Abstract
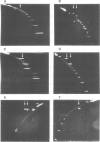
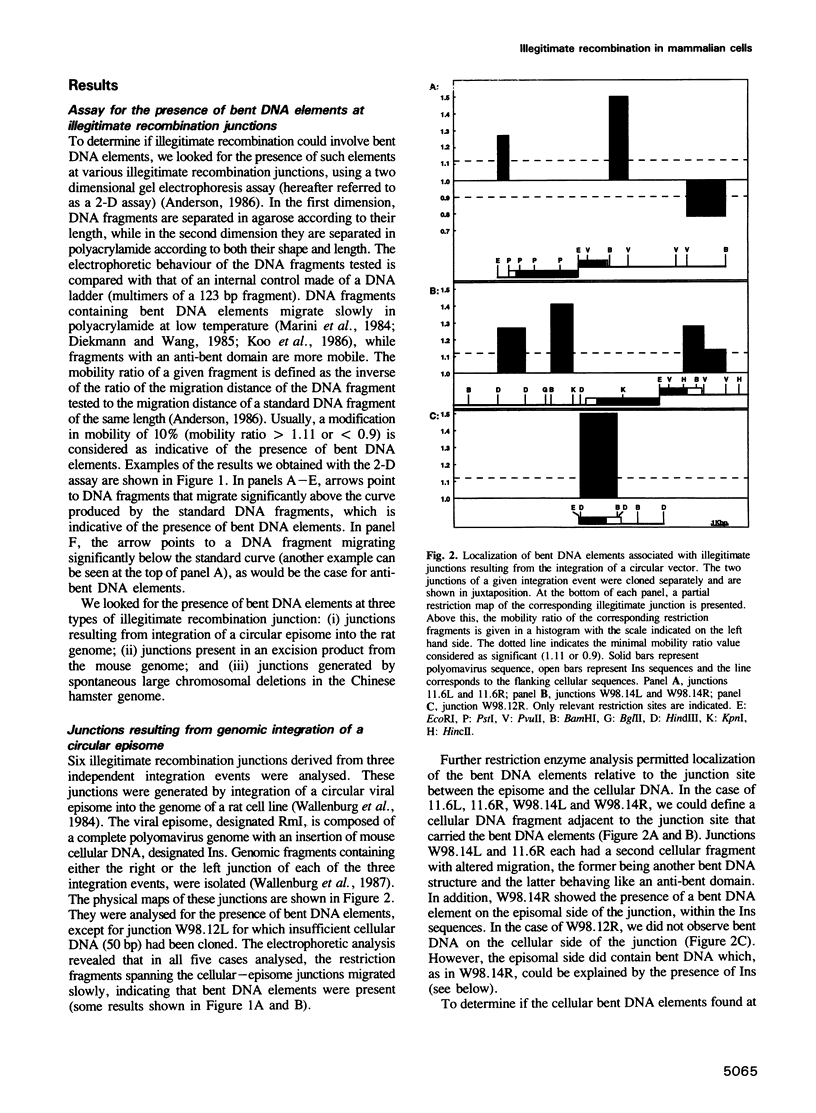
Illegitimate recombination is the most frequent mechanism for chromosomal rearrangements in mammalian cells, yet little is known about this process. Most of the studies to date have looked at the sequences present at illegitimate junctions. These revealed the presence of recurrent DNA motifs, none of which was consistently found. We have undertaken to determine if intrinsic DNA structures such as bent DNA elements could be a major determinant in chromosomal illegitimate recombination. Using a two dimensional electrophoretic assay we found that eight out of eight junctions, resulting from various types of chromosomal rearrangements, had migration behaviour characteristic of DNA containing intrinsically bent DNA elements. In all cases, these occurred within one kilobase of the junctions, and in most cases could be found in both participating DNA segments. We also found that these bent DNA elements were present before the recombination event. When we analysed the frequency of intrinsically bent DNA elements in random chromosomal fragments, we found it to be about one per 11 kilobases. Thus these results suggest that bent DNA is associated with chromosomal illegitimate recombination.
Full text
PDF

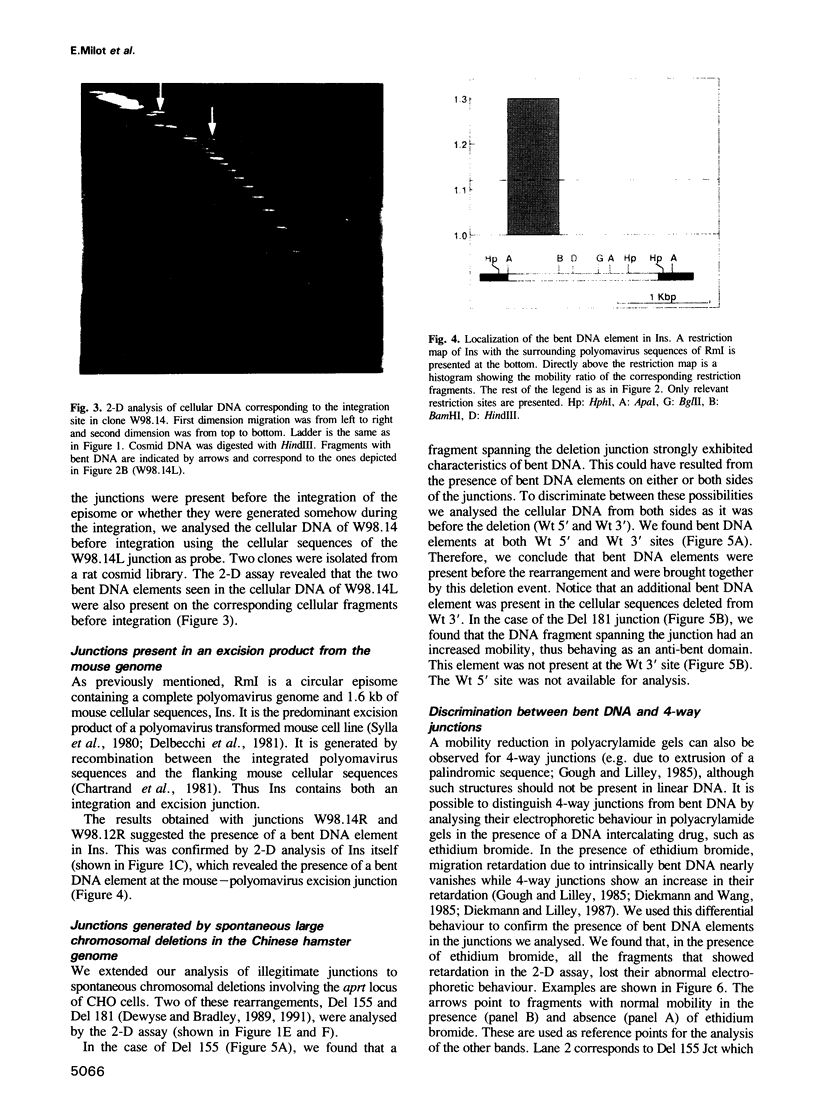
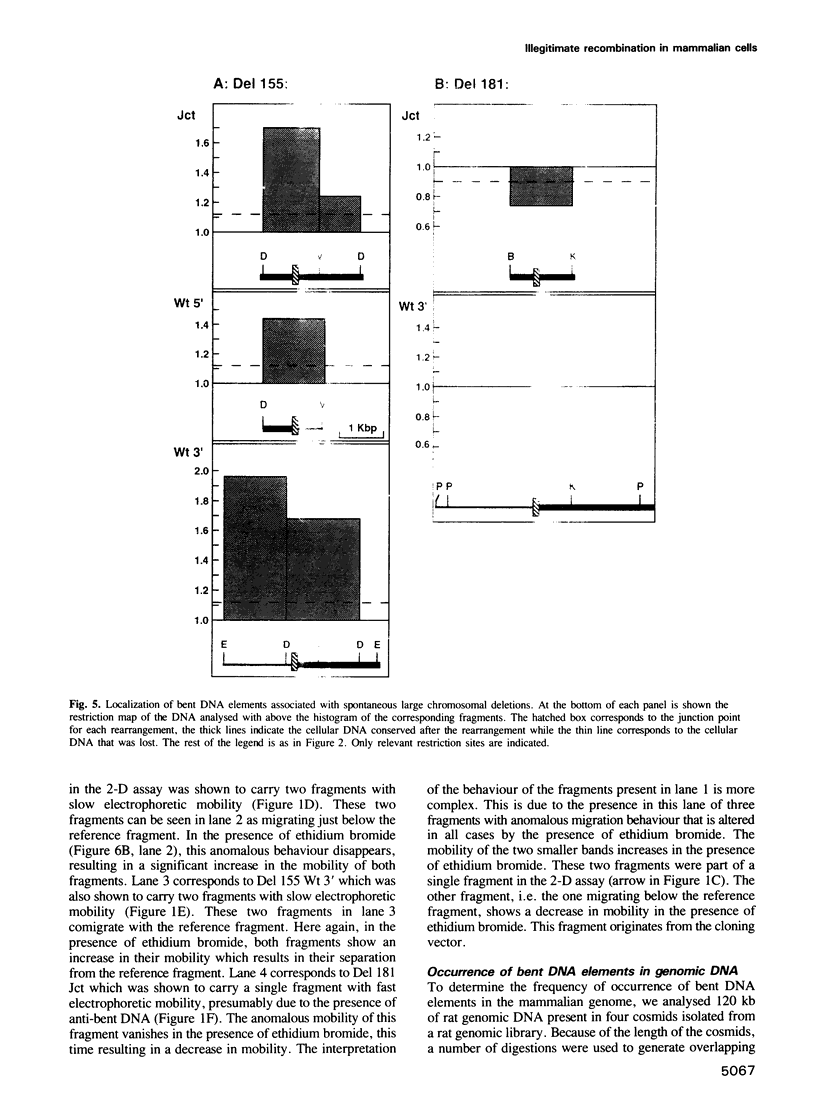



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. N. Detection, sequence patterns and function of unusual DNA structures. Nucleic Acids Res. 1986 Nov 11;14(21):8513–8533. doi: 10.1093/nar/14.21.8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouillette S., Chartrand P. Intermolecular recombination assay for mammalian cells that produces recombinants carrying both homologous and nonhomologous junctions. Mol Cell Biol. 1987 Jun;7(6):2248–2255. doi: 10.1128/mcb.7.6.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddle M. S., Lussier R. H., Heintz N. H. Intramolecular DNA triplexes, bent DNA and DNA unwinding elements in the initiation region of an amplified dihydrofolate reductase replicon. J Mol Biol. 1990 Jan 5;211(1):19–33. doi: 10.1016/0022-2836(90)90008-A. [DOI] [PubMed] [Google Scholar]
- Caserta M., Amadei A., Di Mauro E., Camilloni G. In vitro preferential topoisomerization of bent DNA. Nucleic Acids Res. 1989 Nov 11;17(21):8463–8474. doi: 10.1093/nar/17.21.8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand P., Gusew-Chartrand N., Bourgaux P. Integrated polyoma genomes in inducible permissive transformed cells. J Virol. 1981 Jul;39(1):185–195. doi: 10.1128/jvi.39.1.185-195.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Delbecchi L., Gendron D., Bourgaux P. Inducible permissive cells transformed by a temperature-sensitive polyoma virus: superinfection does not allow excision of the resident viral genome. J Virol. 1981 Jul;39(1):196–206. doi: 10.1128/jvi.39.1.196-206.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewyse P., Bradley W. E. A very large spontaneous deletion at aprt locus in CHO cells: sequence similarities with small aprt deletions. Somat Cell Mol Genet. 1991 Jan;17(1):57–68. doi: 10.1007/BF01233205. [DOI] [PubMed] [Google Scholar]
- Dewyse P., Bradley W. E. High-frequency deletion event at aprt locus of CHO cells: detection and characterization of endpoints. Somat Cell Mol Genet. 1989 Jan;15(1):19–28. doi: 10.1007/BF01534666. [DOI] [PubMed] [Google Scholar]
- Diekmann S., Lilley D. M. The anomalous gel migration of a stable cruciform: temperature and salt dependence, and some comparisons with curved DNA. Nucleic Acids Res. 1987 Jul 24;15(14):5765–5774. doi: 10.1093/nar/15.14.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann S., Wang J. C. On the sequence determinants and flexibility of the kinetoplast DNA fragment with abnormal gel electrophoretic mobilities. J Mol Biol. 1985 Nov 5;186(1):1–11. doi: 10.1016/0022-2836(85)90251-7. [DOI] [PubMed] [Google Scholar]
- Dijkwel P. A., Hamlin J. L. Matrix attachment regions are positioned near replication initiation sites, genes, and an interamplicon junction in the amplified dihydrofolate reductase domain of Chinese hamster ovary cells. Mol Cell Biol. 1988 Dec;8(12):5398–5409. doi: 10.1128/mcb.8.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckdahl T. T., Anderson J. N. Conserved DNA structures in origins of replication. Nucleic Acids Res. 1990 Mar 25;18(6):1609–1612. doi: 10.1093/nar/18.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. The organisation of chromatin loops: characterization of a scaffold attachment site. EMBO J. 1986 Mar;5(3):511–518. doi: 10.1002/j.1460-2075.1986.tb04240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W., Marchionni M., McKnight G. On the antiquity of introns. Cell. 1986 Jul 18;46(2):151–153. doi: 10.1016/0092-8674(86)90730-0. [DOI] [PubMed] [Google Scholar]
- Goodman S. D., Nash H. A. Functional replacement of a protein-induced bend in a DNA recombination site. Nature. 1989 Sep 21;341(6239):251–254. doi: 10.1038/341251a0. [DOI] [PubMed] [Google Scholar]
- Gough G. W., Lilley D. M. DNA bending induced by cruciform formation. Nature. 1985 Jan 10;313(5998):154–156. doi: 10.1038/313154a0. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- Hartley J. L., Gregori T. J. Cloning multiple copies of a DNA segment. Gene. 1981 May;13(4):347–353. doi: 10.1016/0378-1119(81)90014-7. [DOI] [PubMed] [Google Scholar]
- Homberger H. P. Bent DNA is a structural feature of scaffold-attached regions in Drosophila melanogaster interphase nuclei. Chromosoma. 1989 Aug;98(2):99–104. doi: 10.1007/BF00291044. [DOI] [PubMed] [Google Scholar]
- Howard M. T., Lee M. P., Hsieh T. S., Griffith J. D. Drosophila topoisomerase II-DNA interactions are affected by DNA structure. J Mol Biol. 1991 Jan 5;217(1):53–62. doi: 10.1016/0022-2836(91)90610-i. [DOI] [PubMed] [Google Scholar]
- Hyrien O., Debatisse M., Buttin G., de Saint Vincent B. R. A hotspot for novel amplification joints in a mosaic of Alu-like repeats and palindromic A + T-rich DNA. EMBO J. 1987 Aug;6(8):2401–2408. doi: 10.1002/j.1460-2075.1987.tb02518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelewski N. Structure and function of an AT-rich, interspersed repetitive sequence from Chironomus thummi: solenoidal DNA, 142 bp palindrome-frame and homologies with the sequence for site-specific recombination of bacterial transposons. Nucleic Acids Res. 1983 Oct 25;11(20):6985–6996. doi: 10.1093/nar/11.20.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Linial M., Shlomai J. A unique endonuclease from Crithidia fasciculata which recognizes a bend in the DNA helix. Specificity of the cleavage reaction. J Biol Chem. 1988 Jan 5;263(1):290–297. [PubMed] [Google Scholar]
- Linial M., Shlomai J. Bent DNA structures associated with several origins of replication are recognized by a unique enzyme from trypanosomatids. Nucleic Acids Res. 1988 Jul 25;16(14A):6477–6492. doi: 10.1093/nar/16.14.6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linial M., Shlomai J. Sequence-directed bent DNA helix is the specific binding site for Crithidia fasciculata nicking enzyme. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8205–8209. doi: 10.1073/pnas.84.23.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linial M., Shlomai J. The sequence-directed bent structure in kinetoplast DNA is recognized by an enzyme from Crithidia fasciculata. J Biol Chem. 1987 Nov 5;262(31):15194–15201. [PubMed] [Google Scholar]
- Marini J. C., Effron P. N., Goodman T. C., Singleton C. K., Wells R. D., Wartell R. M., Englund P. T. Physical characterization of a kinetoplast DNA fragment with unusual properties. J Biol Chem. 1984 Jul 25;259(14):8974–8979. [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination occurs between the replication origin of the plasmid pC194 and a progressing replication fork. EMBO J. 1986 Dec 20;5(13):3691–3696. doi: 10.1002/j.1460-2075.1986.tb04701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitoso de Vargas L., Kim S., Landy A. DNA looping generated by DNA bending protein IHF and the two domains of lambda integrase. Science. 1989 Jun 23;244(4911):1457–1461. doi: 10.1126/science.2544029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitoso de Vargas L., Landy A. A switch in the formation of alternative DNA loops modulates lambda site-specific recombination. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):588–592. doi: 10.1073/pnas.88.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvo J. J., Grindley N. D. The gamma delta resolvase bends the res site into a recombinogenic complex. EMBO J. 1988 Nov;7(11):3609–3616. doi: 10.1002/j.1460-2075.1988.tb03239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M., Hsieh T. S. Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985 Feb 25;13(4):1057–1072. doi: 10.1093/nar/13.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimke R. T., Sherwood S. W., Hill A. B., Johnston R. N. Overreplication and recombination of DNA in higher eukaryotes: potential consequences and biological implications. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2157–2161. doi: 10.1073/pnas.83.7.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon E., Borrow J., Goddard A. D. Chromosome aberrations and cancer. Science. 1991 Nov 22;254(5035):1153–1160. doi: 10.1126/science.1957167. [DOI] [PubMed] [Google Scholar]
- Sperry A. O., Blasquez V. C., Garrard W. T. Dysfunction of chromosomal loop attachment sites: illegitimate recombination linked to matrix association regions and topoisomerase II. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5497–5501. doi: 10.1073/pnas.86.14.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzner J. R., Chung I. K., Muller M. T. Eukaryotic topoisomerase II preferentially cleaves alternating purine-pyrimidine repeats. Nucleic Acids Res. 1990 Jan 11;18(1):1–11. doi: 10.1093/nar/18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylla B. S., Bourgaux-Ramoisy D., Bourgaux P. Induction of viral DNA synthesis in clonal derivatives of a permissive cell line transformed by a temperature-sensitive polyoma virus. Virology. 1980 Jan 30;100(2):357–369. doi: 10.1016/0042-6822(80)90527-9. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanWye J. D., Bronson E. C., Anderson J. N. Species-specific patterns of DNA bending and sequence. Nucleic Acids Res. 1991 Oct 11;19(19):5253–5261. doi: 10.1093/nar/19.19.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallenburg J. C., Nepveu A., Chartrand P. Integration of a vector containing rodent repetitive elements in the rat genome. Nucleic Acids Res. 1987 Oct 12;15(19):7849–7863. doi: 10.1093/nar/15.19.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallenburg J. C., Nepveu A., Chartrand P. Random and nonrandom integration of a polyomavirus DNA molecule containing highly repetitive cellular sequences. J Virol. 1984 Jun;50(3):678–683. doi: 10.1128/jvi.50.3.678-683.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. DNA topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- von Kries J. P., Phi-Van L., Diekmann S., Strätling W. H. A non-curved chicken lysozyme 5' matrix attachment site is 3' followed by a strongly curved DNA sequence. Nucleic Acids Res. 1990 Jul 11;18(13):3881–3885. doi: 10.1093/nar/18.13.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]