Abstract
The thymus plays a central role in self-tolerance, in part by eliminating precursors with a T cell receptor (TCR) that binds strongly to self-antigens. However, the generation of self-agonist-selected lineages also relies on strong TCR signaling. How thymocytes discriminate between these opposite outcomes remains elusive. Here we identified a human agonist-selected PD-1+ CD8αα+ subset of mature CD8αβ+ T cells that displays an effector phenotype associated with agonist selection. Interestingly, TCR stimulation of immature post-β-selection thymocyte blasts specifically gives rise to this innate subset and fixes early TRAV and TRAJ rearrangements in the TCR repertoire. These findings suggest that the checkpoint for agonist selection precedes conventional selection in human thymus.
Introduction
The generation of a diverse TCR alpha beta (TCRαβ) repertoire in the thymus is crucial for protection against foreign antigens, but at the same time it has to prevent that thymocytes expressing a TCR with strong affinity for self-antigens exit the thymus as naïve T cells. Successful rearrangements of TCR chains are therefore subjected to checkpoints where strength of TCR signaling will determine lineage outcome (1, 2). The majority of mature TCRαβ+ cells generated in the thymus display low affinity for self-peptide MHC complexes and exit the thymus as naïve CD4 or CD8αβ single positive T cells (2). Developing thymocytes with a rearranged TCR that reacts strongly with self-peptide MHC complexes could cause severe autoimmunity if allowed to enter the conventional naïve T cell pool. During thymic selection however, autoreactive immature thymocytes are either clonally deleted during a process of conventional negative selection or alternatively they can be specifically preserved and adopt distinct functional fates when developing along the agonist selection path (3, 4). In contrast to conventional naïve T cells in the spleen and lymph nodes, agonist selected T cells, such as the double negative (DN) intraepithelial T cells (IET) and the NK T cells are predominantly tissue resident cells and they display a full effector phenotype marked by the expression of natural killer (NK) receptors and cytotoxic effector molecules like granzymes and FASL (5, 6). Interestingly, they typically show unconventional MHC-restriction (7), which together with their innate functional phenotype suggests that agonist selected T cells play unique roles in immune function and regulation that are distinct from those of MHC class I- and MHC class II-restricted conventional CD8αβ+ and CD4+ TCRαβ+ subsets.
It is unclear how strong TCR activation in pre-selection thymocytes can lead to such divergent outcomes as apoptosis or agonist-selected maturation. Some studies suggested that the intensity of TCR signaling could lead to differential induction of apoptosis mediators, thereby creating a threshold for clonal deletion (8, 9). An alternative suggestion was that CD28 co-stimulation controlled the outcome of strong TCR signaling in T cell precursors since in the absence of CD28, more agonist-selected DN T cells are generated (10). The proposed mechanisms however all imply that a single progenitor T cell dies by apoptosis or matures to a functional T cell depending on the nature of the (co)stimulus. Alternatively, the precursor might be different, as a pre-selection subset of T cell progenitors in mice was found to specifically give rise to DN T cells (11). This subset is characterized by expression of CD4, CD8αβ and CD8αα (triple positive or TP) and a low level of TCRβ suggesting that these are early post-β-selection precursor cells. Finally, studies in transgenic mice have shown that the development of agonist selected DN TCRαβ+ cells is favored when a pre-rearranged transgenic TCRαβ with high affinity for a cognate self-antigen is expressed ‘pre-maturely’, before conventional selection at the DP stage, indicating that the timing of TCRα rearrangement also influences the outcome of selection (12, 13).
Natural TCR rearrangements occur in an ordered sequence with TCRβ, TCRγ and TCRδ being the first TCR genes to undergo rearrangement. If a functional TCRβ chain is generated, it will associate with an invariant pTα chain and signaling through this pre-TCR complex induces proliferation and developmental progression, a process called β-selection. The phenotypical stage at which β-selection occurs differs between murine and human thymocytes. Whereas TCRβ is first expressed at the CD4 and CD8αβ double negative stage (DN) in murine T cell precursors, human thymocytes rearrange the TCRβ chain at the immature CD4 single positive stage (CD4ISP). Pre-TCR signaled CD4ISP cells are large cycling cells, have upregulated expression of CD28 and enter the CD4 and CD8αβ double positive (DP) stage as large metabolically active CD4hiDP cells (14–16). Both in human and murine thymus the majority of DP precursors are small resting cells with ongoing TCRα rearrangements. Most TCRα rearrangements are found late at the DP stage, but some reports have identified TCRα transcripts at earlier stages as well (17, 18).
Here, we identified a population of mature, PD-1 and CD8αα expressing TCRαβ+ T lymphocytes (6–8, 19) that exhibits innate production of IFN-γ and expresses Hobit, a transcription factor (TF) recently identified as a hallmark TF of tissue-resident effector memory cells and innate T cell subsets (20). We show that the TCRα repertoire of this population is skewed for early TRAV and TRAJ rearrangements. The data clearly show that different pre-selection precursors give rise to either agonist selected or conventionally selected T cells and that the checkpoint in the thymus to divert immature thymocytes to the agonist selection pathway occurs before conventional selection.
Results
Identification of a PD-1+CD8αα+T cell population in human thymus and cord blood
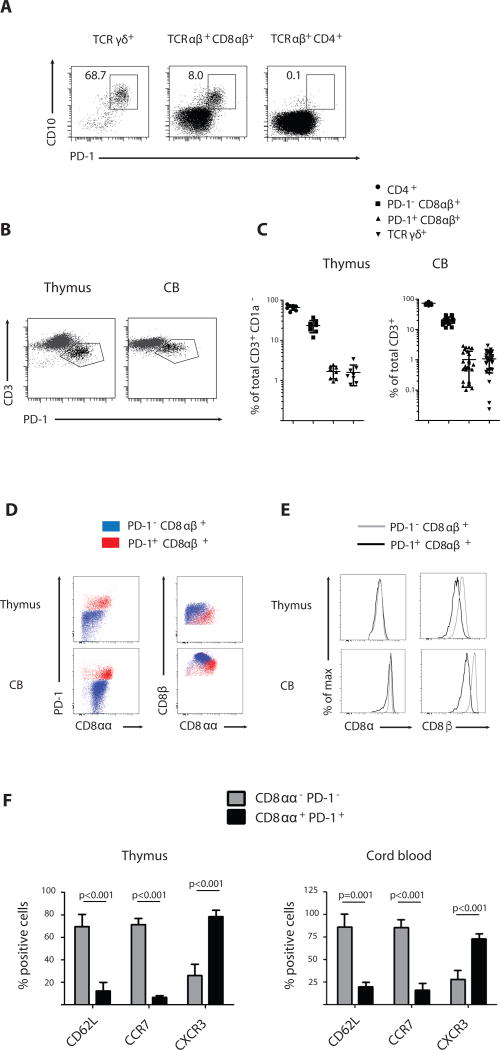
To identify an in vivo equivalent of agonist-selected T cells, which as we showed before can be generated in vitro in an OP9-DL1 culture system (21, 22), we analyzed human postnatal thymocytes ex vivo for expression of PD-1, a recently described marker for agonist selection (7, 8, 10). Expression of CD10, a marker for common lymphoid and more mature B and T cell precursors (23) is lost during thymocyte differentiation but remained selectively expressed on mature PD-1+ thymocytes. Many of the TCRγδ+ thymocytes express both CD10 and PD-1 (Fig. 1a and Fig. S1). However, a clear population of CD10+ PD-1+ cells was also detected within the CD8αβ+ TCRαβ+ subset, but not among the CD4+ TCRαβ+ cells (Fig. 1a and Fig. S2). To check whether these PD-1+ CD8αβ+ TCRαβ+ cells are present in the periphery as well, we screened cord blood from healthy donors and detected a distinct PD-1+ CD8αβ+ TCRαβ+ subset among the CD3+ cells (Fig. 1b and Fig. S3). Both thymus and cord blood PD-1+ CD8αβ+ TCRαβ+ T cells expressed markedly reduced levels of CD3 on their cell surface compared to their PD-1− CD8αβ+ TCRαβ+ counterparts, indicating that PD-1 and CD3low expression characteristically distinguishes this TCRαβ+ T cell subset from mainstream CD8αβ+ TCRαβ+ T cells (Fig. 1b). The PD-1+ CD8αβ+ TCRαβ+ population was present in all thymus and cord blood samples analyzed at a frequency comparable to that of the TCRγδ+ population (Fig. 1c and Table S1–S2). Furthermore, we detected a similar CD3low PD-1+ population among naïve CD45RO− CD8αβ T cells in peripheral blood of young children (Fig. S4), but not of adults. In addition to PD-1, the activation marker CD8αα has also been associated with agonist selection (6, 10, 11). Therefore, we next assessed expression of CD8αα by Thymus Leukemia Antigen (TL)-tetramer binding (24) and found that PD-1+ CD8αβ+ TCRαβ+ cells from thymus and cord blood stained homogeneously with TL tetramer, indicating that they co-expressed CD8αα homodimers together with CD8αβ heterodimers (Fig. 1d). Consistent with that, PD1+ TCRαβ+ cells stained weaker with anti-CD8β antibody as compared with CD8α antibody (Fig. 1e). Together, these data indicate that in the CD8αβ+ TCRαβ+ T cell population in the human thymus and cord blood there is a distinct subset of cells that express PD-1 and CD8αα, both hallmarks of agonist selected cells. Furthermore, these CD8αα+ PD-1+ TCRαβ+ cells express high levels of CXCR3 typically associated with effector and tissue resident memory T cells (25) but low levels of CD62L and CCR7, which mark naïve T cells in lymphoid tissues (26) (Fig. 1f). We will refer to the PD-1+ CD8αα+ CD8αβ+ TCRαβ+ cells as CD8αα+ T cells to distinguish them from the conventional naïve PD-1− CD8αα− CD8αβ+ TCRαβ+ T cells, which we will refer to as CD8αα− T cells.
Figure 1. CD8αα+ CD8αβ+ T cells in human thymus and cord blood.
(A) CD10 and PD-1 expression on human postnatal thymocytes, gated on mature CD1a− CD4− CD8αβ− TCRγδ+ cells, CD8αβ single positive (CD8αβ+) TCRαβ+ and CD4 single positive (CD4+) TCRαβ+ populations. Data are representative of at least 25 samples.
(B) CD3 and PD-1 expression on TCRαβ+ CD8αβ+ cells isolated from human postnatal thymus and cord blood. Representative of at least 25 samples.
(C) Frequency of different T cell subsets as percentage of total mature T cells (CD3+ CD1a−) in thymus and percentage of total T cells in cord blood (CD3+). Subsets are phenotypically defined as CD4+ (CD4+ CD8α−), PD-1− CD8αβ+ (PD-1− CD10− CD8αβ+ in thymus and PD-1− CD3hi CD8αβ+ in cord blood), PD-1+ CD8αβ+ (PD-1+ CD10+ CD8αβ+ in thymus and PD-1+ CD3lo CD8αβ+ in cord blood) and TCRγδ+. Mean +/− SD, n=8 for thymus, n=27 for cord blood
(D) PD-1, CD8β and CD8αα expression on PD-1− CD8αβ+ and PD-1+ CD8αβ+ cells isolated from human postnatal thymus and cord blood. CD8αβ+ populations defined as in (C). CD8αα expression is assessed by thymus leukemia antigen (TL)-tetramer binding. Data are representative of at least 5 samples
(E) CD8α and CD8β expression on PD-1− CD8αβ+ and PD-1+ CD8αβ+ cells isolated from human postnatal thymus and cord blood. CD8αβ+ populations defined as in (C). Data are representative of at least 5 samples
(F) Homing receptor expression determined by flow cytometry on subsets of CD8αβ+ cells isolated from thymus and cord blood. Populations were phenotypically defined as CD8αα− PD-1− (PD-1− CD10− CD8αβ+ in thymus and PD-1− CD3hi CD8αβ+ in cord blood) and CD8αα+ PD-1+ (PD-1+ CD10+ CD8αβ+ in thymus and PD-1+ CD3lo CD8αβ+ in cord blood). Paired samples t-test (2-tailed); p-value is displayed; n=5
Error bars display standard deviation
Hobit marks a distinct transcriptional profile in human CD8αα+ T cells
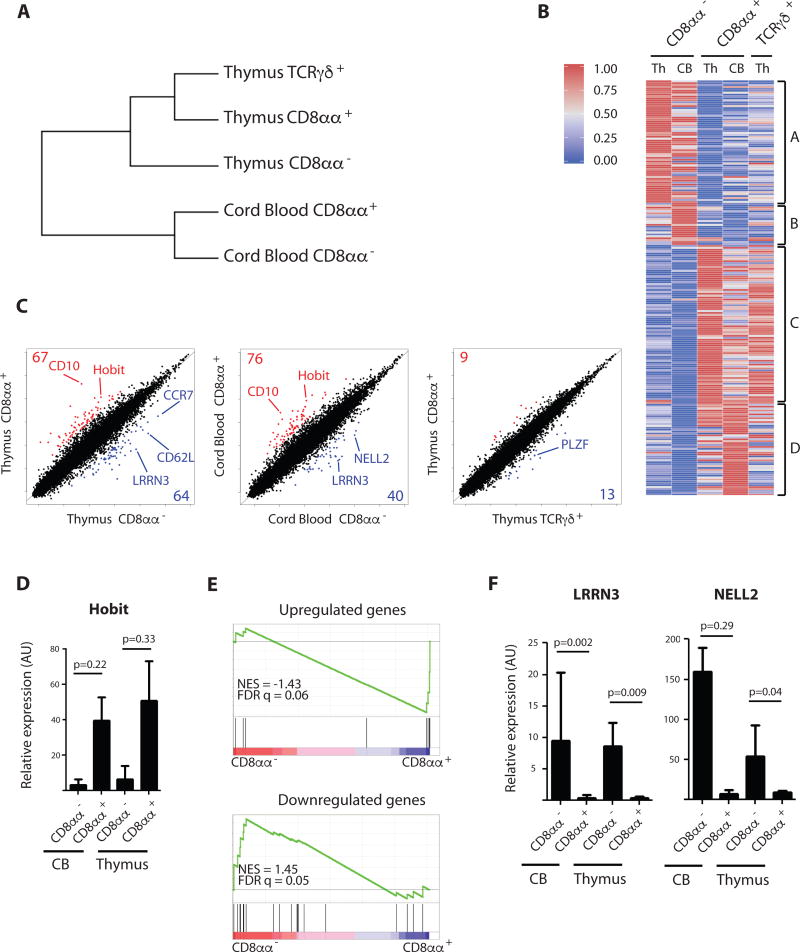
To further characterize and distinguish the CD8αα+ T cells, we analyzed their gene expression profile and compared it with that of CD8αα− T cells isolated from both thymus and cord blood as well as with that of TCRγδ+ thymocytes. Hierarchical clustering indicates that thymic CD8αα+ T cells are more closely related to innate TCRγδ+ thymocytes than to conventional naïve CD8αα− T cells (Fig. 2a). A heatmap representation of the most differentially expressed genes revealed a similar transcriptional signature for thymus and cord blood derived CD8αα+ T cells that is distinct from that of CD8αα− T cells, indicating that thymus and cord blood derived CD8αα+ T cells are closely related populations (Fig. 2b and Table S3). Scatter plot representation of gene expression profiling (Fig. 2c) confirmed the distinct signature in CD8αα− and CD8αα+ T cells and identified Homolog of Blimp-1 in T cells (Hobit) as one of the most differentially expressed genes that distinguishes the two subsets. Hobit was expressed by thymus and cord blood derived CD8αα+ T cells and this was confirmed by qPCR (Fig. 2d). Hobit was recently identified as a hallmark transcription factor expressed by tissue-resident effector memory cells and functionally differentiated innate cells including agonist selected natural killer T (NKT) and liver-resident natural killer (NK) cells(20). Furthermore, a gene set enrichment analysis (GSEA) on the human conventional CD8αα− versus the innate CD8αα+ T cells revealed that CD8αα+ T cells were significantly enriched for those genes that were also associated with the Hobit-expressing cells in mice (20) (Fig. 2e). These data indicate that the CD8αα+ T cells identified in human have already functionally differentiated and activated a transcriptional program for tissue residency shortly after their generation in the thymus. In contrast, CD8αα− T cells express CCR7 and CD62L, typically associated with a naïve and lymph node-homing phenotype (Fig. 2c). NELL2 and LRRN3 are genes that are induced after positive selection (22, 27) and remain highly expressed in naïve TCRαβ+ cells. Accordingly, these genes are not induced in the CD8αα+ subset, supporting the notion that these T cells are derived via a mechanism that is distinct from conventional positive selection (Fig. 2f and 2c). Since thymic CD8αα+ T cells have activated a program for tissue residency, we analyzed human peripheral tissues for the presence of similar innate T cells. A substantial population of small intestine intraepithelial T cells (IET) expresses high levels of CD8α combined with lower levels of CD8β (Fig. S5). Similar to the thymic phenotype, CD8αα+ IETs express PD-1. Analysis of human liver T cells revealed an analogous CD8αα+ population (Fig. S5). Furthermore, we found that NELL2 is not expressed in tissue resident human CD8αα+ T cells, whereas it is highly expressed in peripheral blood CD8αβ+ T cells of both naive (CD45RA+) and memory (CD45RO+) phenotype (Fig. S6).
Figure 2. Distinct transcriptional signature of CD8αα+ T cells.
(A) Hierarchical clustering of CD8αα− (PD-1− CD3hi CD8αβ+) and CD8αα+ (PD-1+ CD3lo CD8αβ+) T cells isolated from cord blood and CD8αα− (PD-1− CD10− CD8αβ+), CD8αα+ (PD-1+ CD10+ CD8αβ+) and TCRγδ+ cells isolated from thymus.
(B) Heatmap representation of genes differentially expressed in CD8αα− (PD-1− CD3hi CD8αβ+) and CD8αα+ (PD-1+ CD3lo CD8αβ+) T cells isolated from cord blood and CD8αα− (PD-1− CD10− CD8αβ+), CD8αα+ (PD-1+ CD10+ CD8αβ+) and TCRγδ+ cells isolated from thymus (list of genes see Table S1). Per gene the expression value is normalized to a value between 0 and 1. The color-coded scale (blue=downregulation and red=upregulation) for the normalized expression value is indicated.
(C) Scatter plots of transcript expression in CD8αα− versus CD8αα+ T cells in human postnatal thymus and cord blood as well as in CD8αα+ versus TCRγδ+ thymocytes. Populations are phenotypically identified as in (A). Numbers in corners indicate total genes overexpressed by logarithmic fold change (log2FC) > −2 in CD8αα+ T cells (red) or underrepresented by log2FC < −2 in CD8αα+ T cells (blue). TCR genes have been excluded from the analysis.
(D) Confirmatory RT-PCR analysis for Hobit transcripts in populations as defined in (A), expressed in arbitrary units (AU) relative to expression in total postnatal human thymus. p-value is displayed; paired samples t-test on log-transformed values (2-tailed), n=3. Error bars display standard deviation.
(E) Gene Set Enrichment Analysis (GSEA) of genes up- and downregulated in tissue resident memory and innate T cells as described by Mackay et al. on CD8αα− (PD-1− CD10− CD8αβ+) versus CD8αα+ (PD-1+ CD10+ CD8αβ+) T cells in human postnatal thymus. Normalized Enrichment Score (NES) and False Discovery Rate (FDR q value) are displayed. (List of genes see Supplemental Methods Table 2)
(F) Confirmatory RT-PCR analysis for LRRN3 and NELL2 transcripts in populations as defined in (A), expressed in arbitrary units (AU) relative to expression in total postnatal human thymus. p-value is displayed; paired samples t-test on log-transformed values (2-tailed), n=3. Error bars display standard deviation.
CD8αα+ T cells display a typical innate functional effector phenotype
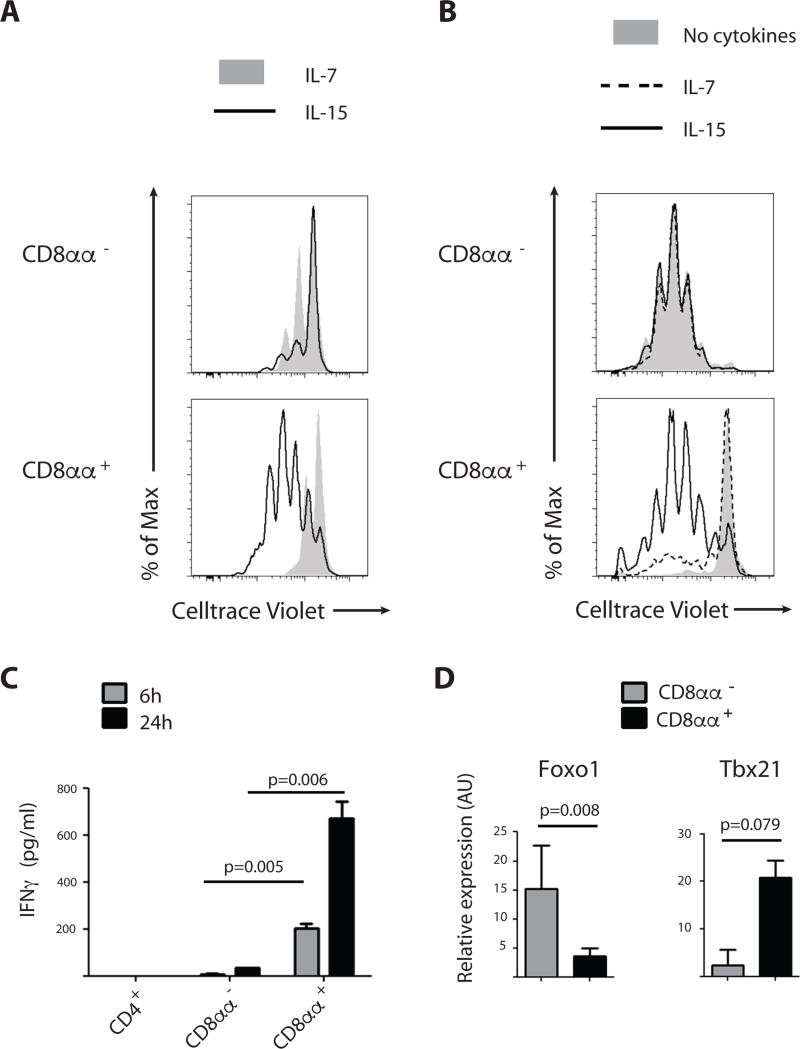
Since innate T cells such as murine CD8αα+ intestinal intraepithelial T cells require interleukin 15 (IL-15) for their homeostasis and activation-induced proliferation (28), whereas naïve TCRαβ+ T cells require IL-7, we compared first the cytokine-driven proliferation of cord blood CD8αα− and CD8αα+ T cells in the presence of IL-7 or IL-15. As expected, naïve CD8αα− T cells showed moderate proliferation in the presence of IL-7 and less with IL-15. In sharp contrast, CD8αα+ TCRαβ+ T cells displayed extensive IL-15-driven proliferation but only a weak response to IL-7 (Fig. 3a and Fig. S7). In addition, IL-15-responsive CD8αα+ cells induced the NK cell-associated receptor CD56, which is characteristic of cytotoxic effector cells (29). In contrast, CD56 induction was limited on the responding CD8αα− T cells (Fig. S7). Next, we tested the TCR-driven proliferation of cord blood CD8αα− and CD8αα+ T cells. CD3/CD28 costimulation of the naïve CD8αα− T cells induced marked proliferation after 5 days of culture in the absence of exogenous common gamma-chain cytokines (Fig. 3b). In contrast, CD3/CD28 costimulation of the already activated CD8αα+ T cells led to cell death in the absence of cytokines. Addition of cytokines, especially IL-15, led to enhanced survival on day 2 and marked proliferation on day 5 (Fig. S8).
Figure 3. CD8αα+ T cells display innate effector function.
(A) Proliferation assessed by Celltrace Violet dye dilution in CD8αα− (PD-1− CD10− CD8αβ+) and CD8αα+ (PD-1+ CD10+ CD8αβ+) isolated from human postnatal thymus after 6 days of incubation in the presence of IL-7 (10ng/ml) or IL-15 (10ng/ml). Data are representative of at least 5 experiments
(B) Proliferation assessed by Celltrace Violet dye dilution in CD8αα− (PD-1− CD3hi CD8αβ+) and CD8αα+ (PD-1+ CD3lo CD8αβ+) T cells isolated from cord blood and activated with anti-CD3 and anti-CD28 for 5 days. Experiments were performed either in the absence of cytokines or in the presence of IL-7 (10ng/ml) or IL-15 (10ng/ml). Data are representative of 3 experiments
(C) IFNγ production, measured by ELISA (pg/ml) of CD4+ (CD4+ CD8α−), CD8αα− (PD-1− CD3hi CD8αβ+) and CD8αα+ (PD-1+ CD3lo CD8αβ+) isolated from cord blood. Cells were incubated with PMA/ionomycin for 6 and 24 hours before harvesting supernatant. Student t-test on duplicates (2-tailed). p-value is displayed; representative of 4 experiments.
(D) Quantitative RT-PCR expression analysis for transcription factors in CD8αα− (PD-1− CD3hi CD8αβ+) and CD8αα+ (PD-1+ CD3lo CD8αβ+) T cells isolated from cord blood, expressed in arbitrary units (AU) relative to expression in total postnatal human thymus. p-value is displayed; paired samples t-test on log-transformed values (2-tailed), n=3.
Error bars display standard deviation.
To assess if like innate T cells, CD8αα+ T cells also display immediate effector functions, we measured IFN-γ production after 6 and 24 hours of stimulation in vitro. Consistent with an innate-like effector profile, CD8αα+ T cells responded rapidly to stimulation and produced significantly higher amounts of IFN-γ after 6 and 24 hours of in vitro stimulation as compared to their CD8αα− or CD4+ counterparts (Fig. 3c). In further agreement with their innate effector phenotype, CD8αα+ T cells also expressed a typical CD8 effector TF profile with high expression of T-bet but low FOXO1 expression (Fig. 3d). In contrast, CD8αα− T cells isolated from cord blood, expressed high levels of FOXO1 but not of T-bet (Fig. 3d).
CD8αα+ T cells express hallmarks of agonist-selected cells
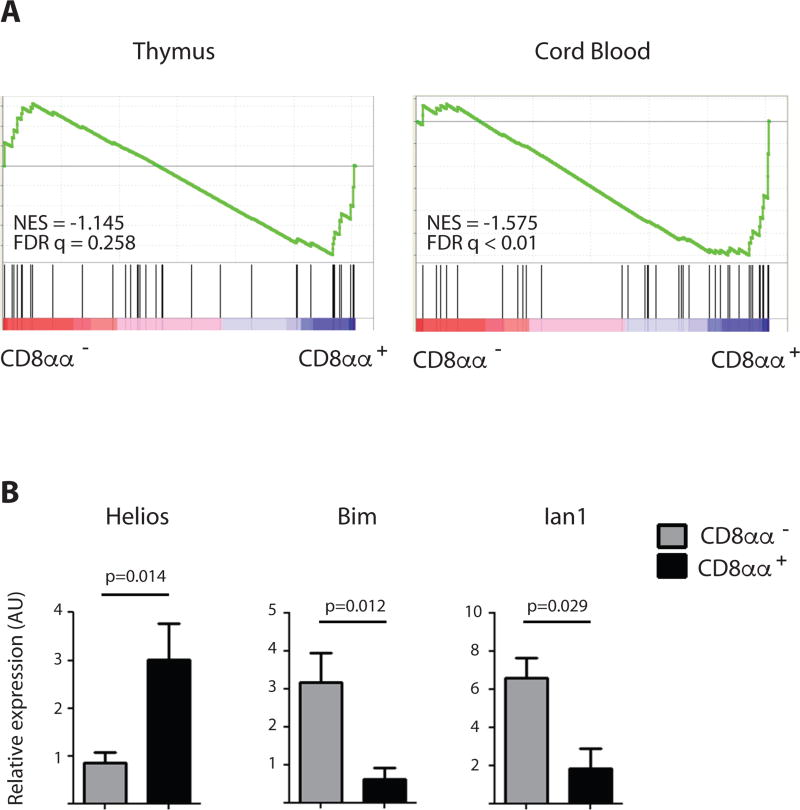
To examine if the innate functional effector phenotype expressed by the mature CD8αα+ T cells might have been imprinted during the selection process in response to self-antigen recognition, we next analyzed the expression of a set of genes that have been associated with post-agonist selected CD8αα+ T cells in mice (6). GSEA on CD8αα− and CD8αα+ T cells showed an enrichment in human CD8αα+ thymocytes for transcripts of genes associated with agonist selection, and this enrichment became even more pronounced in the more mature cord blood CD8αα+ T cells (Fig. 4a). The agonist-selected nature of CD8αα+ T cells was further supported by high levels of Helios, a transcription factor induced upon strong TCR stimulation (30, 31). Remarkably, in spite of an association with strong TCR signals, CD8αα+ T cells expressed significantly lower levels of the clonal deletion-associated genes Bim and Ian1 (32) (Fig. 4b) as compared to the conventional CD8αα− T cells that received positively selecting TCR signals of lower intensity.
Figure 4. Hallmarks of TCR agonist selection in CD8αα+ T cells.
(A) Gene Set Enrichment Analysis (GSEA) of genes upregulated by agonist-selection as described by Yamagata et al. on CD8αα− (PD-1− CD10− CD8αβ+) versus CD8αα+ (PD-1+ CD10+ CD8αβ+) T cells isolated from thymus and CD8αα− (PD-1− CD3hi CD8αβ+) versus CD8αα+ (PD-1+ CD3lo CD8αβ+) T cells isolated from cord blood. Normalized Enrichment Score (NES) and False Discovery Rate (FDR q value) are displayed. (List of genes see Supplemental Methods Table 2)
(B) Confirmatory RT-PCR analysis for Helios, Bim and Ian1 transcripts in CD8αα− (PD-1− CD10− CD8αβ+) and CD8αα+ (PD-1+ CD10+ CD8αβ+) T cells from human postnatal thymus, expressed in arbitrary units (AU) relative to expression in total postnatal human thymus. p-value is displayed; paired samples t-test on log-transformed values (2-tailed), n=3. Error bars display standard deviation.
CD8αα expression on early post-β selection thymocytes marks a distinct precursor subset
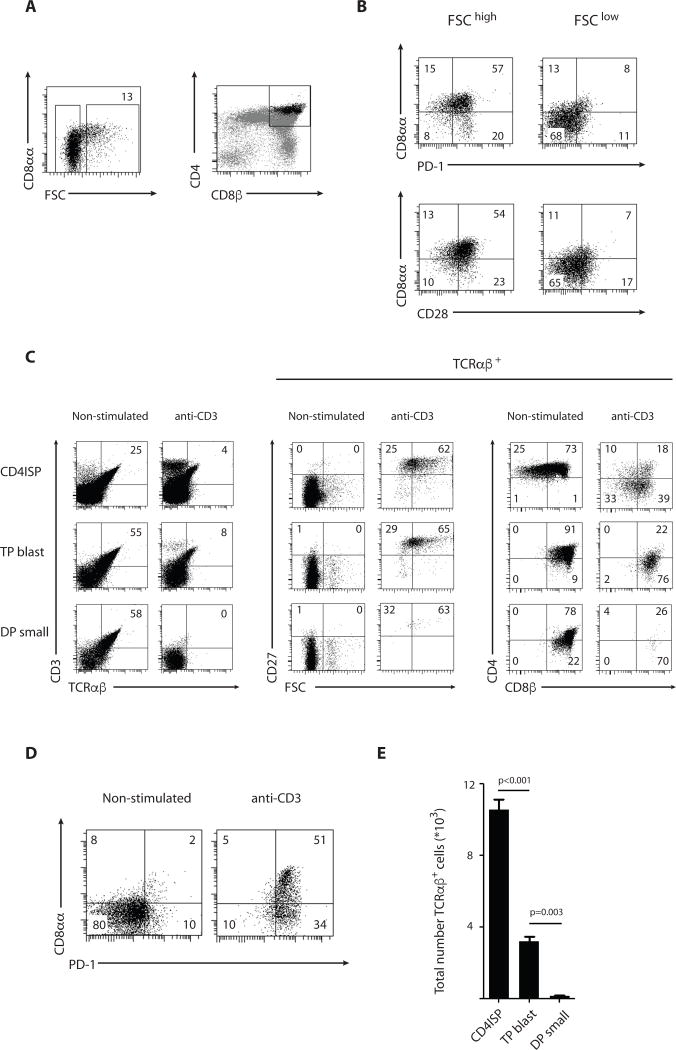
In mice, triple positive thymocytes (TP) that co-express CD8αα together with CD8αβ and CD4 are able to survive strong TCR signals and differentiate into mature DN or CD8αα+ T cells (11). To examine whether a similar subset exists among human DP thymocytes, we stained with TL tetramer and found that CD8αα expression marks large early post-β-selection DP precursors similar to the CD4hiDP population previously described (16) (Fig. 5a). In addition to CD8αα, these thymocytes also express PD-1 and CD28, both markers induced by pre-TCR signaling (Fig. 5b) (14, 33–35). Together, these observations indicate that among the DP progenitors, the expression of CD8αα, PD-1 and CD28 marks a subset that we will refer to as ‘TP blasts’. This population is distinct from the vast majority of DP thymocytes that constitute the ‘DP small’ subset and do not co-express CD8αα, PD-1 and CD28 (Fig. 5a–b). In addition, the expression level of several hallmark genes associated with DP thymocytes is in agreement with the notion that the TP blast and DP small populations represent early and late post-β selection stages (36) (Fig. S9).
Figure 5. CD8αα expression on early post-β selection thymocytes marks a distinct precursor subset.
(A) CD3− CD4+ CD8β+ DP thymocytes were analyzed for size (FSC) and CD8αα expression as assessed by TL tetramer binding. Right panel displays expression levels of CD4 and CD8β on total thymocytes (gray) and FSChigh DP thymocytes (black). Data are representative of at least 5 experiments.
(B) CD3− CD4+ CD8β+ DP blasts (FSChigh) and small (FSClow) thymocytes were analyzed for PD-1, CD28 and CD8αα expression as assessed by TL tetramer binding. Data are representative of at least 5 experiments.
(C) Phenotypic analysis of cells present in vitro after 3+3 days in culture with IL-7 and IL-15 in presence or absence of stimulation with anti-CD3 for the first 3 days. Cultures were initiated with post-β-selection CD4ISP (FSChigh CD4+ CD8α− CD3− CD28+), TP blast (FSChigh CD4+ CD8α+ CD69− CD27−) and DP small (FSClow CD4+ CD8α+ CD69− CD27−), isolated from human thymus. Middle and right panels are gated on TCRαβ+ cells displayed in left panel. Data are representative of 2 experiments.
(D) PD-1 and CD8αα expression as assessed by TL tetramer binding on the progeny of TP blast thymocytes either non-stimulated (left) or stimulated with anti-CD3 (right).
(E) Absolute number of TCRαβ+ cells present in vitro after 3+3 days in culture with IL-7 and IL-15 in presence or absence of stimulation with anti-CD3 for the first 3 days. Cultures were initiated with 105 post-β-selection CD4ISP (FSChigh CD4+ CD8α− CD3− CD28+), TP blast (FSChigh CD4+ CD8α+ CD69− CD27−) and DP small (FSClow CD4+ CD8α+ CD69− CD27−), isolated from human thymus. Numbers are adjusted for cell proliferation levels and TCRαβ+ cells present after 3 days in absence of stimulation (7% TCRαβ+ cells for CD4ISP; 44% TCRαβ+ cells for TP blast and 66% TCRαβ+ cells for DP small; see materials and methods). p-values are displayed; Student t-test on triplicates (2-tailed). Data are representative of 2 experiments. Error bars display standard deviation.
To test whether early post-β-selection progenitors have a higher potential to survive strong TCR signals associated with agonist selection, we compared the differentiation potential of TP blast and DP small, along with CD4ISP as the most immature post-β-selection population identified in human. We stimulated these pre-selection populations with anti-CD3 and monitored their ability to mature into post-selection T cells. Although unstimulated populations survive rather well in culture (Fig. S10), stimulation with a high dose of anti-CD3 resulted in the death of virtually all TCRαβ+ DP small cells whereas TCRαβ+ progeny of the stimulated CD4ISP and TP blast survived (Fig. 5c). The surviving anti-CD3 stimulated thymocytes upregulated the maturation marker CD27, downregulated CD4 expression but remained CD8αβ+ (Fig. 5c) and more importantly, expressed high levels of the activation markers PD-1 and CD8αα (Fig. 5d) similar to the CD8αα+ T cells isolated ex vivo from the human thymus and cord blood. Mature TCRαβ+ cells generated from CD4ISP precursors also expressed CD8αβ, suggesting that precursors developed along a DP intermediate before rearranging a TCRα chain that allows agonist selection. Indeed, already after 3 days of in vitro culture of CD4ISP, TCRαβ+ DP precursors were present (Fig. S11). CD4ISP and TP blast not only survived strong TCR stimulation but also proliferated (Fig. S12). In fig 5e, we corrected for differential proliferation as well as for the differential degree of rearranged TCRαβ+ cells between the 3 populations (Fig. S11) to calculate the agonist selection potential of each progenitor population (Fig. 5e). This clearly shows the loss in agonist selection potential as the precursor population progresses from early to late post-β-selection.
Early TCRα rearrangements favor development of agonist selection associated innate CD8αα+ T cells
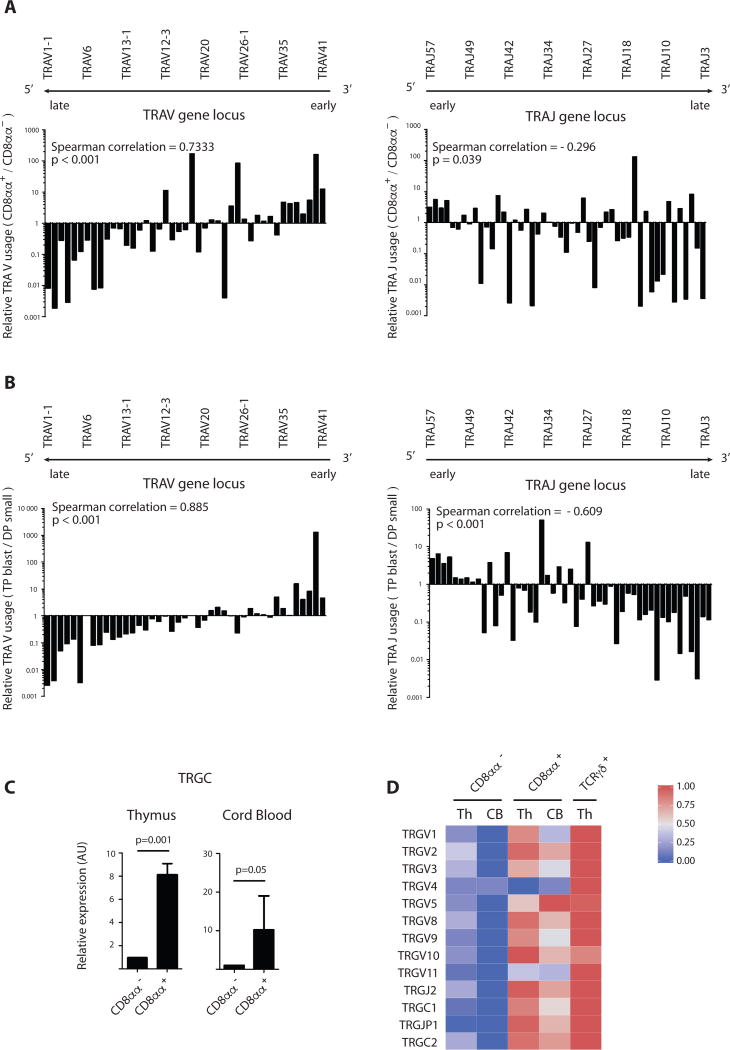
To investigate whether the footprint of agonist selection is reflected in the human post-selection TCR repertoire, we determined the TCRα rearrangements in mature ex vivo-isolated conventional CD8αα− and agonist selection-associated CD8αα+ thymocytes. Since it is known that human TCRα rearrangements occur sequentially in time from J-proximal to J-distal for the TRAV gene segments and from V-proximal to V-distal for the TRAJ gene segments, we analyzed TCRVα and Jα usage for both populations (Fig. 6a, Fig. S13–S14 and Table S4). Next generation sequencing of TCRα transcripts was performed on both populations of 4 donors and revealed that the agonist selection-associated CD8αα+ T cells preferentially use early J-proximal TCRVα rearrangements (TRAV41-TRAV40-…), whereas conventional positive selected CD8αα− T cells contained a relatively higher frequency of late J-distal TRAV1-1 and TRAV1–2 gene segments: evaluation of the distribution of all TRAV and TRAJ genes showed a significant enrichment for early rearrangements in CD8αα+ T cells as evidenced by the spearman correlation coefficient >0 for the V gene segment usage and <0 for the J segment usage. This pattern was also observed in post-thymic cord blood CD8αα+ T cells, as assessed by microarray expression analysis (Fig. S14). To investigate at what point the repertoire imprinting occurred during development, we analyzed TCRα rearrangements of the TP blast and DP small thymocyte subsets. We identified the same bias for early TCRVα and Jα usage in the TP blasts when compared with the DP small precursor cells (Fig. 6b). These findings constitute strong evidence for the concept that the developmental stage of the precursor at which the TCRα rearrangements occur, allowing expression of the full TCRαβ, plays a decisive role in determining the fate of the maturing thymocytes in response to strong agonist selection signals.
Figure 6. Early TCRα rearrangements favor development of innate CD8αα+ T cells.
(A and B) Ratio of TRAV gene usage (left) and TRAJ gene usage (right) in CD8αα+ (PD-1+ CD10+ CD8αβ+) to CD8αα− T cells (PD-1− CD10− CD8αβ+) (A) and in TP blast (FSChigh CD4+CD8β+CD69−) to DP small (FSClow CD4+CD8β+CD69−CD28−) (B) isolated from human postnatal thymus. TRAV (TRAJ) gene usage was determined by next-generation sequencing and calculated as % of total TRAV (TRAJ) gene reads. All genes are displayed from 5’ to 3’ along the TRA locus. Ranked Spearman correlation coefficient was calculated on log-transformed values of ratios, p-value of correlation is displayed. Graph shows results from 1 donor, representative of 4 donors (see supplementary table 2).
(C) Quantitative RT-PCR analysis for TRGC transcripts (primers amplify both TRGC1 and TRGC2) in CD8αα− (PD-1− CD10− CD8αβ+) and CD8αα+ (PD-1+ CD10+ CD8αβ+) T cells isolated from thymus and in CD8αα− (PD-1− CD3hi CD8αβ+) and CD8αα+ (PD-1+ CD3lo CD8αβ+) T cells isolated from cord blood. Expressed in arbitrary units (AU) relative to expression in CD8αα+ T cells. P-values are displayed; paired samples t-test on log-transformed values (2-tailed), n=3
(D) Heatmap representation of TRGV, TRGJ and TRGC gene segments in CD8αα− and CD8αα+ populations from cord blood and CD8αα−, CD8αα+ and TCRγδ+ T cells from thymus as determined by microarray expression analysis. Populations are phenotypically defined as in (C). The color-coded scale (blue=downregulation and red=upregulation) for the normalized expression value is indicated. Error bars display standard deviation.
In addition to the differential timing for TCRα rearrangements, we observed that TCRγ transcripts were also differentially expressed in CD8αα+ and CD8αα− T cells. As demonstrated by qPCR for TRGC and microarray analysis for the different TRGV, TRGJ and TRGC transcripts, CD8αα+ T cells do not silence TCRγ transcripts as conventional T cells do (Fig. 6c–d). A similar high level of TCRγ expression was previously described for agonist selection-associated innate T cells in mice, including NKT and intestinal epithelial CD8αα+ T cells (37). Together, we conclude that human CD8αα+ T cells are preferentially selected by strong TCR engagement on a subset of progenitors that express a full TCRαβ early on, leading to the generation of a post-selection T cell population with innate functional capacity and a markedly distinct TCR repertoire.
Discussion
Strong TCR signals result in the elimination of self-reactive thymocytes by clonal deletion or the induction of a specialized functional program characteristic of agonist-selected T cells. The mechanisms underlying the different outcomes of strong TCR activation on thymocytes are still poorly understood. We characterized an agonist-selected T cell population in human that expresses CD8αα homodimers. Thymic selection of this subset induces an innate effector phenotype hallmarked by the expression of a Hobit-associated gene expression profile. This study’s main finding is that early TCRα rearrangements favor the generation of this lineage in an unmanipulated setting in human, indicating that the checkpoint in the thymus to divert immature thymocytes to the agonist selection pathway occurs early before conventional selection of DP cells.
Human CD8αα+ T cells are present in thymus, cord blood and peripheral blood of young children but not of adults. This suggests that this innate T cell population is mainly generated early in life, but does not remain present in the circulation thereafter. In contrast, CD8αα+ T cells are found in adult human small intestine epithelium and liver and, like thymic CD8αα+ T cells, lack expression for NELL2. The ability of the CD8αα+ T cells to home to the tissues seems to be acquired at the time of agonist selection as Hobit is sharply upregulated in the thymic CD8αα+ T cells. Hobit is a Blimp-1 homolog that is expressed in long-lived resident effector memory T cells that combine long-term quiescent maintenance with the immediate capacity to induce effector molecules (20, 38, 39). Such a functional program makes these cells ideally equipped for immune surveillance in the tissues as has been suggested for both murine DN T cells as well as human CD8αα+ tissue-resident T cells in several models of infection (40–43). An important role in tumor immunosurveillance was recently described for murine innate lymphoid cells and innate-like T cells that comprise of both TCRαβ+ and TCRγδ+ cells with similar functional properties (44). Furthermore, murine DN TCRαβ+ cells can recognize peptides processed in a TAP-independent fashion or by non-classical MHC molecules, suggesting that these cells are designed to target cells when conventional antigen processing or presentation is impaired, such as viral infection or cellular transformation (7).
The differential TCRα usage between agonist-selected and conventional T cells reveals that the decision of agonist-selected differentiation or apoptosis upon strong TCR stimulation is based on the timing and developmental stage at which a complete TCRαβ complex engages its ligand. We showed evidence that only early CD4ISP or TP blasts survive strong TCR activation whereas small DPs undergo apoptosis. Alternatively, small DPs clearly represent the main precursor population of conventionally selected cells since small DPs are more enriched for late-rearranging TRAVs and TRAJs. Our findings link together several observations made in TCR transgenic and wild-type mice. First, a prominent DN T cell population is generated upon cognate TCR ligand interactions in TCR transgenic mice and this phenomenon has been attributed to premature expression of the transgenic TCRα chain (12). Also in non-TCR transgenic mice, thymocytes with extremely early spontaneous TCRα rearrangements at pre-DP stages preferentially give rise to T cells that accumulate in the small intestinal epithelium (45). However, lineage fate mapping showed that CD8αα+ intraepithelial T cells arise from RORγt+ precursors, suggesting a DP intermediate (46). Such an intermediate population was identified by Gangadharan et al. (11) as CD8αα+ DPs or TPs, and confirmed to be the direct progenitors of agonist-selected T cells in mice. Our findings here unite these previous observations and define TP thymocyte precursors as early post-β-selection blasts that express an early rearranged TCRα repertoire that is maintained in the repertoire of post-selection CD8αα+ T cells and that is clearly different from that of conventional selected T cells.
Finally, we found that TP blast and small DP thymocytes not only differ in the pattern of TCRα rearrangements, but also respond differently to strong TCR signaling. It has been shown that CD8αα induction on strongly activated conventional CD8αβ+ T cells can mediate their selective survival and differentiation into effector memory T cells that preferentially accumulate in peripheral tissues (47). Activation-induced CD8αα expression can dampen TCR signaling and disrupt lipid rafts, but could potentially also sequester or redirect lymphocyte-specific protein tyrosine kinase (p56lck) and LAT toward other receptors located outside the rafts, like IL-15R (19). We found indeed that what enables TP blasts to survive strong TCR activation and avoid expression of Bim is not only the downmodulation of the TCR signal as the levels of Bim expression in agonist-selected cells are surprisingly low compared to those in the weakly signaled conventional thymocytes. Alternatively, the ability to upregulate Bim upon strong TCR activation could be a property absent in early thymocytes and acquired during progression to small DP stages.
Our data demonstrate that the checkpoint for agonist selection precedes conventional selection in human thymus. Analysis of the TCRα repertoire provides a useful tool in studying human T cell development as it naturally reflects developmental progression where genetic manipulations are not possible. However, it neither detects at what point during thymic development the deviation occurs nor does it provide insight into the mechanisms that mediate this early deviation.
The picture emerges that during thymic development there is a phase immediately after β-selection where TCRα rearrangement and expression of a full TCRαβ early on deviates precursors to develop along the agonist selection pathway and to functionally differentiate to innate-like T cells in response to strong agonist selection signals. In contrast, precursors that undergo E-protein mediated progression to late DP stages (48) are destined to either be positively selected as stem cell-like naive conventional T cells (49) or deleted in response to strong TCR signals during conventional negative selection. It is currently not clear yet which molecular processes determine the onset and timing of TCRα rearrangements at the single cell level. However, it was shown that this regulation is distinct from that of DP progression, as only the upregulation of RORγt but not the onset of TCRα rearrangement is dependent on proliferation and expression of Myc (50). Recent work has suggested that heterogeneity among DPs could be introduced at the pre-TCR checkpoint since some pre-TCR complexes can interact with thymic ligands and this interaction was shown to enhance pre-TCR signaling (51). The current findings turn the spotlight on early developmental stages as a critical checkpoint in self-reactive T cell development and present agonist selection as an instructed program rather than the result of an escape from negative selection.
Materials and Methods
Study Design
This study was initiated to identify agonist-selected T cells in human thymus and peripheral blood and tissues as well as to gain insight in their developmental pathways. Human post-natal thymus was obtained from children (0–8 years) that underwent cardiac surgery and cord blood was obtained from healthy donors. These cells were used following the guidelines of the Medical Ethical Committee of Ghent University Hospital, after informed consent had been obtained in accordance with the declaration of Helsinki. For the purpose of phenotypical characterization, 8 donors were used for thymic samples and 27 donors were used for cord blood samples since these numbers reflect the availability of both samples throughout the timeframe of the study. For the microarray analysis, 3 donors were used for both thymus and cord blood samples and mixed in an effort to increase RNA yield and minimize bias caused by inter-donor variability. For the TRA next generation sequencing, 4 donors were used and individually analyzed for TRA sequence since the assay was technically achievable with lower RNA yields when compared with microarray analysis. Peripheral blood of young children of 1–2 years old (6 donors), adult liver (3 donors) or adult small intestine (1 donor) was obtained according to the national legislation concerning the use of human rest material and ethical approval was obtained from the Medical Ethical Committee of Ghent University Hospital. The number of donors reflects the availability of the samples throughout the timeframe of the study. The study was unblinded and not randomized.
Cells and Purification
CD8αβ+ and CD8αα+ T cells were isolated from post-natal thymus after prior depletion of CD4+ fractions using FITC-conjugated mouse anti-human CD4 antibodies (OKT4) in combination with sheep anti-mouse IgG-coated Dynabeads (Life Technologies). TCRγδ+ cells were isolated from post-natal thymus by anti-TCRγδ magnetic-activated cell sorting (Miltenyi). CD8αβ+ and CD8αα+ T cells were isolated from cord blood after prior enrichment with biotin-conjugated anti-CD8α antibodies (OKT8) in combination with Streptavidin microbeads (Miltenyi). Obtained cell population were subsequently labeled with fluorochrome-conjugated antibodies according to the experimental design and sorted with a fluorescent-activated cell sorting Aria II (BD Biosciences).
Isolation of IET from human small intestine
Human ileum was obtained from a patient who underwent right hemicolectomy. The ileum was dissected from the colon and cut in pieces of 1–5 mm2. Intraepithelial cells were isolated by incubation in isolation buffer (IMDM + 10% FCS +1mM EDTA + 1mM dithiothreitol) during mechanical rotation at 37°C for 20 minutes. This isolation step was performed twice and isolated cells in suspension were next subjected to percoll centrifugation (44% and 67% percoll) at 1800 rpm for 20 minutes to purify the lymphocytic fraction.
Isolation of liver T cells
Human liver samples were obtained from patients who underwent partial hepatectomy. The healthy liver tissue was dissected and cut in pieces of 1–5 mm2. Liver-resident cells were enzymatically isolated (0.5mg/ml collagenase, 0.02 mg/ml DNase, 2% FCS and 0.6% BSA in HBSS) at 37°C for 20 minutes. Isolated cells in suspension were next subjected to percoll centrifugation (30% and 70% percoll) at 1800 rpm for 20 minutes to purify the lymphocytic fraction.
Flow cytometry and antibodies
The following anti-human monoclonal antibodies were used : FITC conjugated CD4, TCRγδ (BD Biosciences), PE-conjugated PD-1, TCRαβ (Miltenyi), CD62L, CCR7 (BD Pharmingen), CD28 (BD Biosciences), PE-cy7 conjugated CD8β (ebiosciences), CD10 (BD biosciences), APC-conjugated CD3, CD5, CD8α, CD56 (BD Biosciences), TCRαβ (Miltenyi), CXCR3 (Biolegend), APC-cy7 conjugated CD3, CD27 (ebiosciences), CD8α (BD biosciences), efluor 450-conjugated CD1a (ebiosciences), Alexafluor 700 conjugated CD4 (BD Pharmingen), biotin-conjugated PD-1 (Miltenyi), PERCP-cy5.5 conjugated Streptavidin (ebiosciences). CD8αα expression was assessed by staining with PE-conjugated TL-tetramer (provided by H. Cheroutre) after prior staining with anti-CD8β antibodies. Annexin V apoptosis detection kit APC (ebiosciences) was used for quantification of apoptosis. Flow cytometric analysis was performed using a LSR II Cytometer (BD Biosciences).
Real-time quantitative PCR (RT-qPCR)
Total RNA was prepared from FACS-sorted samples, using the miRNeasy microkit (Qiagen). cDNA synthesis was performed by the Superscript III Reverse Transcriptase for RT-PCR kit (Invitrogen). RT-qPCR with the SYBR Green I technology was performed using the LightCycler 480 SYBR Green I Master kit on a LightCycler 480 II (both Roche) according to the manufacturer’s protocol. Primer sequences (Integrated DNA Technologies, Heverlee, Belgium) are listed in Supplemental Methods Table 1. Primers for Ian1 (alias Gimap4) were purchased from Biorad.
T cell stimulation and IFN-γ ELISA
For ELISA, 20–50*103 cells were stimulated for 6 and 24 hours with PMA (60 ng/ml) and ionomycin (6 µg/ml), followed by collection of 50 µl supernatant. ELISA was performed according to the manufacturer’s protocol using the PeliPair human IFNγ ELISA reagent set and PeliKine-Tool set (Sanquin, Amsterdam, Netherlands) in 96 well half-area ELISA plates (Greiner Bio-one, Wemmel, Belgium).
CellTrace proliferation assay
Cells were labeled with the CellTrace™ Violet cell proliferation kit (Invitrogen) following the standard method for labeling cells in suspension as described by the manufacturer.
Microarray RNA expression analysis
RNA was isolated using the micro-RNA isolation kit (Qiagen) and sent to the Nucleomics facility, VIB Leuven, Belgium where the microarrays were performed using the GeneChip Human Gene 2.0 ST arrays (Affymetrix). Samples were subsequently analysed using R/Bioconductor. All samples passed quality control, and the Robust Multi-array Average procedure was used to normalize data within arrays (probeset summarization, background correction and log2-transformation) and between arrays (quantile normalization). Only probesets that mapped uniquely to one gene were kept, and for each gene, the probeset with the highest expression level was kept. Due to the high stringency of this method, some genes of interest were lost. These genes were added manually as long as one probeset is linked to only one gene of interest.
TCRα next generation sequencing
RNA was isolated from sorted (50,000 – 1 million cells) thymocyte subsets with the RNeasy microkit (Qiagen) followed by template-switch anchored RT-PCR (52, 53). More specifically, a template-switch adaptor, AAGCAGTGGTATCAACGCAGAGTACATrGrGrG, was ligated at the 5’ end of mRNA during cDNA generation using the Superscript II RT enzyme (Invitrogen). The cDNA product was purified using AMPure XP Beads (Agencourt). Then, PCR amplification (Lightcycler, Roche) was performed using a Cα specific primer containing an adapter used in subsequent sequencing (5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTCTCAGCTGGTACACGGCAGGGTCAGGGT-3’, adapter in italic) and a primer complementary to the template-switch adapter (5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAAGCAGTGGTATCAACGCAG-3’, adapter in italic) with the KAPA Real-Time Library Amplification Kit (Kapa Biosystems). The PCR was stopped when the amplified PCR product, as measured with SYBRGreen, was within the standards 2 and 3. After purification with AMPure XP Beads, an index PCR with Illumina sequencing adapters was performed using the Nextera XT Index Kit. This second PCR product was again purified with AMPure XP beads. High-throughput sequencing of the generated amplicon products containing the TRA sequences was performed on an Illumina MiSeq platform using the V2 300 kit, with 200bp at the 3’end (read 2) and 100bp at the 5’end (read 1) (at the GIGA center, University of Liège, Belgium). The percentages of Vα and Jα usage of each thymocyte subset were obtained after analysis of the FASTAQ files (read 2) with the MiTCR software (54) using the default settings for the correction level (level 2) and quality threshold values (PHRED value 25). The ratio of the percentage Vα and Jα usage in thymocytes populations was determined and to avoid division by “0”, 0.00625% was added to the calculated % of each TRAV or TRAJ gene (corresponds to approximately 0.5 sequence counts).
In vitro TCR agonist selection
105 cells of thymic precursor populations were cultured for 3 days in the presence of IL-7 (10 ng/ml) and IL-15 (10 ng/ml) on BSA or anti-CD3 (10 µg/ml) coated wells of 96 well plate. After 3 days cells were washed and cultured in absence of anti-CD3 stimulation in U-bottom 96-well plate in the presence of IL-7 (10 ng/ml) and IL-15 (10 ng/ml) for another 3 days to allow re-expression of the TCR complex before flow cytometry analysis. To determine the agonist selection potential of each progenitor population, we calculated the ratio of mature TCRαβ+ cells present on D6 to the total number of TCRαβ+ cells present or generated in each culture during the first 3 days of culture that cells have the opportunity to be activated by plastic-coated anti-CD3 (7% TCRαβ+ cells for CD4ISP; 44% TCRαβ+ cells for TP blast and 66% TCRαβ+ cells for DP small). This number is corrected for the level of proliferation in each culture as determined by Flowjo software in a separate proliferation assay using Celltrace Violet dilution.
Statistical analysis
Student t-tests and Paired samples t-tests (2-tailed) were performed using SPSS software. For RT-qPCR experiments, the mean of duplicates from different donor samples were first Log-transformed before performing paired samples t-tests (2-tailed) across different donor samples using SPSS software. Gene Set Enrichment Analysis (GSEA) was performed using the GSEA tool v2.2.2 of the Broad Institute (http://software.broadinstitute.org/gsea/index.jsp). The ‘GseaPreranked’ module was run using standard parameters and 1000 permutations. Different gene sets (grp extension), preranked lists (rnk extension) and the chip annoation file were uploaded. For analysis of TRAV and TRAJ usage, 44 TRAV genes and 51 TRAJ genes were ranked according to their location within the TRA locus (1–44 for TRAV and 1–51 for TRAJ) and Ranked Spearman correlation coefficients as well as corresponding p-values were calculated on log-transformed values of ratios corresponding to a single TRAV or TRAJ gene using SPSS software.
Supplementary Material
Figure S1 : PD-1+ T cell subsets in human thymus.
Figure S2 : Gating strategy for PD-1+ CD8αβ+ (CD8αα+) T cells in human thymus.
Figure S3 : Gating strategy for PD-1+ CD8αβ+ (CD8αα+) T cells in human cord blood.
Figure S4 : CD8αα+ CD8αβ+ T cells in human peripheral blood in infants.
Figure S5 : CD8αα+ CD8αβ+ T cells in human small intestinal epithelium and liver.
Figure S6 : NELL2 expression on tissue and circulating T cell subsets.
Figure S7 : Cytokine-induced proliferation of CD8αα+ T cells.
Figure S8 : Differential survival upon TCR activation for CD8αα− and CD8αα+ T cells.
Figure S9 : TP blast express low levels of hallmark DP genes.
Figure S10 : Survival of post-β-selection subsets in vitro.
Figure S11 : In vitro differentiation of post-β-selection subsets in absence of stimulation.
Figure S12 : Proliferation of post-β-selection subsets in vitro.
Figure S13 : TRAV gene usage in thymic CD8αα+ and CD8αα− T cells.
Figure S14 : TRAV gene usage in cord blood CD8αα+ and CD8αα− T cells.
Table S1 : CD8αβ+ T cell fractions in human thymus.
Table S2 : CD8αβ+ T cell fractions in human cord blood.
Table S3 : CD8αα+ T cell associated gene sets.
Table S4 : CD8αα+ T cells and TP blast precursors show enrichment for early TRAV and TRAJ genes.
Supplemental methods Table 1 : primer sequences
Supplemental methods Table 2 : Gene sets used for GSEA analyses
Acknowledgments
The authors would like to thank Dr. K Francois of the Department of Cardiac Surgery of Ghent University Hospital for providing thymus samples and Dr C Matthys (Cord Blood Bank UZ Gent) for providing human cord blood samples. We thank Daniel Douek (NIH, US) for sharing their NGS/TCR protocol, Tom Boterberg for the irradiation procedures and Sophie Vermaut for help with flow cytometry and cell sorting. Funding: This work was supported by the Agency for Innovation by Science and Technology (Agentschap voor Innovatie door Wetenschap en Technologie or IWT), the Research Foundation - Flanders (Fonds voor Wetenschappelijk Onderzoek Vlaanderen, FWO), the Interuniversity Attraction Poles Program (BELSPO), Stichting tegen Kanker, Kinderkankerfonds, Fonds National de la Recherche Scientifique (FRS-FNRS) and the Fonds Gaston Ithier. GV and YVC are supported by the Agency for Innovation by Science and Technology (IWT), SDM is supported by the Research Foundation – Flanders (FWO). Y.S. is an ISAC Marylou Ingram Scholar.
Footnotes
Author contributions: GV performed the experiments, designed the research, analyzed and interpreted the data and wrote the manuscript; DV, LM, NT and YS analyzed the data; GG and MB performed the experiments; YVC, SDM, KW, SB, GL, TT and TK provided reagents and participated in discussions; JVD ensured supply of human tissue samples; HC provided reagents, interpreted the data and wrote the manuscript; BV designed the research, interpreted the data and wrote the manuscript.
Competing interests: The authors declare that they have no conflicting financial interests.
Data and materials availability: Microarray data are available in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/gds) under the accession number GSE81098.
References
- 1.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annual Review of Immunology. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 2.Gascoigne NRJ, Palmer E. Signaling in thymic selection. Current Opinion in Immunology. 2011;23:207–212. doi: 10.1016/j.coi.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hogquist KA, Jameson SC. The self-obsession of T cells: how TCR signaling thresholds affect fate 'decisions' and effector function. Nat Immunol. 2014;15:815–823. doi: 10.1038/ni.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stritesky GL, Jameson SC, Hogquist KA. In: Annual Review of Immunology. Paul WE, editor. Vol. 30. Vol. 30. 2012. pp. 95–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denning TL, Granger S, Mucida D, Graddy R, Leclercq G, Zhang W, Honey K, Rasmussen JP, Cheroutre H, Rudensky AY, Kronenberg M. Mouse TCR alpha beta(+)CD8 alpha intraepithelial lymphocytes express genes that down-regulate their antigen reactivity and suppress immune responses. (vol 178, pg 4230, 2007) Journal of Immunology. 2007;178:6654–6654. doi: 10.4049/jimmunol.178.7.4230. [DOI] [PubMed] [Google Scholar]
- 6.Yamagata T, Mathis D, Benoist C. Self-reactivity in thymic double-positive cells commits cells to a CD8 alpha lineage with characteristics of innate immune cells. Nature Immunology. 2004;5:597–605. doi: 10.1038/ni1070. [DOI] [PubMed] [Google Scholar]
- 7.Mayans S, Stepniak D, Palida SF, Larange A, Dreux J, Arlian BM, Shinnakasu R, Kronenberg M, Cheroutre H, Lambolez F. alpha beta T Cell Receptors Expressed by CD4(−)CD8 alpha beta Intraepithelial T Cells Drive Their Fate into a Unique Lineage with Unusual MHC Reactivities. Immunity. 2014;41:207–218. doi: 10.1016/j.immuni.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonald BD, Bunker JJ, Ishizuka IE, Jabri B, Bendelac A. Elevated T Cell Receptor Signaling Identifies a Thymic Precursor to the TCR alpha beta(+)CD4(−)CD8 beta(−) Intraepithelial Lymphocyte Lineage. Immunity. 2014;41:219–229. doi: 10.1016/j.immuni.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malhotra D, Linehan JL, Dileepan T, Lee YJ, Purtha WE, Lu JV, Nelson RW, Fife BT, Orr HT, Anderson MS, Hogquist KA, Jenkins MK. Tolerance is established in polyclonal CD4(+) T cells by distinct mechanisms, according to self-peptide expression patterns. Nature Immunology. 2016;17:187–195. doi: 10.1038/ni.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pobezinsky LA, Angelov GS, Tai XG, Jeurling S, Van Laethem F, Feigenbaum L, Park JH, Singer A. Clonal deletion and the fate of autoreactive thymocytes that survive negative selection. Nature Immunology. 2012;13:569-+. doi: 10.1038/ni.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gangadharan D, Lambolez F, Attinger A, Wang-Zhu Y, Sullivan BA, Cheroutre H. Identification of pre- and postselection TCR alpha beta(+) intraepithelial lymphocyte precursors in the thymus. Immunity. 2006;25:631–641. doi: 10.1016/j.immuni.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 12.Baldwin TA, Sandau MM, Jameson SC, Hogquist KA. The timing of TCR alpha expression critically influences T cell development and selection. Journal of Experimental Medicine. 2005;202:111–121. doi: 10.1084/jem.20050359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leishman AJ, Gapin L, Capone M, Palmer E, MacDonald HR, Kronenberg M, Cheroutre H. Precursors of functional MHC class I- or class II-restricted CD8 alpha alpha(+) T cells are positively selected in the thymus by agonist self-peptides. Immunity. 2002;16:355–364. doi: 10.1016/s1074-7613(02)00284-4. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell JL, Seng A, Yankee TM. Ikaros, Helios, and Aiolos protein levels increase in human thymocytes after beta selection. Immunol Res. 2016;64:565–575. doi: 10.1007/s12026-015-8754-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taghon T, Van de Walle I, De Smet G, De Smedt M, Leclercq G, Vandekerckhove B, Plum J. Notch signaling is required for proliferation but not for differentiation at a well-defined beta-selection checkpoint during human T-cell development. Blood. 2009;113:3254–3263. doi: 10.1182/blood-2008-07-168906. [DOI] [PubMed] [Google Scholar]
- 16.Swainson L, Kinet S, Manel N, Battini JL, Sitbon M, Taylor N. Glucose transporter 1 expression identifies a population of cycling CD4+ CD8+ human thymocytes with high CXCR4-induced chemotaxis. Proc Natl Acad Sci U S A. 2005;102:12867–12872. doi: 10.1073/pnas.0503603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernandez-Munain C, Sleckman BP, Krangel MS. A developmental switch from TCR delta enhancer to TCR alpha enhancer function during thymocyte maturation. Immunity. 1999;10:723–733. doi: 10.1016/s1074-7613(00)80071-0. [DOI] [PubMed] [Google Scholar]
- 18.Aifantis I, Bassing CH, Garbe AI, Sawai K, Alt FW, von Boehmer H. The E delta enhancer controls the generation of CD4- CD8- alphabeta TCR-expressing T cells that can give rise to different lineages of alphabeta T cells. J Exp Med. 2006;203:1543–1550. doi: 10.1084/jem.20051711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheroutre H, Lambolez F. Doubting the TCR coreceptor function of CD8alphaalpha. Immunity. 2008;28:149–159. doi: 10.1016/j.immuni.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Mackay LK, Minnich M, Kragten NA, Liao Y, Nota B, Seillet C, Zaid A, Man K, Preston S, Freestone D, Braun A, Wynne-Jones E, Behr FM, Stark R, Pellicci DG, Godfrey DI, Belz GT, Pellegrini M, Gebhardt T, Busslinger M, Shi W, Carbone FR, van Lier RA, Kallies A, van Gisbergen KP. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 2016;352:459–463. doi: 10.1126/science.aad2035. [DOI] [PubMed] [Google Scholar]
- 21.Van Coppernolle S, Verstichel G, Timmermans F, Velghe I, Vermijlen D, De Smedt M, Leclercq G, Plum J, Taghon T, Vandekerckhove B, Kerre T. Functionally Mature CD4 and CD8 TCR alpha beta Cells Are Generated in OP9-DL1 Cultures from Human CD34(+) Hematopoietic Cells. Journal of Immunology. 2009;183:4859–4870. doi: 10.4049/jimmunol.0900714. [DOI] [PubMed] [Google Scholar]
- 22.Snauwaert S, Verstichel G, Bonte S, Goetgeluk G, Vanhee S, Van Caeneghem Y, De Mulder K, Heirman C, Stauss H, Heemskerk MHM, Taghon T, Leclercq G, Plum J, Langerak AW, Thielemans K, Kerre T, Vandekerckhove B. In vitro generation of mature, naive antigen-specific CD8 (+) T cells with a single T-cell receptor by agonist selection. Leukemia. 2014;28:830–841. doi: 10.1038/leu.2013.285. [DOI] [PubMed] [Google Scholar]
- 23.Galy A, Travis M, Cen D, Chen B, Human T. B, natural killer, and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity. 1995;3:459–473. doi: 10.1016/1074-7613(95)90175-2. [DOI] [PubMed] [Google Scholar]
- 24.Leishman AJ, Naidenko OV, Attinger A, Koning F, Lena CJ, Xiong Y, Chang HC, Reinherz E, Kronenberg M, Cheroutre H. T cell responses modulated through interaction between CD8 alpha and the nonclassical MHC class I molecule, TL. Science. 2001;294:1936–1939. doi: 10.1126/science.1063564. [DOI] [PubMed] [Google Scholar]
- 25.Ariotti S, Beltman JB, Chodaczek G, Hoekstra ME, van Beek AE, Gomez-Eerland R, Ritsma L, van Rheenen J, Maree AF, Zal T, de Boer RJ, Haanen JB, Schumacher TN. Tissue-resident memory CD8+ T cells continuously patrol skin epithelia to quickly recognize local antigen. Proc Natl Acad Sci U S A. 2012;109:19739–19744. doi: 10.1073/pnas.1208927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 27.Dik WA, Pike-Overzet K, Weerkamp F, de Ridder D, de Haas EFE, Baert MRM, van der Spek EEL, Reinders MJT, van Dongen JJM, Langerak AW, Staal FJT. New insights on human T cell development by quantitative T cell receptor gene rearrangement studies and gene expression profiling. Journal of Experimental Medicine. 2005;201:1715–1723. doi: 10.1084/jem.20042524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki H, Duncan GS, Takimoto H, Mak TW. Abnormal development of intestinal intraepithelial lymphocytes and peripheral natural killer cells in mice lacking the IL-2 receptor beta chain. Journal of Experimental Medicine. 1997;185:499–505. doi: 10.1084/jem.185.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pittet MJ, Speiser DE, Valmori D, Cerottini JC, Romero P. Cutting edge: Cytolytic effector function in human circulating CD8(+) T cells closely correlates with CD56 surface expression. Journal of Immunology. 2000;164:1148–1152. doi: 10.4049/jimmunol.164.3.1148. [DOI] [PubMed] [Google Scholar]
- 30.Daley SR, Hu DY, Goodnow CC. Helios marks strongly autoreactive CD4(+) T cells in two major waves of thymic deletion distinguished by induction of PD-1 or NF-kappa B. Journal of Experimental Medicine. 2013;210:269–285. doi: 10.1084/jem.20121458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Getnet D, Grosso JF, Goldberg MV, Harris TJ, Yen HR, Bruno TC, Durham NM, Hipkiss EL, Pyle KJ, Wada S, Pan F, Pardoll DM, Drake CG. A role for the transcription factor Helios in human CD4(+)CD25(+) regulatory T cells. Molecular Immunology. 2010;47:1595–1600. doi: 10.1016/j.molimm.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baldwin TA, Hogquist KA. Transcriptional analysis of clonal deletion in vivo. Journal of Immunology. 2007;179:837–844. doi: 10.4049/jimmunol.179.2.837. [DOI] [PubMed] [Google Scholar]
- 33.Williams JA, Hathcock KS, Klug D, Harada Y, Choudhury B, Allison JP, Abe R, Hodes RJ. Regulated costimulation in the thymus is critical for T cell development: dysregulated CD28 costimulation can bypass the pre-TCR checkpoint. J Immunol. 2005;175:4199–4207. doi: 10.4049/jimmunol.175.7.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taghon T, Van de Walle I, De Smet G, Leclercq G, Vandekerckhove B, Plum J. Notch signaling is required for proliferation but not for differentiation at a well-defined beta-selection checkpoint during human T-cell development. Blood. 2009;113:3254–3263. doi: 10.1182/blood-2008-07-168906. [DOI] [PubMed] [Google Scholar]
- 35.Nishimura H, Honjo T, Minato N. Facilitation of beta selection and modification of positive selection in the thymus of PD-1-deficient mice. J Exp Med. 2000;191:891–898. doi: 10.1084/jem.191.5.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, Ericson J, Bendall S, Spitzer M, Nolan G, Kobayashi K, von Boehmer H, Mathis D, Benoist C, Best AJ, Knell J, Goldrath A, Jojic V, Koller D, Shay T, Regev A, Cohen N, Brennan P, Brenner M, Kim F, Rao TN, Wagers A, Heng T, Ericson J, Rothamel K, Ortiz-Lopez A, Mathis D, Bezman NA, Sun JC, Min-Oo G, Kim CC, Lanier LL, Miller J, Brown B, Merad M, Gautier EL, Jakubzick C, Randolph GJ, Monach P, Blair DA, Dustin ML, Shinton SA, Hardy RR, Laidlaw D, Collins J, Gazit R, Rossi DJ, Malhotra N, Sylvia K, Kang J, Kreslavsky T, Fletcher A, Elpek K, Bellemare-Pelletier A, Malhotra D, Turley C. Immunological Genome, The transcriptional landscape of alpha beta T cell differentiation. Nature Immunology. 2013;14:619-+. doi: 10.1038/ni.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kisielow J, Tortola L, Weber J, Karjalainen K, Kopf M. Evidence for the divergence of innate and adaptive T-cell precursors before commitment to the alpha beta and gamma delta lineages. Blood. 2011;118:6591–6600. doi: 10.1182/blood-2011-05-352732. [DOI] [PubMed] [Google Scholar]
- 38.Braga FAV, Hertoghs KML, Kragten NAM, Doody GM, Barnes NA, Remmerswaal EBM, Hsiao CC, Moerland PD, Wouters D, Derks IAM, van Stijn A, Demkes M, Hamann J, Eldering E, Nolte MA, Tooze RM, ten Berge IJM, van Gisbergen K, van Lier RAW. Blimp-1 homolog Hobit identifies effector-type lymphocytes in humans. European Journal of Immunology. 2015;45:2945–2958. doi: 10.1002/eji.201545650. [DOI] [PubMed] [Google Scholar]
- 39.Braun J, Frentsch M, Thiel A. Hobit and human effector T-cell differentiation: The beginning of a long journey. European Journal of Immunology. 2015;45:2762–2765. doi: 10.1002/eji.201545959. [DOI] [PubMed] [Google Scholar]
- 40.Hossain MS, Takimoto H, Ninomiya T, Yoshida H, Kishihara K, Matsuzaki G, Kimura G, Nomoto K. Characterization of CD4(−) CD8(−) CD3(+) T-cell receptor-alpha beta(+) T cells in murine cytomegalovirus infection. Immunology. 2000;101:19–29. doi: 10.1046/j.1365-2567.2000.00052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cowley SC, Hamilton E, Frelinger JA, Su J, Forman J, Elkins KL. CD4(−)CD8(−) T cells control intracellular bacterial infections both in vitro and in vivo. Journal of Experimental Medicine. 2005;202:309–319. doi: 10.1084/jem.20050569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu J, Peng T, Johnston C, Phasouk K, Kask AS, Klock A, Jin L, Diem K, Koelle DM, Wald A, Robins H, Corey L. Immune surveillance by CD8 alpha alpha(+) skin-resident T cells in human herpes virus infection (vol 497, pg 494, 2013) Nature. 2013;500 doi: 10.1038/nature12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D’Acquisto F, Crompton T. CD3(+)CD4(−)CD8(−) (double negative) T cells: Saviours or villains of the immune response? Biochemical Pharmacology. 2011;82:333–340. doi: 10.1016/j.bcp.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 44.Dadi S, Chhangawala S, Whitlock BM, Franklin RA, Luo CT, Oh SA, Toure A, Pritykin Y, Huse M, Leslie CS, Li MO. Cancer Immunosurveillance by Tissue-Resident Innate Lymphoid Cells and Innate-like T Cells. Cell. 2016;164:365–377. doi: 10.1016/j.cell.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hendricks DW, Fink PJ. Uneven colonization of the lymphoid periphery by T cells that undergo early TCR{alpha} rearrangements. J Immunol. 2009;182:4267–4274. doi: 10.4049/jimmunol.0804180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eberl G, Littman DR. Thymic origin of intestinal alphabeta T cells revealed by fate mapping of RORgammat+ cells. Science. 2004;305:248–251. doi: 10.1126/science.1096472. [DOI] [PubMed] [Google Scholar]
- 47.Madakamutil LT, Christen U, Lena CJ, Wang-Zhu Y, Attinger A, Sundarrajan M, Ellmeier W, von Herrath MG, Jensen P, Littman DR, Cheroutre H. CD8alphaalpha-mediated survival and differentiation of CD8 memory T cell precursors. Science. 2004;304:590–593. doi: 10.1126/science.1092316. [DOI] [PubMed] [Google Scholar]
- 48.Jones ME, Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;27:860–870. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bedoui S, Gebhardt T, Gasteiger G, Kastenmuller W. Parallels and differences between innate and adaptive lymphocytes. Nat Immunol. 2016;17:490–494. doi: 10.1038/ni.3432. [DOI] [PubMed] [Google Scholar]
- 50.Kreslavsky T, Gleimer M, Miyazaki M, Choi Y, Gagnon E, Murre C, Sicinski P, von Boehmer H. beta-Selection-induced proliferation is required for alphabeta T cell differentiation. Immunity. 2012;37:840–853. doi: 10.1016/j.immuni.2012.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mallis RJ, Bai K, Arthanari H, Hussey RE, Handley M, Li Z, Chingozha L, Duke-Cohan JS, Lu H, Wang JH, Zhu C, Wagner G, Reinherz EL. Pre-TCR ligand binding impacts thymocyte development before alphabetaTCR expression. Proc Natl Acad Sci U S A. 2015;112:8373–8378. doi: 10.1073/pnas.1504971112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gros A, Robbins PF, Yao X, Li YF, Turcotte S, Tran E, Wunderlich JR, Mixon A, Farid S, Dudley ME, Hanada K, Almeida JR, Darko S, Douek DC, Yang JC, Rosenberg SA. PD-1 identifies the patient-specific CD8(+) tumor-reactive repertoire infiltrating human tumors. Journal of Clinical Investigation. 2014;124:2246–2259. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quigley MF, Almeida JR, Price DA, Douek DC. Unbiased molecular analysis of T cell receptor expression using template-switch anchored RT-PCR. Curr Protoc Immunol. 2011:33. doi: 10.1002/0471142735.im1033s94. Chapter 10, Unit10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bolotin DA, Shugay M, Mamedov IZ, Putintseva EV, Turchaninova MA, Zvyagin IV, Britanova OV, Chudakov DM. MiTCR: software for T-cell receptor sequencing data analysis. Nature Methods. 2013;10:813–814. doi: 10.1038/nmeth.2555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 : PD-1+ T cell subsets in human thymus.
Figure S2 : Gating strategy for PD-1+ CD8αβ+ (CD8αα+) T cells in human thymus.
Figure S3 : Gating strategy for PD-1+ CD8αβ+ (CD8αα+) T cells in human cord blood.
Figure S4 : CD8αα+ CD8αβ+ T cells in human peripheral blood in infants.
Figure S5 : CD8αα+ CD8αβ+ T cells in human small intestinal epithelium and liver.
Figure S6 : NELL2 expression on tissue and circulating T cell subsets.
Figure S7 : Cytokine-induced proliferation of CD8αα+ T cells.
Figure S8 : Differential survival upon TCR activation for CD8αα− and CD8αα+ T cells.
Figure S9 : TP blast express low levels of hallmark DP genes.
Figure S10 : Survival of post-β-selection subsets in vitro.
Figure S11 : In vitro differentiation of post-β-selection subsets in absence of stimulation.
Figure S12 : Proliferation of post-β-selection subsets in vitro.
Figure S13 : TRAV gene usage in thymic CD8αα+ and CD8αα− T cells.
Figure S14 : TRAV gene usage in cord blood CD8αα+ and CD8αα− T cells.
Table S1 : CD8αβ+ T cell fractions in human thymus.
Table S2 : CD8αβ+ T cell fractions in human cord blood.
Table S3 : CD8αα+ T cell associated gene sets.
Table S4 : CD8αα+ T cells and TP blast precursors show enrichment for early TRAV and TRAJ genes.
Supplemental methods Table 1 : primer sequences
Supplemental methods Table 2 : Gene sets used for GSEA analyses