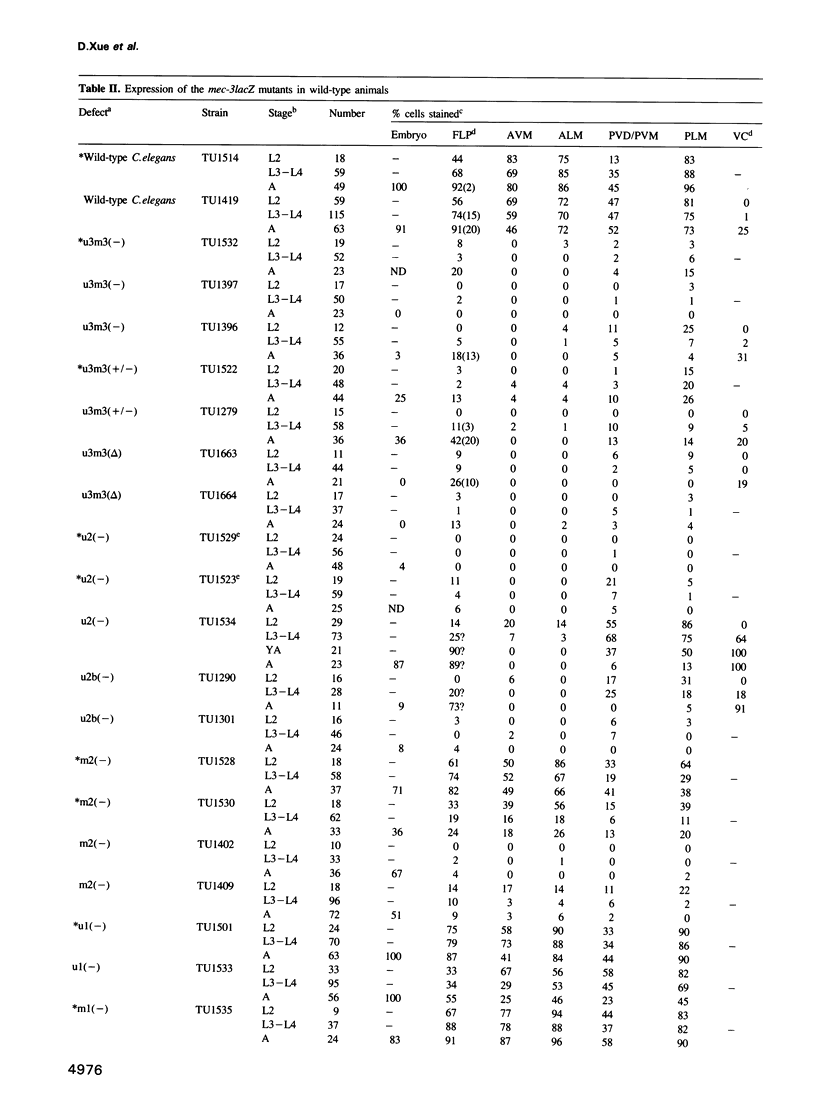
The mec-3 gene encodes a homeodomain protein with LIM repeats that is required for the specification of touch cell fate in Caenorhabditis elegans. Previous experiments suggested that mec-3 expression requires the product of the unc-86 gene, a POU-type homeoprotein, and mec-3 itself. We have analyzed the control of mec-3 expression by identifying potential cis regulatory elements in the mec-3 gene (by conservation in a related nematode and by DNase I footprinting using unc-86 and mec-3 proteins) and testing their importance by transforming C.elegans with mec-3lacZ fusions in which these sites have been mutagenized in vitro. Both unc-86 and mec-3 proteins bind specifically to the promoter of the mec-3 gene, suggesting that both proteins may be directly involved in the regulation of the mec-3 gene. In addition, the footprint pattern with mec-3 protein is altered in the presence of unc-86 protein. In vivo transformation experiments reveal that some of the binding regions of the two proteins are needed for general positive control and maintenance of mec-3 expression while others have no detectable, unique function. Interestingly, the unc-86 gene appears to be required not only to initiate mec-3 expression but also to maintain it.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974 May;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M., Au M. Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science. 1989 Feb 24;243(4894 Pt 1):1027–1033. doi: 10.1126/science.2646709. [DOI] [PubMed] [Google Scholar]
- Chalfie M., Horvitz H. R., Sulston J. E. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 1981 Apr;24(1):59–69. doi: 10.1016/0092-8674(81)90501-8. [DOI] [PubMed] [Google Scholar]
- Chalfie M., Sulston J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol. 1981 Mar;82(2):358–370. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- Driscoll M., Chalfie M. The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration. Nature. 1991 Feb 14;349(6310):588–593. doi: 10.1038/349588a0. [DOI] [PubMed] [Google Scholar]
- Emerson C. P. Myogenesis and developmental control genes. Curr Opin Cell Biol. 1990 Dec;2(6):1065–1075. doi: 10.1016/0955-0674(90)90157-a. [DOI] [PubMed] [Google Scholar]
- Finney M., Ruvkun G., Horvitz H. R. The C. elegans cell lineage and differentiation gene unc-86 encodes a protein with a homeodomain and extended similarity to transcription factors. Cell. 1988 Dec 2;55(5):757–769. doi: 10.1016/0092-8674(88)90132-8. [DOI] [PubMed] [Google Scholar]
- Finney M., Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990 Nov 30;63(5):895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- Fire A., Harrison S. W., Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990 Sep 14;93(2):189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- Freyd G., Kim S. K., Horvitz H. R. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature. 1990 Apr 26;344(6269):876–879. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- García-Blanco M. A., Clerc R. G., Sharp P. A. The DNA-binding homeo domain of the Oct-2 protein. Genes Dev. 1989 Jun;3(6):739–745. doi: 10.1101/gad.3.6.739. [DOI] [PubMed] [Google Scholar]
- Hanes S. D., Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989 Jun 30;57(7):1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- Hoey T., Levine M. Divergent homeo box proteins recognize similar DNA sequences in Drosophila. Nature. 1988 Apr 28;332(6167):858–861. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- Ip Y. T., Park R. E., Kosman D., Yazdanbakhsh K., Levine M. dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes Dev. 1992 Aug;6(8):1518–1530. doi: 10.1101/gad.6.8.1518. [DOI] [PubMed] [Google Scholar]
- Karin M. Transcriptional control and the integration of cell-autonomous and environmental cues during development. Curr Opin Cell Biol. 1990 Dec;2(6):996–1002. doi: 10.1016/0955-0674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- Karlsson O., Thor S., Norberg T., Ohlsson H., Edlund T. Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature. 1990 Apr 26;344(6269):879–882. doi: 10.1038/344879a0. [DOI] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989 Feb;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Chalfie M. Organogenesis in C. elegans: positioning of neurons and muscles in the egg-laying system. Neuron. 1990 May;4(5):681–695. doi: 10.1016/0896-6273(90)90195-l. [DOI] [PubMed] [Google Scholar]
- Li P. M., Reichert J., Freyd G., Horvitz H. R., Walsh C. T. The LIM region of a presumptive Caenorhabditis elegans transcription factor is an iron-sulfur- and zinc-containing metallodomain. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9210–9213. doi: 10.1073/pnas.88.20.9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991 Dec;10(12):3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzolo M. J., Hamilton B. A., Ding D. L., Martin C. H., Mead D. A., Mierendorf R. C., Raghavan K. V., Meyerowitz E. M., Lipshitz H. D. Phage lambda cDNA cloning vectors for subtractive hybridization, fusion-protein synthesis and Cre-loxP automatic plasmid subcloning. Gene. 1990 Mar 30;88(1):25–36. doi: 10.1016/0378-1119(90)90056-w. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg A. H., Lade B. N., Chui D. S., Lin S. W., Dunn J. J., Studier F. W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56(1):125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M. G. POU-domain transcription factors: pou-er-ful developmental regulators. Genes Dev. 1991 Jun;5(6):897–907. doi: 10.1101/gad.5.6.897. [DOI] [PubMed] [Google Scholar]
- Ruvkun G., Finney M. Regulation of transcription and cell identity by POU domain proteins. Cell. 1991 Feb 8;64(3):475–478. doi: 10.1016/0092-8674(91)90227-p. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage C., Hamelin M., Culotti J. G., Coulson A., Albertson D. G., Chalfie M. mec-7 is a beta-tubulin gene required for the production of 15-protofilament microtubules in Caenorhabditis elegans. Genes Dev. 1989 Jun;3(6):870–881. doi: 10.1101/gad.3.6.870. [DOI] [PubMed] [Google Scholar]
- Serfling E. Autoregulation--a common property of eukaryotic transcription factors? Trends Genet. 1989 May;5(5):131–133. doi: 10.1016/0168-9525(89)90049-8. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Treisman J., Gönczy P., Vashishtha M., Harris E., Desplan C. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell. 1989 Nov 3;59(3):553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
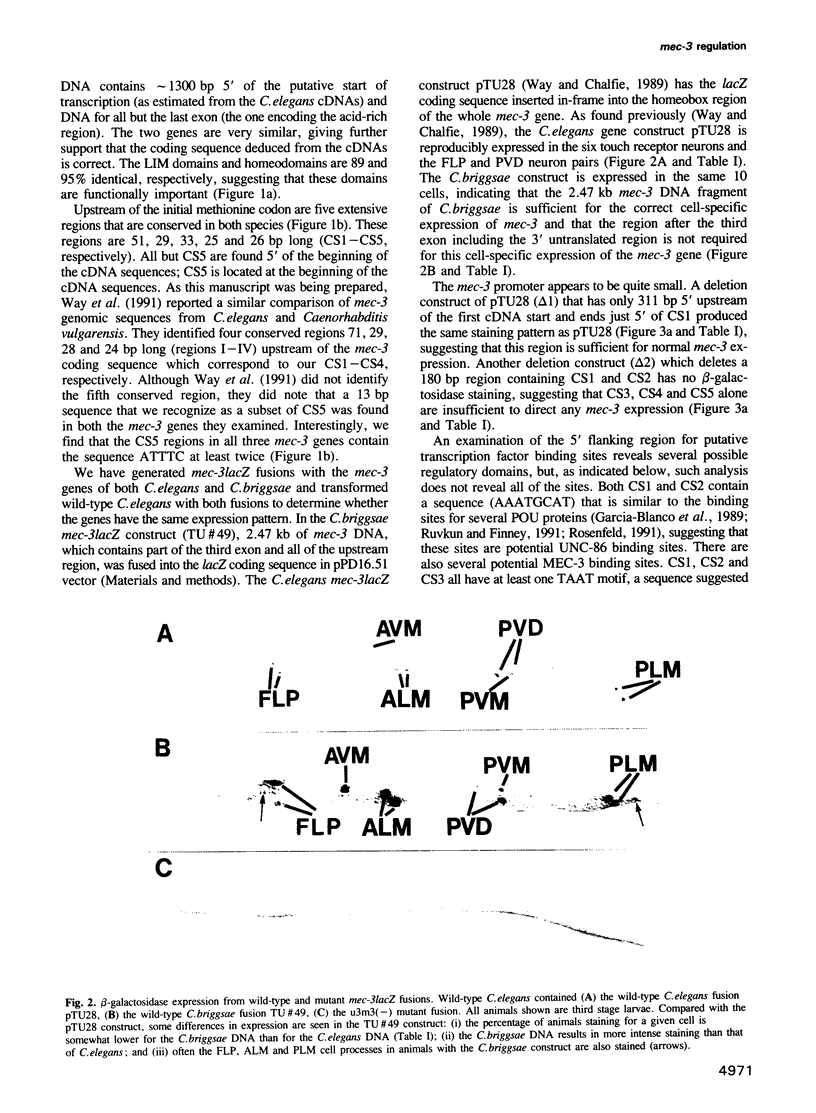
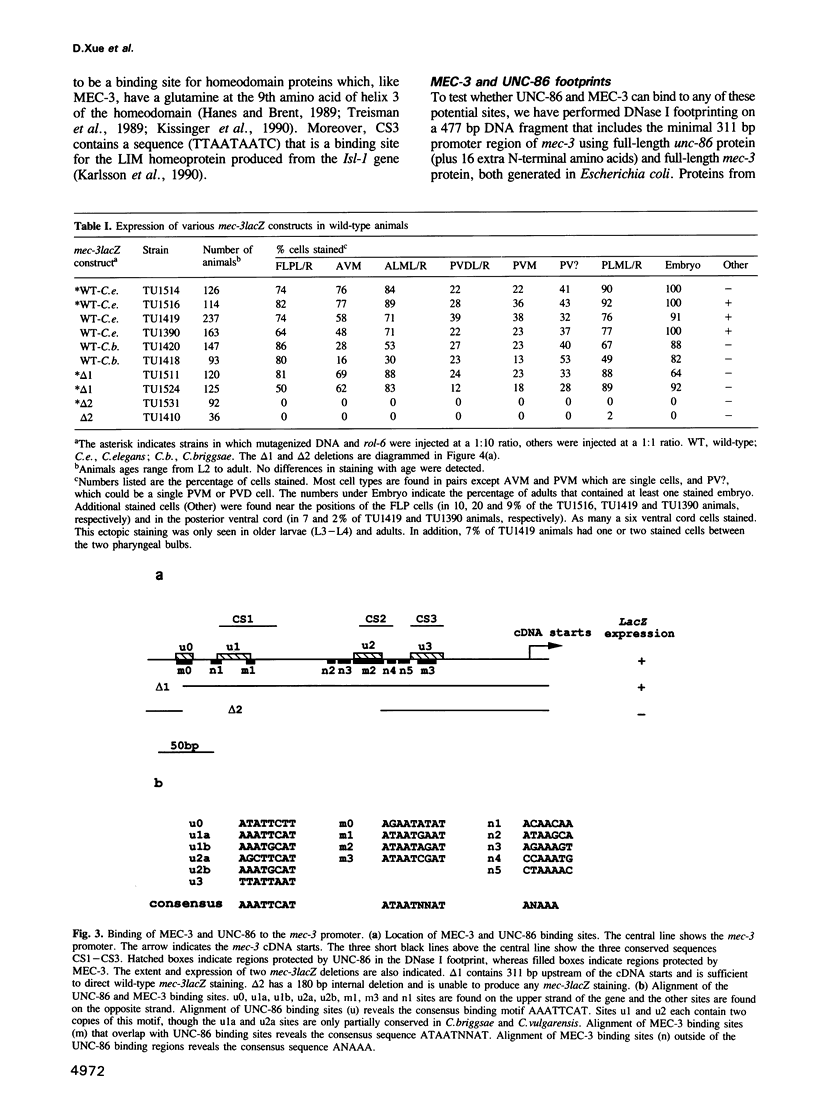
- Way J. C., Chalfie M. The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes Dev. 1989 Dec;3(12A):1823–1833. doi: 10.1101/gad.3.12a.1823. [DOI] [PubMed] [Google Scholar]
- Way J. C., Chalfie M. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell. 1988 Jul 1;54(1):5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]
- Way J. C., Wang L., Run J. Q., Wang A. The mec-3 gene contains cis-acting elements mediating positive and negative regulation in cells produced by asymmetric cell division in Caenorhabditis elegans. Genes Dev. 1991 Dec;5(12A):2199–2211. doi: 10.1101/gad.5.12a.2199. [DOI] [PubMed] [Google Scholar]