Abstract
The mechanism of mitochondrial DNA (mtDNA) replication in Saccharomyces cerevisiae is controversial. Evidence exists for double-strand break (DSB) mediated recombination-dependent replication at mitochondrial replication origin ori5 in hypersuppressive ρ− cells. However, it is not clear if this replication mode operates in ρ+ cells. To understand this, we targeted bacterial Ku (bKu), a DSB binding protein, to the mitochondria of ρ+ cells with the hypothesis that bKu would bind persistently to mtDNA DSBs, thereby preventing mtDNA replication or repair. Here, we show that mitochondrial-targeted bKu binds to ori5 and that inducible expression of bKu triggers petite formation preferentially in daughter cells. bKu expression also induces mtDNA depletion that eventually results in the formation of ρ0 cells. This data supports the idea that yeast mtDNA replication is initiated by a DSB and bKu inhibits mtDNA replication by binding to a DSB at ori5, preventing mtDNA segregation to daughter cells. Interestingly, we find that mitochondrial-targeted bKu does not decrease mtDNA content in human MCF7 cells. This finding is in agreement with the fact that human mtDNA replication, typically, is not initiated by a DSB. Therefore, this study provides evidence that DSB-mediated replication is the predominant form of mtDNA replication in ρ+ yeast cells.
INTRODUCTION
Mitochondria are double-membrane organelles that generally harbor multiple copies of mitochondrial DNA (mtDNA) (1), which are organized as nucleoprotein complexes called nucleoids (2). Different eukaryotic species exhibit heterogeneity in size and physical form of their mtDNA molecules. For example, the ∼16.6 kb human mtDNA, which encodes protein subunits of the oxidative phosphorylation (OXPHOS) system, typically forms double-stranded, closed circles of one genome length (1,3). In contrast, mtDNA in the budding yeast Saccharomyces cerevisiae exists predominantly as head-to-tail linear multimers (concatemers) of several genome units, with a minor proportion occurring in a circular form (4,5). The mtDNA monomer in S. cerevisiae (strain S288C) is ∼85.8 kb that, like human mtDNA, encodes protein components of the OXPHOS system [(6) and references therein]. The topological variation of mtDNA between human (typically closed-circular) and S. cerevisiae (mainly polydisperse linear) is likely due to the differences in the mode of mtDNA replication. However, the mechanisms by which these species replicate their mitochondrial genome are only partially understood and multiple models have been proposed for human [for review see (7)] as well as for S. cerevisiae (discussed below).
Budding yeast mtDNA contains eight replication origin-like sequences (oris 1–8), of which at least three are believed to be active origins (oris 2, 3 and 5) (6). The active oris contain three GC clusters (A–C), each separated by stretches of AT sequence. A transcription promoter (r) is present upstream of the GC cluster C that contains the consensus sequence for mitochondrial RNA polymerase (Rpo41) binding (6,8,9). Two distinct mechanisms for mtDNA replication have been proposed in S. cerevisiae: transcription-dependent replication and recombination-dependent replication (10). According to the transcription-dependent model, Rpo41 recognizes transcriptional promoters at the active oris and produces RNA transcripts, which are further processed to generate primers for mtDNA replication [for review see (9)]. However, the transcription-dependent replication is largely hypothetical and evidence supporting this model is limited only to the initiation steps (11–13). Furthermore, Rpo41 is not essential for mtDNA maintenance (14,15), clearly indicating the existence of a transcription-independent mtDNA replication mechanism.
Recombinant-dependent replication is the more popular mtDNA replication model in yeast. This model was proposed to explain the observation that the major form of mtDNA in mother cells is concatemers, whereas circular monomers prevail in growing buds (16). According to this model, recombination-dependent rolling circle replication (RDR) initiates at ori5. This replication origin has a bubble-like structure and bases on the opposite strands of ori5 are preferentially damaged by reactive oxygen species (ROS). These base damages are eventually converted into a double-strand break (DSB) by the base-excision repair enzyme Ntg1. The DSB is then processed to generate a 3΄-tail, which is bound by the mitochondrial homologous recombinase (Mhr1). The nucleoprotein filament then invades a template circular mtDNA molecule to form a heteroduplex joint and initiation of rolling circle replication ensues. This process can generate mtDNA concatemers of multiple genome units that are selectively transmitted to growing buds, where concatemers are circularized into monomers (16–21). In addition to explaining the existence of mtDNA in different topologies (linear and circular), this model also accounts for a characteristic feature of the S. cerevisiae mitochondrial system: its tendency to maintain the state of homoplasmy (22). The evidence for ROS-instigated DSB at ori5 and its involvement in mtDNA replication was obtained from the hypersuppressive (HS) ρ− mitochondrial genomes (18,19) [for review of ρ+, ρ−, HS ρ−, and ρ0 see (23)]. Even though exposure of ρ+ cells to low levels of ROS increases mtDNA copy number in an Ntg1- and Mhr1-dependent manner (19), it remains to be determined whether a DSB at ori5 is also involved in mtDNA replication in ρ+ cells. Uncertainty also exists as to which mode(s) of replication is/are primarily utilized by ρ+ cells. To gain insight into these issues, we targeted a DSB DNA binding protein (bacterial Ku) to the mitochondria of ρ+ cells.
Ku is a protein involved in non-homologous end joining (NHEJ) DSB repair [for review see (24,25)] and exists predominantly as a heterodimeric Ku70-Ku80 complex in eukaryotes. Each eukaryotic Ku subunit is composed of three distinct regions: an N-terminal domain, a central ‘core’ domain, and a C-terminal domain. While the ‘core’ domain is required for dimerization as well as for the formation of a ring-like structure that wraps around DNA ends, the N- and C-terminal domains are involved in recruiting and interacting with downstream NHEJ factors to mediate DSB repair (24,25). Analogous to eukaryotes, homologs of Ku and an intact NHEJ system have been identified in certain bacteria including multiple species of mycobacterium [for review see (26)]. Bacterial Ku (bKu) proteins form small homodimers that possess an evolutionarily conserved ‘core’ domain but lack the additional N- and C-terminal domains present in eukaryotes (27–29). Because these terminal domains are required for communication with eukaryotic NHEJ proteins, expression of bKu in eukaryotic cells should, in theory, bind to DNA DSBs and prevent repair due to lack of communication between bKu and eukaryotic repair proteins. Indeed, we showed previously that nuclear-targeted Mycobacterium tuberculosis Ku (MtKu) binds to laser-induced DSBs and increases sensitivity of human cancer cells to bleomycin sulfate, a DSB inducing agent (30). Nuclear-targeted MtKu also attenuates homologous recombination in mammalian cells (31). Analogous to M. tuberculosis, M. marinum was recently shown to possess a proficient NHEJ system (32). We have determined that M. marinum Ku (MmKu) is expressed at a higher level than MtKu in bacterial (32) and mammalian cells (this study). Hence, we selected mitochondrial-targeted MmKu as a molecular tool to bind to DSBs in ρ+ mtDNA to test the hypothesis that DSB-bound MmKu could prevent mtDNA replication or repair.
Here, we report that expression of mitochondrial-targeted MmKu induces petite formation, which occurs selectively in daughter cells. We show that MmKu binds to ori5 in the yeast mtDNA and that MmKu expression triggers mtDNA depletion. Since MmKu possesses a DSB-binding domain but lacks domains to communicate with eukaryotic repair factors, we conclude that binding of MmKu to ori5 inhibits mtDNA RDR, thereby preventing transmission of mtDNA from mother cells to daughter cells. We also demonstrate that mitochondrial-targeted MmKu does not decrease mtDNA content in human MCF7 cells. This observation is in agreement with the current knowledge on human mtDNA replication, which typically does not involve DSBs (7,33). These findings indicate that mitochondrial-targeted MmKu impairs mtDNA homeostasis only when DSBs are involved in the perpetuation of the mtDNA. Finally, our data suggest that DSB-mediated replication is the predominant form of mtDNA replication in the ρ+ yeast cells since binding of MmKu to ori5 can result in the complete loss of mtDNA.
MATERIALS AND METHODS
Oligodeoxyribonucleotides
Oligodeoxyribonucleotides (oligonucleotides) were purchased from Operon Technologies Inc. (Alameda, CA, USA), the DNA Facility at Iowa State University (Ames, IA, USA) or the DNA Sequence/Synthesis Facility at University of Wisconsin Biotechnology Center (Madison, WI, USA). The sequences of the oligonucleotides used in this study are listed in Supplemental Table S1.
DNA manipulation and plasmid construction
For detailed description of plasmid construction see Supplemental Materials and Methods. The bKu coding sequences in all plasmids were sequenced by the DNA Facility at Iowa State University to ensure correct reading frame and correct orientation of inserts.
The pTRE-Tight vector system (Clontech, Mountain View, CA, USA) was used to express the MmKu and MtKu in MCF7 Tet-On cells (Clontech, Mountain View, CA, USA). These proteins were expressed as fusion proteins from plasmids pTREcmKuFlag and pTREctKuFlag, respectively. The mitochondrial targeting sequence from human cytochrome C oxidase subunit VIII was ligated at the 5΄ end of the MmKu and MtKu coding sequences and the DNA sequence encoding two Flag peptides was ligated 3΄ to the coding sequences.
In yeast, the mitochondrial targeting sequence of F0-ATPase subunit 9 (Su9) of Neurospora crassa was used to target proteins to mitochondria (34) and the MmKu was expressed as a Flag-tagged protein; two Flag peptides were fused to the C-terminus of the protein. The pGal plasmid (35) (Supplemental Figure S1A) was used to express Su9-RFP (pGalSu9RFP) and Su9-MmKuFlag (pGalSu9MmKu) from the GAL1 promoter. Expression was induced by galactose (Gal), repressed by glucose (D), and growth in raffinose (Raff) medium neither induced nor repressed the promoter.
pRS305 (36), a yeast integrating plasmid, was used to generate pRS305MmKu. This plasmid expressed mitochondrial-targeted Flag-tagged MmKu from the yeast nuclear genome in Gal-inducible manner.
Bacteria carrying plasmids were grown in LB media containing carbenicillin (100 μg/ml; Life Technologies, Carlsbad, CA, USA). Plasmids were harvested from bacteria and purified using the QIAfilter Plasmid Maxi Kit (QIAGEN GmbH, Hilden, Germany).
Yeast strains, media and transformation
Yeast strains used in this study were congenic to strain KT1112 [nuclear genotype: MATaleu2 ura3 his3; mitochondrial genotype: ρ+] (37), unless stated otherwise. The ntg1 null mutant (KT1112/Δntg1) was constructed by amplifying the G418 cassette from the ntg1 deletion panel strain (38) with NTG1F and NTG1R primers. Purified PCR products were introduced into KT1112 and mutants were selected in medium containing G418 (200 μg/ml; Life Technologies, Grand Island, NY, USA). Positive clones were confirmed for the absence of the NTG1 gene by PCR using NTG1F and deltaNTG1R primers.
Strain KT1112 pRS305MmKu (nuclear genotype: MATaleu2:pRS305MmKu:LEU2 ura3 his3; mitochondrial genotype: ρ+) was constructed by transforming KT1112 with pRS305MmKu linearized with EcoRV within LEU2. Transformants were selected in medium lacking leucine (–Leu).
Strain KT1177 (nuclear genotype: bar1::LEU2 ura3 his4; mitochondrial genotype: ρ0), was engineered as described previously (39).
Yeast transformation was performed by a LiAc procedure as described previously (40) with the following modifications: (i) cells were grown in YPD to OD600 of 0.6-0.9, and (ii) carrier DNA was from salmon testes (Sigma, Saint Louis, MO, USA). Yeast transformed with pGalSu9RFP or pGalSu9MmKu were selected and grown in medium lacking uracil (–Ura).
YPD, YPGal, and YPRaff contained 1% yeast extract and 2% bacto peptone with 2% glucose, galactose, and raffinose, respectively. YPEG medium contained 1% yeast extract, 2% bacto peptone, 2% ethanol, and 3% glycerol. Selective media were prepared by combining complete supplement mixture lacking uracil (–Ura) or leucine (–Leu) with yeast nitrogen base (Sunrise Science Products, San Diego, CA, USA) and this was supplemented with 2% carbon source (–Ura D or Gal or Raff, –Leu D or Gal or Raff). Synthetic complete medium contained complete supplement mixture (Sunrise Science Products) and yeast nitrogen base (Sunrise Science Products) with 2% carbon source.
Human cell culture
MCF7 Tet-On cells were co-transfected with pTK-Hyg (Clontech, Mountain View, CA, USA) and pTRE-Tight, pTREcmKuFlag or pTREctKuFlag using Lipofectamine (Invitrogen, ThermoFisher Scientific) and selected with hygromycin B (200 μg/ml; Sigma, Saint Louis, MO, USA) as well as G418 (200 μg/ml). The hygromycin B selection was removed after the individual colonies were isolated for expansion into cell lines. Cell lines containing pTRE-Tight, pTREcmKuFlag or pTREctKuFlag (named MCF7 TreTight, MCF7 cMmKu, and MCF7 cMtKu, respectively) were identified by PCR. Doxycycline (0.5 μg/ml; Sigma, Saint Louis, MO, USA) dependent expression of MmKu and MtKu was confirmed by western analyses. All MCF7 Tet-On cell lines were grown in DMEM containing stable glutamine and sodium pyruvate (Corning Life Sciences, Manassas, VA, USA) supplemented with 10% fetal bovine serum (Gemini Bio-Products, West Sacramento, CA, USA) and G418 (200 μg/ml) at 37°C and 10% CO2. The medium was further supplemented with 50 μg/ml uridine (Sigma, Saint Louis, MO, USA) to make it suitable for growth of respiratory deficient and/or ρ0 human cells.
Spot dilution analysis
Stationary phase yeast cultures pre-grown in –Ura Raff liquid medium were 25-fold serially diluted using sterile water and 6 μl of each dilution was applied to a –Ura Gal plate. Cells were grown at 30°C for 48 h, following which colonies were replica-plated to YPEG and grown at 30°C for 36 h.
Microscopy
A mid-log phase culture of KT1112 harboring pGalSu9RFP was grown in Gal for 6 h, following which cells were incubated with MitoTracker® Green FM (Molecular Probes, Eugene, OR, USA) according to the manufacturer's instructions. Cells were then examined by fluorescence microscopy using the TRITC filter set (for RFP) and GFP filter set (for MitoTracker® Green FM). For mtDNA visualization, live yeast cells treated with DAPI (0.2 μg/ml; Sigma, Saint Louis, MO, USA) were incubated at 30°C for 30 min and imaged for DAPI. Fluorescence images (TRITC, GFP, and DAPI filter sets; Olympus, Japan) and DIC images were acquired using SlideBook Digital Microscopy Imaging Software (Intelligent Imaging Innovations, Denver, CO, USA).
Determination of percentage of ρ+ colonies
Determination of percentage of ρ+ colonies was performed as described below, unless stated otherwise. Stationary phase yeast cultures harboring pGalSu9MmKu or pRS305MmKu were pre-grown in –Ura Raff or –Leu Raff, respectively, and diluted to a final concentration of OD600 of 0.04. The cultures were grown at 30°C for 3 h on a rotary shaker (200 rpm), following which 2% Gal or 2% Raff was added. The experimental procedure up to this stage is referred to as the initial culture set up. The cultures were then returned to the rotary shaker, and aliquots were removed at the indicated time points. Aliquots were then serially diluted in sterile water to plate ∼200 cells on –Ura D or –Leu D agar plates and grown to obtain colonies at 30°C for 72 h. The colonies were counted, replica-plated to YPEG and incubated at 30°C for 36 h, before counting YPEG-positive colonies. The percentage of ρ+ colonies was calculated using the following formula:
% ρ+ colonies = (number of colonies on YPEG/ number of colonies on –Ura D or –Leu D) × 100.
Immunoblot analysis
Yeast total protein extracts were prepared by lysing yeast with glass beads in trichloroacetic acid as described previously (41). Yeast mitochondrial extract (Mito) was prepared from cells using the Yeast Mitochondria Isolation Kit according to manufacturer's instruction (Bio Vision, Milpitas, CA, USA). Protein extracts were subjected to SDS-PAGE and transferred to nitrocellulose (Bio-Rad Laboratories, Inc., Germany). Membranes were probed with the indicated primary antibodies: for anti-Flag M2 antibody (1:2000; Sigma, Saint Louis, MO, USA), a sheep anti-mouse secondary horseradish peroxidase antibody (1:8000; Amersham Biosciences, Buckinghamshire, UK) was used, whereas for anti-Cox4 (1:2000; MitoSciences, Eugene, OR, USA) and anti-Pgk1 (1:30000; Invitrogen, Frederick, MD, USA) antibodies, a goat anti-mouse secondary horseradish peroxidase antibody (1:20 000 and 1:31 250, respectively; Bio-Rad, Hercules, CA, USA) was used.
Human total protein extract was prepared as previously described (42). Mitochondrial and cytosolic extracts were obtained using the Mitochondria Isolation Kit for Mammalian Cells according to the manufacturer's instructions (Thermo Scientific, Rockford, IL, USA). Protein extracts were subjected to SDS-PAGE and transferred to nitrocellulose. Membranes were probed with the following antibodies: for anti-Flag M2 (1:4000) and anti-Actin (1:4000; BD Biosciences Pharmingen, San Diego, CA) antibodies, a sheep anti-mouse secondary horseradish peroxidase antibody (1:5000) was used, whereas for anti-SDHB antibody (1:2000; Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA), an anti-rabbit secondary horseradish peroxidase antibody (1:2000; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) was used.
For all immunoblots, antibody binding was detected by enhanced chemiluminescence (ECL-Prime; GE Healthcare, Buckinghamshire, UK) and visualized using autoradiography and/or a Chemi Doc MP Imaging System (Bio-Rad, USA). Image Lab Software 5.2 (Bio-Rad, USA) was used to quantitate the signal on immunoblots.
Total DNA isolation
Isolation of yeast total DNA was performed as described previously (43). Briefly, yeast cells were lysed with glass beads for 5 min in lysis buffer and phenol/chloroform/isoamylalcohol (25:24:1; Fisher Scientific, Fair Lawn, NJ, USA). The pellet obtained after ethanol precipitation was resuspended in TE and treated with DNase-free RNase (Sigma, Saint Louis, MO, USA) for 1 h at 37°C. A final DNA precipitation was performed with ice-cold absolute ethanol in the presence of ammonium acetate. The DNA pellet was dried and resuspended in TE. Total DNA from human cells was isolated using the QIAamp DNA Mini Kit according to the manufacturer's instructions (QIAGEN GmbH, Hilden, Germany). DNA was quantitated using a Nanodrop ND-1000 spectrophotometer (Wilmington, DE, USA).
End-point PCR and real-time quantitative PCR (qPCR)
End-point PCR was performed with 250 nM of each primer, 20 ng template DNA, 3 mM MgCl2, 200 μM (each) dATP, dGTP, dCTP and dTTP, and 0.5 U Go Taq Flexi DNA polymerase (Promega, Madison, WI, USA) in a MyCycler Thermal Cycler (Bio-Rad, USA). To determine mtDNA content, mtDNA amplification was performed using XhoIF and XhoIR primers for 25 cycles with an annealing temperature of 50°C, whereas nuclear DNA (nDNA) amplification was performed using Y2Oli2 and Y2Oli7 primers for 20 cycles with an annealing temperature of 42°C. To examine the PCR product generated across ori5, mtDNA amplification was performed using ORI5AcrF and ORI5R for 23 cycles, and XhoIUpInF and XhoIUpInR for 18 cycles. Both primer sets used an annealing temperature of 53.5°C. PCR products were subjected to electrophoresis, visualized using a Chemi Doc MP Imaging System, and quantitated with Image Lab Software 5.2.
For real-time qPCR, Power SYBR Master Mix (Applied Biosystems, Warrington, UK), 300 nM of each primer and 1 ng of DNA template were mixed in a 15 μl reaction. All qPCR was performed in a 7500 Fast Real-Time PCR systems (Applied Biosystems) using the following cycling conditions: 50°C for 2 min, 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 1 min, followed by a dissociation curve analysis to ascertain amplicon specificity. Nuclease-free water was substituted for template in negative control reactions. mtDNA amplification was performed using COX1F and COX1R primers (amplicon length 147 bp) for yeast cells, and Human-mtDNAF and Human-mtDNAR primers (amplicon length 125 bp) for human cells. Similarly, nDNA amplification was performed using ACT1F and ACT1R primers (amplicon length 89 bp) for yeast cells, and Human18SF and Human18SR primers (amplicon length 100 bp) for human cells. Relative DNA (mtDNA and nDNA) levels were determined by using a standard curve, which was generated with serially diluted total DNA (ranging from 10 000 pg to 1 pg) obtained from KT1112 (for yeast) or MCF7 (for human cells). Primer efficiencies were established from the slope of the standard curve and hence the efficiencies were taken into account in the determination of the relative amounts of DNA. mtDNA amplification was normalized to nDNA amplification to obtain the relative mtDNA copy number.
Determination of ρ° formation
Following the initial culture set up, KT1112 cells containing pGalSu9MmKu were grown in Raff or Gal for 8 h and ∼200 cells were plated on –Ura D to obtain colonies. The 8 h cultures were diluted to OD600 of 0.04 and grown for another 24 h so that the total growth period in the indicated sugar was 32 h. ∼200 cells were then plated on –Ura D to obtain colonies. Colonies were then replica-plated to YPEG and –Ura D. Petite colonies were identified as those unable to grow on YPEG. At each time point, 5 ρ+ colonies from the Raff culture and 12 petite colonies from the Gal culture were randomly selected and used to initiate cultures in –Ura D medium. Total DNA was isolated from each culture and relative mtDNA copy number was determined by real-time qPCR. Genomic DNA from KT1177 (ρ0 strain) was used in negative control reactions for mtDNA amplification. The mtDNA status in petite colonies that showed no mtDNA amplification was verified with DAPI.
mtDNA immunoprecipitation (mtDNA IP)
The procedure for mtDNA IP was adapted from a protocol described originally for chromatin immunoprecipitation (44). A detailed protocol is provided in the Supplemental Materials and Methods. Briefly, following the initial culture set up, yeast cells containing pGalSu9MmKu were grown in Gal for 5 h. DNA was crosslinked to proteins by the addition of formaldehyde, cells were harvested, resuspended in lysis buffer and lysed with glass beads by vigorous shaking. The sample was sonicated to obtain a mean DNA length of ∼0.3 kb and DNA–protein complexes were immunoprecipitated with 1.5 μg of anti-Flag M2 antibody or mouse IgG1. The purified immunoprecipitated DNA was subjected to real-time qPCR using ori5 specific primers (ORI5F and ORI5R) and COX1 specific primers. A standard curve specific for each amplicon was used to determine the amount of mtDNA present in each immunoprecipitate and background signal obtained from the IgG1 control was subtracted. The net immunoprecipitate obtained was used to calculate the percentage of input immunoprecipitate (IPT) using the following formula, where total input represents the total amount of DNA-protein complex used in the immunoprecipitation:
% Input IPT = (amount of mtDNA in net IP/amount of mtDNA in total input) × 100.
End-point PCR across the ori5 DSB region
Cultures of KT1112 and KT1112/Δntg1 were grown as described for the mtDNA IP assay. End-point PCR was performed from total cellular DNA as described above using primers ORI5AcrF and ORI5R to amplify a 562 bp region that encompassed the ori5 DSB region, and XhoIUpInF and XhoIUpInR to amplify a 281 bp segment of mtDNA that lies >24 kb away from ori5 and is a control region on mtDNA. PCR products were subjected to electrophoresis, visualized using a Chemi Doc MP Imaging System, and quantitated with Image Lab Software 5.2. A ratio of the amount of PCR product across the ori5 DSB region/PCR product from the control region was calculated.
Pedigree analysis
Pedigree analysis was adapted from (45). Briefly, pedigree plates consisted of two segments of agar containing YPGal and YPD separated by ∼5 mm. Early log phase yeast cells grown in YPRaff were streaked on the YPGal segment, from which unbudded cells were transferred to discrete positions on YPGal by micromanipulation. These cells were designated as mother cells for the pedigree. After cell division, the daughter cell from each mother cell was transferred from YPGal to YPD. This process was continued for three daughter generations from each mother cell, following which the mother cell was placed on YPD. During transfer, each pedigree was observed regularly such that the interval of observation was always <1 h. After completion of cell transfer, cells were incubated at 30°C for 72 h. The colonies were replica plated to YPEG and incubated at 30°C for 36 h. The percentage of ρ+ colonies for each daughter generation and the mother generation was calculated as follows:
% of ρ+ colonies = (number of colonies on YPEG/number of colonies on YPD) × 100.
Statistical analysis
Data are presented as mean ± standard deviation (SD) of three independent experiments, unless stated otherwise. Statistical comparisons were performed either with Student's t-test (between two groups) or with ANOVA (for more than two groups) followed by post hoc analysis using Tukey's multiple comparison test. Differences are considered statistically significant at P < 0.05.
RESULTS
Expression of mitochondrial-targeted proteins from pGal plasmids
To express mitochondrial-targeted proteins in yeast cells, we used the presequence of the F0-ATPase subunit 9 (Su9) of Neurospora crassa as a mitochondrial targeting sequence (34). pGalSu9RFP contains the Su9 in frame with RFP. Induction of the promoter with Gal produces a fusion protein in which the Su9 sequence is linked to the N-terminus of RFP (Supplemental Figure S1B, upper panel). Induction of pGalSu9MmKu with Gal results in expression of a fusion protein in which MmKu has the Su9 sequence at the N-terminus and two Flag epitopes at the C-terminus (Supplemental Figure S1B, lower panel). Su9 contains 69 amino acid residues and possesses a mitochondrial matrix processing peptidase site at the interface between the 66th and 67th amino acids, hence the last three amino acids at the C-terminus of Su9 remain attached to the N-terminus of downstream proteins following the activity of the mitochondrial matrix processing peptidase (46,47). To confirm the ability of Su9 to deliver proteins into yeast mitochondria, KT1112 harboring pGalSu9RFP was grown in the presence of Gal for 6 h, following which cells were treated with MitoTracker® Green. Visualization of cells using fluorescence microscopy revealed that the RFP and the MitoTracker® Green localized to the same mitochondrial structures (Supplemental Figure S1C). To assess expression of MmKu, total cell extracts and mitochondrial extracts from KT1112 carrying pGalSu9MmKu were subjected to western analysis. Growth of cells in Gal for 8 h triggered expression of MmKu, which localized to yeast mitochondria (Supplemental Figure S1D).
Mitochondrial-targeted MmKu triggers petite formation
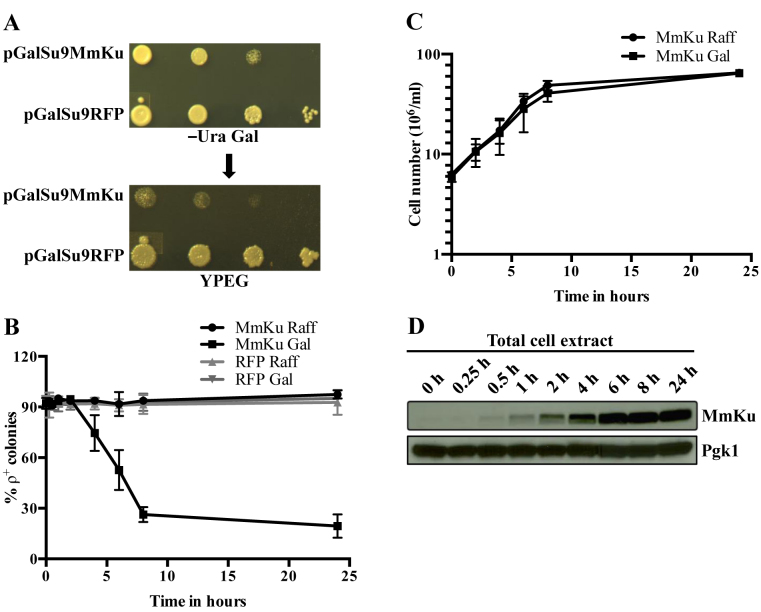
Petite formation was determined by the ability of yeast cells to perform mitochondrial respiration. KT1112 cells carrying pGalSu9MmKu or pGalSu9RFP were serially diluted and applied to Gal medium to induce expression of the mitochondrial-targeted proteins. Cells carrying pGalSu9RFP grew normally on the medium, whereas cells harboring pGalSu9MmKu grew slowly (Figure 1A, upper panel). The colonies were then replica plated to YPEG, a medium on which only respiratory competent yeast cells can grow. Although cells expressing RFP grew well on YPEG, cells harboring pGalSu9MmKu formed small or no colonies (Figure 1A, lower panel), indicating a compromised mitochondrial electron transport chain. To quantitate the respiration defect, yeast cells transformed with MmKu or RFP expression plasmid were grown in Raff or Gal liquid medium for increasing time intervals, following which cells were plated on solid medium containing glucose. The colonies were then replica plated to YPEG and the percentage of ρ+ colonies was determined. The percentage of ρ+ colonies in cells expressing MmKu decreased with time; only ∼26% and ∼19% of colonies were ρ+ at 8 and 24 h, respectively (Figure 1B). In contrast, colonies obtained from control cultures maintained the ρ+ phenotype. The percentage of ρ+ colonies for cells expressing MmKu plateaus after 8 h, and this corresponded to the time that the yeast culture exited exponential growth (Figure 1C). When the 8 h Gal culture was diluted with medium containing Gal and grown for another 24 h, the percentage of ρ+ colonies further decreased (Figure 2C). Western analysis detected MmKu within 30 min of Gal treatment, and expression was maximal at ∼6–8 h (Figure 1D). These results indicate that mitochondrial-targeted MmKu impairs mitochondrial respiration and induces petite formation.
Figure 1.
Mitochondrial-targeted MmKu impairs mitochondrial respiration. (A) Following serial dilution, cells containing pGalSu9MmKu or pGalSu9RFP were spotted on –Ura Gal, and imaged after 48 h (upper panel). The colonies were replica-plated to YPEG and imaged after 36 h (lower panel). Images are representative of at least three independent experiments. (B) For determination of the percentage of ρ+ colonies, the procedure for the initial culture set up (see the Materials and Methods) was modified slightly such that the stationary phase yeast cultures carrying MmKu or RFP expression plasmid pre-grown in –Ura Raff were diluted 20-fold in the same medium. After addition of Raff or Gal for the indicated time periods, cells were plated on –Ura D. Colonies were replica-plated to YPEG and the percentage of ρ+ colonies was calculated. Each curve depicts average measurements from three independent experiments. (C) Growth of cultures containing pGalSu9MmKu was followed over the indicated time period. Each curve represents average measurements from two independent experiments. (D) Cells carrying pGalSu9MmKu were grown in Gal and samples were taken at indicated time points to obtain total cell extracts, which were probed with anti-Flag or anti-Pgk1 antibody. The immunoblot shown is representative of two independent experiments. Pgk1, phosphoglycerate kinase, is used as a loading control.
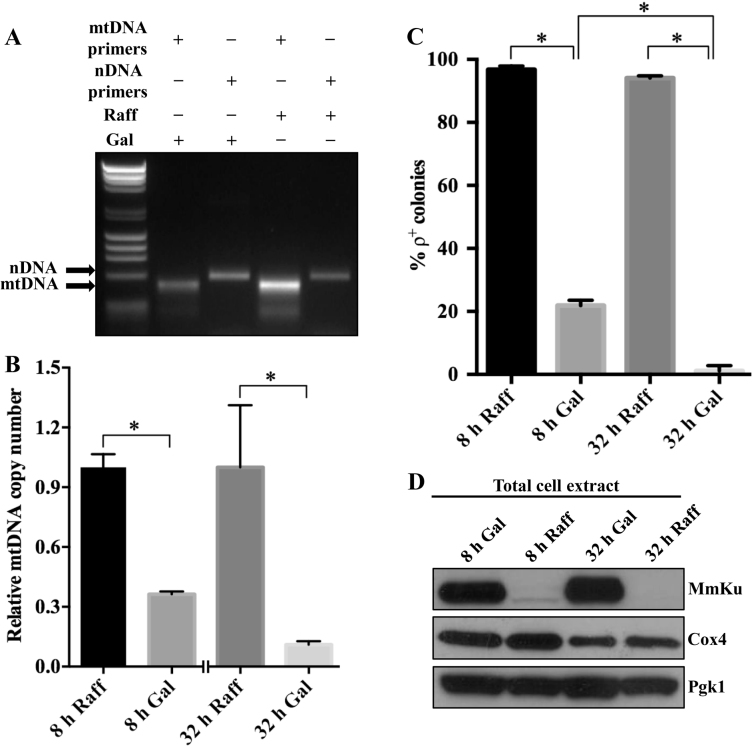
Figure 2.
Mitochondrial-targeted MmKu decreases mtDNA content in yeast cells. Total DNA was isolated from KT1112 harboring pGalSu9MmKu grown in Raff or Gal for 8 or 32 h and end-point PCR (A) or real-time qPCR (B) was performed to amplify segments of mtDNA and nDNA. An image (A) representative of at least three independent end-point PCR experiments is shown. For real-time qPCR (B), data are represented graphically as the mean ± SD from three independent experiments. * represents P < 0.05. (C) Part of the cultures used for DNA isolation were used to quantitate the percentage of ρ+ colonies, and the averages are shown graphically. Error bars represent SD and * represents P < 0.05. (D) MmKu, Cox4 and Pgk1 expression in KT1112 carrying pGalSu9MmKu grown in Raff or Gal for 8 h and 32 h. An immunoblot representative of three independent experiments is shown. Cox4, cytochrome C oxidase subunit 4, is a mitochondrial protein.
Mitochondrial-targeted MmKu induces mtDNA depletion
As MmKu possesses a DSB DNA binding domain, we hypothesized that mitochondrial-targeted MmKu triggers petite formation by disturbing mtDNA homeostasis. To test this, we determined the approximate mtDNA content of pGalSu9MmKu-containing cells by examining the mtDNA/nDNA ratio using end-point PCR (Figure 2A). Total DNA isolated from cells grown in Raff or Gal for 8 h was used as the template for PCR. Quantitation of EtBr-stained amplicons revealed ∼4-fold decrease in the mtDNA/nDNA ratio for cells grown in Gal compared to Raff (data not shown). To verify this effect of MmKu, we assessed the relative mtDNA copy number using real-time qPCR. As shown in Figure 2B, there was ∼3-fold decrease in relative mtDNA copy number in cells grown in Gal for 8 h. The 8 h cultures were diluted and grown for another 24 h in Gal or Raff such that the total period of growth was 32 h. After 32 h, there was further decrease in the relative mtDNA copy number in Gal-grown cells compared to Raff-grown cells. The respiratory competence of these cells was also tested and only ∼22% and ∼1% of colonies from cells grown in Gal maintained ρ+ phenotype at 8 h and 32 h, respectively (Figure 2C). In contrast, the percentage of ρ+ colonies for Raff-grown cells was ∼94% at 8 and 32 h. The expression of MmKu at 8 and 32 h was confirmed by western analysis (Figure 2D).
As a control, we determined the relative mtDNA copy number from total DNA obtained from cells containing pGalSu9RFP grown in Raff or Gal for 8 or 32 h (Supplemental Figure S2A). The relative mtDNA copy number did not vary between the 8 h treatment groups and there was only an ∼9% reduction in the relative mtDNA copy number in 32 h Gal-grown cells compared to 32 h Raff control. This indicates expression of a foreign mitochondrial-targeted protein alone or growth in Gal does not dramatically reduce the mtDNA copy number.
To visualize the mtDNA, cells containing pGalSu9RFP or pGalSu9MmKu grown in Raff or Gal for 32 h were stained with DAPI. Whereas bright punctate mtDNA nucleoids were observed towards the cell periphery in 32 h Raff-grown cells and 32 h Gal-grown pGalSu9RFP-containing cells, the punctate staining was markedly reduced in 32 h Gal-grown pGalSu9MmKu-containing cells (Supplemental Figures S2 and S3). The loss of punctate staining therefore confirmed the reduction in mtDNA in the Gal-grown pGalSu9MmKu-containing cells.
Further examination of the western analysis in Figure 2D revealed there was a relatively low Cox4 signal in Gal samples compared to Raff samples. Quantitation of Cox4 relative to the loading control Pgk1 revealed a significantly decreased level of Cox4 in 32 h Gal-grown cells compared to cells grown in Raff for the same period (Supplemental Table S2). A small reduction in Cox4 level was also observed for cells grown in Gal for 8 h compared to the 8 h Raff control (Supplemental Table S2). We therefore examined the Cox4 levels in other western analyses performed during these studies using strains carrying the pGalSu9MmKu and pRS305MmKu after 8 h of Gal treatment (Supplemental Figures S1D and S7A). A 9–12% reduction of Cox4 was also detected in these experiments in Gal-grown compared to Raff-grown cells. This suggests MmKu resulted in a reduction of expression of nuclear-encoded Cox4.
Mitochondrial-targeted MmKu triggers ρ° formation
To determine if MmKu induces complete loss of mtDNA, we used real-time qPCR to analyze the mtDNA/nDNA ratio for individual ρ+ colonies obtained from Raff cultures and petite colonies from Gal cultures. As shown in Table 1, the mtDNA/nDNA ratio was >1 for almost all colonies from the Raff culture. In contrast, 33% of the colonies from the 8 h Gal culture and 50% of the colonies from the 32 h Gal culture had undetectable levels of mtDNA. These colonies with undetectable levels of mtDNA were verified for the complete loss of mtDNA by DAPI staining (Supplemental Figure S4).
Table 1. Mitochondrial-targeted MmKu induces ρ° formation.
| Culture in Raff | mtDNA/nDNA ratio in 5 randomly chosen ρ+ colonies | |||
|---|---|---|---|---|
| <x–0.005 | 0.0051–0.8 | 0.81–1.0 | 1.01–1.25 | |
| 8 h | 0 | 0 | 0 | 5 (100%) |
| 32 h | 0 | 0 | 1 (20%) | 4 (80%) |
| Culture in Gal | mtDNA/nDNA ratio in 12 randomly chosen petite colonies | |||
| <x | >x and <0.005 | 0.005–0.8 | 0.81–1.0 | |
| 8 h | 4 (33%) | 4 (33%) | 3 (25%) | 1 (8%) |
| 32 h | 6 (50%) | 6 (50%) | 0 | 0 |
KT1112 harboring pGalSu9MmKu were grown in Raff or Gal for 8 h and cells were plated on –Ura D. 8 h cultures were then diluted and grown for another 24 h, following which cells were plated on –Ura D. The colonies were replica plated to YPEG and –Ura D. After identifying ρ+ and petite colonies, total DNA was isolated from randomly selected ρ+ and petite colonies to determine the mtDNA/nDNA ratio. In Table 1, x is the ratio for the border between the undetectable level and the least detectable level.
MmKu binds to the mtDNA replication origin ori5
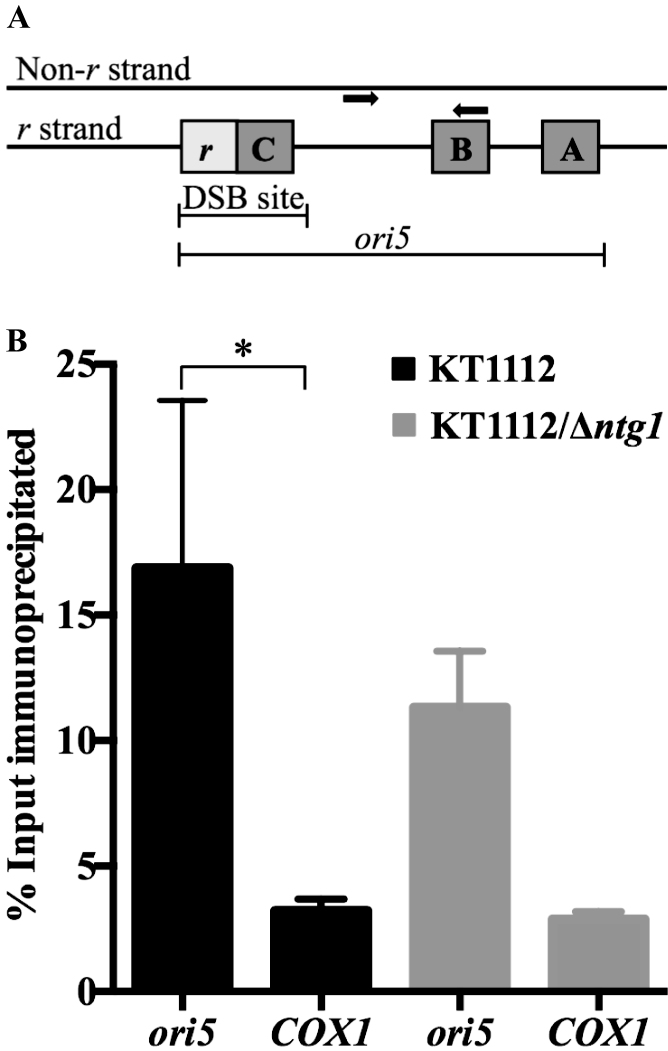
A DSB at yeast mitochondrial replication origin ori5 (Figure 3A) has been implicated in initiation of mtDNA RDR (18,19). As MmKu possesses a DSB binding domain, we hypothesized that MmKu binds to DSBs at ori5 and inhibits mtDNA replication, thereby decreasing total mtDNA content. To test this, mtDNA IP assays were performed using Flag-tagged MmKu expressed in KT1112. An anti-Flag antibody or an IgG1 control antibody was used for the IP. Real-time qPCR with purified immunoprecipitated DNA was performed using ori5-specific primers or COX1-specific primers. The COX1 mitochondrial gene was used as a control because this gene is situated at least 23 kb away from ori5 region. As shown in Figure 3B, binding of MmKu was significantly higher to ori5 compared to the COX1 region, which supports the idea of MmKu binding to DSBs at ori5.
Figure 3.
MmKu binds preferentially to ori5 in the yeast mitochondrial genome. (A) Schematic representation of ori5. The replication origin consists of 3 GC clusters (A–C) and a transcriptional promoter r. A DSB at ori5 can occur in a region that spans the promoter r and ∼5 bp downstream of GC cluster C (18). The ori5-specific primers (bold arrows) amplify a region of ori5 that is downstream of the DSB-occurring region. Figure 3A is adapted from (9) and (18). (B) The indicated strains harboring pGalSu9MmKu were grown in Gal for 5 h, following which DNA–protein complexes were crosslinked with formaldehyde, sheared by sonication and immunoprecipitated with either anti-Flag antibody or IgG1 control. Crosslinks were reversed and purified immunoprecipitated DNA samples were quantitated by real-time qPCR using ori5-specific primers and COX1-specific primers. The graph shows mean ± SD from three independent experiments. P values are as follows: for KT1112 ori5 versus KT1112 COX1 P = 0.0064; for KT1112/Δntg1 ori5 versus KT1112/Δntg1 COX1 P = 0.0741; and for KT1112 ori5 versus KT1112/Δntg1 ori5 P = 0.2892.
MmKu expression in an ntg1 null mutant
Ntg1 has been shown to play a role in instigating DSBs at ori5. In the absence of Ntg1 in hypersuppressive ρ− yeast, the extent of DSBs at ori5 decreases (18,19) and hence MmKu binding at ori5 would be expected to be reduced in a strain deleted in ntg1. The mtDNA IP was therefore repeated in an ntg1 null mutant (KT1112/Δntg1). Binding of MmKu to ori5 was not significantly different from the COX1 region in this strain, which suggests reduced binding to ori5 in the absence of Ntg1-induced DSBs. However, when the results from independent experiments were compared for KT1112/Δntg1 and KT1112, even though the average MmKu binding to ori5 was lower in KT1112/Δntg1, the difference was not statistically significant (Figure 3B). The binding of MmKu to the COX1 control region in the two strains was comparable and also not statistically significant (Figure 3B).
The effect of MmKu expression on mitochondrial respiration was also examined in KT1112/Δntg1 and KT1112. In both strains, the percentage of ρ+ colonies was dramatically decreased by growth in Gal (Supplemental Figure S5). There was also a trend for decreased formation of ρ+ colonies in KT1112/Δntg1 compared to KT1112 when the strains were grown in Gal for 8 h, but there was no statistical difference. At 32 h in Gal, the percentage of ρ+ formation was comparable between the two strains.
Since our data indicated that binding of MmKu to ori5 was not dramatically different in the absence of Ntg1, end-point PCR across the ori5 DSB region (Figure 3A) was used to compare the induction of DSBs at ori5 in KT1112/Δntg1 and KT1112. An increase in PCR product was used as the indicator of a decrease in DSB induction. In each of three independent experiments, there was a small increase in product generated from PCR across the ori5 DSB region in KT1112/Δntg1 compared to KT1112 (Supplemental Figure S6A). Even though this suggests there was reduced DSB formation at ori5 in KT1112/Δntg1, there was no statistical difference between the average amounts of PCR product from the three experiments (Supplemental Figure S6B).
MmKu triggers petite formation preferentially in daughter cells
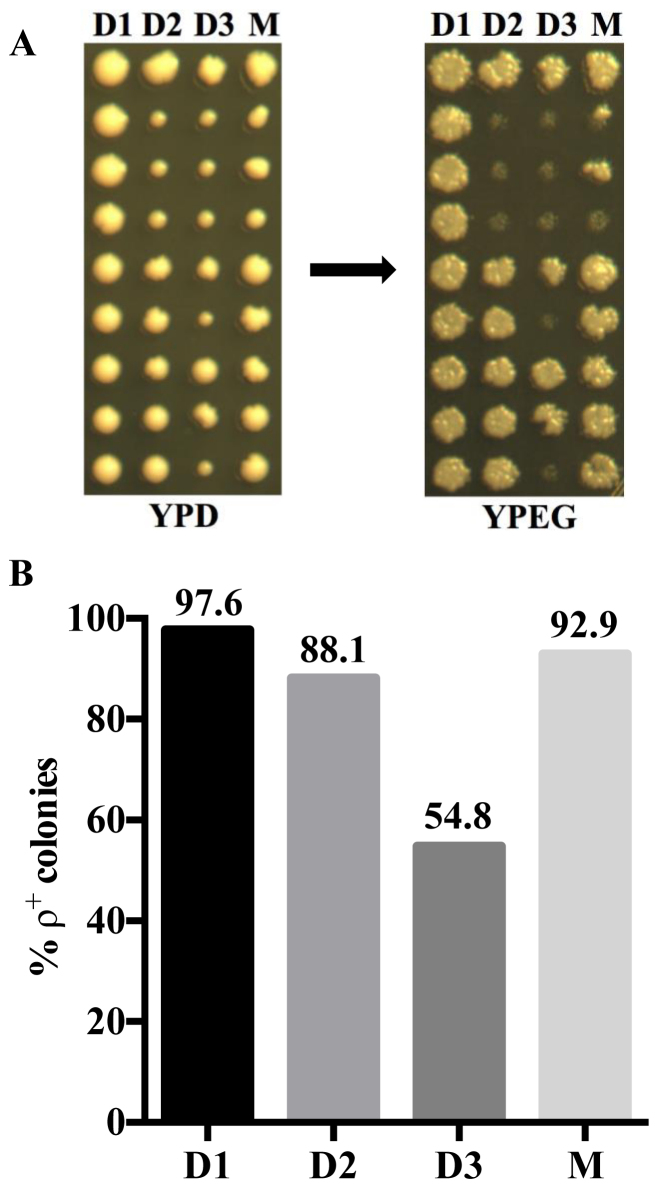
Since the products of mtDNA RDR are selectively transmitted to the daughter cells (17), we reasoned that mtDNA depletion and accompanying petite formation due to MmKu expression occurs preferentially in daughter cells. To test this, we constructed a yeast strain (KT1112 pRS305MmKu) with a nuclear-integrated Gal-inducible MmKu expression plasmid. Following confirmation of expression and the petite generating activity of mitochondrial-targeted MmKu (Supplemental Figure S7), pedigree analyses were performed on KT1112 pRS305MmKu on plates containing YPGal and YPD agar segments. Yeast cells were first placed on YPGal and after cell division, daughter cells were transferred to YPD. Following transfer of three successive daughters, the respective mother was transferred to YPD. It should be noted that each mother cell was exposed to the YPGal medium for ∼7 h, whereas daughter cells were transferred immediately to YPD after cell division. As shown in Figure 4A, petite formation was observed particularly from the second daughter generation. We analyzed a total of 42 colonies for each generation and determined the percentage of ρ+ formation (Figure 4B). We found that ∼45% of colonies from the third daughter (D3) generation were petites, in contrast to ∼7% petite colonies from the mother (M) generation. As a control, we repeated the procedure to analyze the pedigree of the parental strain KT1112 and observed that colonies from all three daughter generations and the mother generation essentially maintained the ρ+ phenotype (Supplemental Figure S8). These data reveal that MmKu induces petite formation preferentially in daughter generations, which is indicative of mtDNA loss selectively in daughter cells.
Figure 4.
Pedigree analyses indicate that MmKu expression inhibits mtDNA segregation into daughter cells. (A) Unbudded early log phase yeast cells were streaked on YPGal segment of a plate. After cell division, a daughter cell from each mother cell was transferred from YPGal to YPD segment of the plate. This process was continued for 3 daughter generations, following which the mother cell was transferred to YPD. The colonies were replica-plated to YPEG to determine the percentage of ρ+ colonies. D1, D2 and D3 represent first, second and third daughter generations, respectively, from the mother M. (B) The data from five independent experiments for KT1112 pRS305MmKu pedigree were pooled to determine the percentage of ρ+ colonies for each daughter generation and the mother generation.
Mitochondrial-targeted bKu proteins do not decrease mtDNA content in human cells
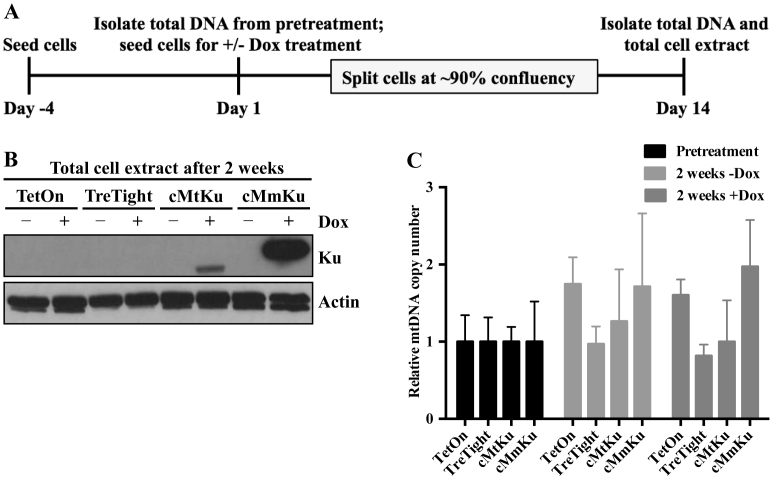
To understand if bKu affects mtDNA metabolism in human cells, MCF7 Tet-On cell lines were generated that expressed mitochondrial-targeted MmKu or MtKu under doxycycline (Dox) control. These cell lines, together with the vector control cell line, MCF7 TreTight, were grown in the presence or absence of Dox for 48 h, following which total cell extracts were isolated. Western analysis revealed Dox-dependent expression of bKu proteins (Supplemental Figure S9A). To confirm the subcellular localization of bKu in human cells, mitochondrial and cytosolic extracts were obtained from MCF7 cMtKu and MCF7 cMmKu cells. Because the expression level of MtKu was small compared to MmKu, 35 μg of MCF7 cMtKu extracts and 3.5 μg of MCF7 cMmKu extracts were used for immunoblot analysis. Both MtKu and MmKu localized to cytosolic as well as mitochondrial fractions in human cells (Supplemental Figure S9B).
To understand if mitochondrial-targeted bKu depletes human mtDNA content, the relative mtDNA copy number was determined in the MCF7 Tet-On cell lines. As illustrated in Figure 5A, cells were harvested after 4 days of growth in culture medium. From half of the harvested cells, total DNA (pretreatment) was isolated, whereas the other half was used to passage cells in the presence or absence of Dox for 2 weeks. After the 2-week period, cells were harvested to obtain total DNA and total cell extracts. Western analysis demonstrated expression of bKu after growth for 2 weeks only in cells treated with Dox (Figure 5B). However, the relative mtDNA copy number in cells expressing bKu did not change compared to Dox-untreated controls (Figure 5C). Human ρ° cells require uridine and pyruvate for normal growth (48). To exclude the possibility that we were losing respiratory deficient and/or ρ0 MCF7 cells following bKu expression, we repeated the experiment by growing MCF7 TreTight and MCF7 cMmKu in medium containing uridine and pyruvate. Analogous to Figure 5C, the relative mtDNA copy number in cells treated with Dox did not vary compared to Dox-untreated control (Supplemental Figure S10). Hence, mitochondrial-targeted bKu does not decrease mtDNA copy number in human MCF7 cells.
Figure 5.
Mitochondrial-targeted bKu proteins do not induce mtDNA depletion in human MCF7 cells. (A) MCF7 cell lines were grown for 4 days prior to treatment with Dox (pretreatment) and then harvested. Total DNA was isolated from a portion of the cells, and the remaining cells were propagated in the presence or absence of Dox for 2 weeks. During the 2-week period, each cell line was passaged after reaching ∼90% confluency. At the end of 2 weeks, cells were harvested to obtain total DNA and total cell extract (TCE). (B) Western analysis of TCEs was performed and the membranes were probed with anti-Flag or anti-actin antibody. An immunoblot representative of two independent experiments is shown. TetOn: MCF7 Tet-On; TreTight: MCF7 TreTight; cMtKu: MCF7 cMtKu; cMmKu: MCF7 cMmKu. (C) From total cell DNA, the relative mtDNA copy number was determined for each cell line in each group. The relative mtDNA copy number of each cell line in the treatment groups (+ and −Dox) was normalized to the respective cell line in the pretreatment group. Data are represented graphically as mean ± SD from three independent experiments.
DISCUSSION
In this study, we have utilized the DSB binding property of bacterial Ku to probe for DSB-mediated mtDNA replication in yeast and human cells. The work presented here provides evidence for DSB-mediated mtDNA replication in ρ+ yeast cells but not in human MCF7 cells.
Ori5 has been frequently studied as a model replication origin in yeast mtDNA (11,13,18,19) and evidence indicates that ori5 initiates mtDNA replication in a DSB-induced manner in HS ρ− cells (18,19). Previously, it was not clear whether such DSB-mediated mtDNA replication also operates in ρ+ cells. Here, we show that MmKu binds preferentially to ori5 compared to a control region (COX1) in ρ+ mtDNA, indicating that DSBs are present at ori5. This finding is in agreement with studies that showed the presence of DSBs at ori5 in HS ρ− mtDNA (18,19). The low level of MmKu binding to the COX1 region could be due to MmKu binding to undamaged mtDNA. A recent study found that M. smegmatis Ku can bind to DNA without free ends and this property was attributed to its lysine-rich C-terminal extension (49). MmKu has a similar C-terminal extension (32,50).
Ntg1 has been shown to play an important role in generating DSBs at ori5 (18,19) as seen from the observation that ntg1 null HS ρ− mutants have about half the level of DSBs at ori5 compared to the isogenic NTG1 HS ρ− cells (19). Although we see a small decrease in binding of MmKu to ori5 in KT1112/Δntg1, no statistical difference was found compared to KT1112 for the mtDNA IP assay and for the end-point PCR assay used to determine whether less DSBs were induced in normal growing ρ+ntg1 null cultures. This suggests there is also an Ntg1-independent mechanism for DSB generation at ori5. The DSBs at ori5 could be generated via the action of an unidentified mitochondrial enzyme, as previously suggested (19). Alternatively, ROS could abstract a hydrogen atom from any of the carbons of the deoxyribose in the DNA backbone, subsequently leading to the formation of a single-strand break (51–54). This could occur on both strands of the damage-prone bubble-like ori5 DNA structure resulting in the formation of a DSB.
The observation that MmKu expression triggers mtDNA depletion supports our proposal that binding of MmKu to DSBs at ori5 inhibits mtDNA RDR. Mhr1 is regarded as a central molecule in mtDNA RDR and evidence indicates that Mhr1 facilitates heteroduplex joint formation between a linearized mtDNA molecule and its complementary region in a template circular mtDNA for the initiation of RDR (16,17,19) (Figure 6A). A study with mhr1 temperature-sensitive mutants showed that growth of mhr1 mutants at the non-permissive temperature induces mtDNA loss as well as petite formation (55). In addition, the degree of mtDNA loss and the extent of petite formation in mhr1 mutants increased progressively with time at the non-permissive temperature (55). Analogous to that study, we observe that MmKu expression induces mtDNA loss and petite formation in yeast cells. In addition, the degree of mtDNA depletion and the frequency of petite formation increased progressively with the duration of MmKu expression. The extent of mtDNA depletion was such that ρ0 yeast colonies were obtained from MmKu-expressing cells grown in Gal for 8 h; after 32 h in Gal, the percentage of ρ0 formation further increased. MmKu-triggered mtDNA depletion also accompanied reduction in the level of Cox4, a nuclear-encoded mitochondrial protein that forms a subunit of complex IV of the mitochondrial respiratory chain (56). The reduced level of Cox4 could be due to diminished COX4 gene transcription, a phenomenon that has been documented previously in ρ0 yeast cells (57).
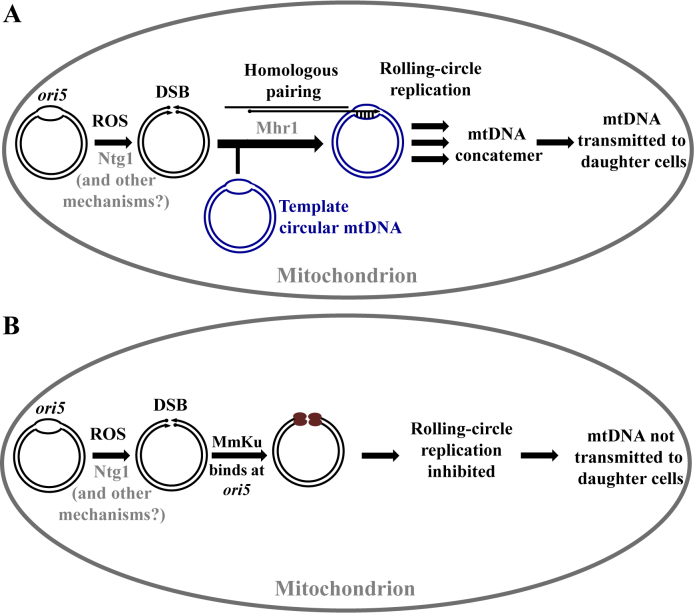
Figure 6.
A model explaining the effect of MmKu on S. cerevisiae mtDNA. (A) Under normal physiological conditions, ROS oxidizes yeast mtDNA at ori5. Ntg1 recognizes these modifications and introduces a DSB. Other unidentified mechanisms can also account for DSBs at ori5. A 3΄ single-stranded tail is generated, to which Mhr1 binds. This nucleoprotein filament invades a template circular mtDNA to mediate the initiation of mtDNA RDR. This results in formation of mtDNA concatemers, which are selectively transmitted to daughter cells. Figure 6A is adapted from (19). (B) Yeast cells expressing mitochondrial-targeted MmKu have the Ku protein bound to the DSB at ori5. This inhibits mtDNA RDR, so new mtDNA molecules are not synthesized. As a consequence, mtDNA molecules are not segregated to daughter cells.
The products of mtDNA RDR are concatemers, which occur predominantly in mother cells, and these multi-genome units are segregated into growing buds where they are processed into genome size monomers (16,17). With this knowledge, we hypothesized that MmKu-induced mtDNA depletion and petite formation occur preferentially in daughter cells. As predicted, we observed that MmKu triggers petite formation preferentially in daughter generations indicating loss of mtDNA in daughter cells. The extent of petite formation was such that ∼45% of colonies from the third daughter (D3) generation formed petites. This is indicative of improper mtDNA segregation to almost half of the D3 generation. Even though mother cells spent more time than any daughter cell of the same pedigree in the Gal medium that induced MmKu expression, only ∼7% of colonies from the mother generation (M) formed petites. This observation supports our proposal that MmKu inhibits mtDNA replication, thereby preventing transmission of mtDNA from mother cells into the daughter cells. To account for the effect of MmKu on yeast mtDNA content, we propose a model (Figure 6B) in which the DSB binding domain of MmKu binds to DSBs at ori5 in the yeast mitochondrial genome. Since it lacks domains to communicate with eukaryotic proteins, MmKu persistently occupies ori5. This results in inhibition of mtDNA RDR, thereby preventing the formation of concatemers. As a consequence, there is reduced/no mtDNA segregation into the daughter cells, which eventually form ρ0.
Evidence for transcription-dependent mtDNA replication also exists for yeast cells (11–13); however, those studies were performed either in hypersuppressive ρ− mtDNA (11,13) or with cloned replication origins examined in vitro (12). It is currently not known if such transcription-dependent mechanisms actually operate in ρ+ cells. Instead, transcription-independent mtDNA replication seems more probable because of the observation that yeast cells with disrupted RPO41 gene can still maintain mtDNA (14,15). In addition, yeast cells that completely lack ori sequences also propagate their mitochondrial genomes (58). The DSB-mediated mtDNA RDR has gained increased attention over the last few years and this mechanism can explain many conundrums, including: (i) a mechanism for maintenance of mtDNA in Rpo41 deficient cells and cells lacking any replication origin; (ii) a mechanism for maintenance of the state of homoplasmy; (iii) a mechanism by which mtDNA concatemers and circular monomers co-exist in yeast cells.
The exact mode of human mtDNA replication is still vigorously debated and multiple replication models have been proposed. These replication modes appear to occur by a transcription-dependent mechanism and do not seem to involve DSBs (7,33). In agreement with this, we find that expression of mitochondrial-targeted bKu proteins (MmKu and MtKu) do not decrease mtDNA content in human MCF7 cells. This observation supports the idea that MmKu impairs mtDNA homeostasis only when DSBs are involved in the perpetuation of the mitochondrial genome. Even though DSBs and other DNA damages are generally considered deleterious for cellular health, studies have indicated that controlled damage of mtDNA is important for its maintenance, both in yeast (18,19) as well as in mammalian cells (59). Recently, oxidative damage to the mtDNA regulatory region (D-loop) under hypoxic conditions has been correlated with an increase in mtDNA copy number in mammalian cells (59). Another study has revealed that exposure of MELAS patient-derived primary fibroblasts to exogenous ROS triggers the formation of mtDNA concatemers via rolling circle replication, likely initiated from ROS-induced DSBs (60). Although beyond the scope of this study, it would be interesting to determine if the expression of mitochondrial-targeted MmKu in ROS-treated MELAS patient-derived fibroblasts inhibits mtDNA replication and concatemer formation. Given the complex diversity of cells in mammals, it is possible that multiple mtDNA replication mechanisms coexist in higher organisms and that certain cell types utilize particular replication mechanisms. With respect to a single-celled S. cerevisiae, however, DSB-mediated mtDNA replication appears to be the prime mode. In this regard, we consider ori5 as a hot spot for DSB-mediated recombination. The concept of DSB-mediated replication is not new and was originally proposed for yeast mtDNA in 1991 by Maleszka et al. (4) and for E. coli DNA replication in 1994 by Asai et al. (61). It is currently not known if other replication origins in yeast mtDNA initiate replication in a DSB-mediated manner and our system could be used to address this in future studies. At present, our study provides crucial evidence that ρ+ yeast cells rely on DSB-induced mtDNA replication for propagation of mtDNA, as perturbation of this pathway can result in the complete loss of their mitochondrial genome.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr David S. Gross (LSUHSC-Shreveport) and Dr Anandhakumar Jayamani (LSUHSC-Shreveport) for technical assistance with mtDNA IP. We thank Reneau Castore (LSUHSC-Shreveport) for her help with MCF7 cell lines.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Feist-Weiller Cancer Center (LSUHSC-Shreveport) for providing a Carroll Feist Predoctoral Fellowship (to K.P.); National Institutes of Health R21 grant [1R21 CA167796 to L.H.]. Funding for open access charge: LSU.
Conflict of interest statement. None declared.
REFERENCES
- 1. Taanman J.W. The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta. 1999; 1410:103–123. [DOI] [PubMed] [Google Scholar]
- 2. Kucej M., Butow R.A.. Evolutionary tinkering with mitochondrial nucleoids. Trends Cell Biol. 2007; 17:586–592. [DOI] [PubMed] [Google Scholar]
- 3. Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F. et al. . Sequence and organization of the human mitochondrial genome. Nature. 1981; 290:457–465. [DOI] [PubMed] [Google Scholar]
- 4. Maleszka R., Skelly P.J., Clark-Walker G.D.. Rolling circle replication of DNA in yeast mitochondria. EMBO J. 1991; 10:3923–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bendich A.J. Structural analysis of mitochondrial DNA molecules from fungi and plants using moving pictures and pulsed-field gel electrophoresis. J. Mol. Biol. 1996; 255:564–588. [DOI] [PubMed] [Google Scholar]
- 6. Foury F., Roganti T., Lecrenier N., Purnelle B.. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 1998; 440:325–331. [DOI] [PubMed] [Google Scholar]
- 7. Holt I.J., Reyes A.. Human mitochondrial DNA replication. Cold Spring Harbor Perspect. Biol. 2012; 4:a012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baldacci G., Bernardi G.. Replication origins are associated with transcription initiation sequences in the mitochondrial genome of yeast. EMBO J. 1982; 1:987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lecrenier N., Foury F.. New features of mitochondrial DNA replication system in yeast and man. Gene. 2000; 246:37–48. [DOI] [PubMed] [Google Scholar]
- 10. Kaniak-Golik A., Skoneczna A.. Mitochondria-nucleus network for genome stability. Free Rad. Biol. Med. 2015; 82:73–104. [DOI] [PubMed] [Google Scholar]
- 11. Baldacci G., Cherif-Zahar B., Bernardi G.. The initiation of DNA replication in the mitochondrial genome of yeast. EMBO J. 1984; 3:2115–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu B., Clayton D.A.. A persistent RNA-DNA hybrid is formed during transcription at a phylogenetically conserved mitochondrial DNA sequence. Mol. Cell. Biol. 1995; 15:580–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Graves T., Dante M., Eisenhour L., Christianson T.W.. Precise mapping and characterization of the RNA primers of DNA replication for a yeast hypersuppressive petite by in vitro capping with guanylyltransferase. Nucleic Acids Res. 1998; 26:1309–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fangman W.L., Henly J.W., Brewer B.J.. RPO41-independent maintenance of [rho-] mitochondrial DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990; 10:10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lorimer H.E., Brewer B.J., Fangman W.L.. A test of the transcription model for biased inheritance of yeast mitochondrial DNA. Mol. Cell. Biol. 1995; 15:4803–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ling F., Shibata T.. Recombination-dependent mtDNA partitioning: in vivo role of Mhr1p to promote pairing of homologous DNA. EMBO J. 2002; 21:4730–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ling F., Shibata T.. Mhr1p-dependent concatemeric mitochondrial DNA formation for generating yeast mitochondrial homoplasmic cells. Mol. Biol. Cell. 2004; 15:310–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ling F., Hori A., Shibata T.. DNA recombination-initiation plays a role in the extremely biased inheritance of yeast [rho-] mitochondrial DNA that contains the replication origin ori5. Mol. Cell. Biol. 2007; 27:1133–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hori A., Yoshida M., Shibata T., Ling F.. Reactive oxygen species regulate DNA copy number in isolated yeast mitochondria by triggering recombination-mediated replication. Nucleic Acids Res. 2009; 37:749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ling F., Hori A., Yoshitani A., Niu R., Yoshida M., Shibata T.. Din7 and Mhr1 expression levels regulate double-strand-break-induced replication and recombination of mtDNA at ori5 in yeast. Nucleic Acids Res. 2013; 41:5799–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shibata T., Ling F.. DNA recombination protein-dependent mechanism of homoplasmy and its proposed functions. Mitochondrion. 2007; 7:17–23. [DOI] [PubMed] [Google Scholar]
- 22. Dujon B. Strathern JN, James EWJ, Broach R. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. 1981; NY: Cold Spring Harbor Laboratory Press; 505–635. [Google Scholar]
- 23. Williamson D. The curious history of yeast mitochondrial DNA. Nat. Rev. Genet. 2002; 3:475–481. [DOI] [PubMed] [Google Scholar]
- 24. Downs J.A., Jackson S.P.. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 2004; 5:367–378. [DOI] [PubMed] [Google Scholar]
- 25. Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010; 79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shuman S., Glickman M.S.. Bacterial DNA repair by non-homologous end joining. Nat. Rev. Microbiol. 2007; 5:852–861. [DOI] [PubMed] [Google Scholar]
- 27. Doherty A.J., Jackson S.P., Weller G.R.. Identification of bacterial homologues of the Ku DNA repair proteins. FEBS Lett. 2001; 500:186–188. [DOI] [PubMed] [Google Scholar]
- 28. Weller G.R., Kysela B., Roy R., Tonkin L.M., Scanlan E., Della M., Devine S.K., Day J.P., Wilkinson A., d’Adda di Fagagna F. et al. . Identification of a DNA nonhomologous end-joining complex in bacteria. Science. 2002; 297:1686–1689. [DOI] [PubMed] [Google Scholar]
- 29. Aravind L., Koonin E.V.. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001; 11:1365–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Castore R., Hughes C., Debeaux A., Sun J., Zeng C., Wang S.Y., Tatchell K., Shi R., Lee K.J., Chen D.J. et al. . Mycobacterium tuberculosis Ku can bind to nuclear DNA damage and sensitize mammalian cells to bleomycin sulfate. Mutagenesis. 2011; 26:795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shao Z., Davis A.J., Fattah K.R., So S., Sun J., Lee K.J., Harrison L., Yang J., Chen D.J.. Persistently bound Ku at DNA ends attenuates DNA end resection and homologous recombination. DNA Repair. 2012; 11:310–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wright D.G., Castore R., Shi R., Mallick A., Ennis D.G., Harrison L.. Mycobacterium tuberculosis and Mycobacterium marinum non-homologous end-joining proteins can function together to join DNA ends in Escherichia coli. Mutagenesis. 2017; 32:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vega R.B., Horton J.L., Kelly D.P.. Maintaining ancient organelles: mitochondrial biogenesis and maturation. Circ. Res. 2015; 116:1820–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rapaport D., Brunner M., Neupert W., Westermann B.. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 1998; 273:20150–20155. [DOI] [PubMed] [Google Scholar]
- 35. Babu P., Bryan J.D., Panek H.R., Jordan S.L., Forbrich B.M., Kelley S.C., Colvin R.T., Robinson L.C.. Plasma membrane localization of the Yck2p yeast casein kinase 1 isoform requires the C-terminal extension and secretory pathway function. J. Cell Sci. 2002; 115:4957–4968. [DOI] [PubMed] [Google Scholar]
- 36. Sikorski R.S., Hieter P.. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989; 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stuart J.S., Frederick D.L., Varner C.M., Tatchell K.. The mutant type 1 protein phosphatase encoded by glc7-1 from Saccharomyces cerevisiae fails to interact productively with the GAC1-encoded regulatory subunit. Mol. Cell. Biol. 1994; 14:896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H. et al. . Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999; 285:901–906. [DOI] [PubMed] [Google Scholar]
- 39. Goldring E.S., Grossman L.I., Krupnick D., Cryer D.R., Marmur J.. The petite mutation in yeast. Loss of mitochondrial deoxyribonucleic acid during induction of petites with ethidium bromide. J. Mol. Biol. 1970; 52:323–335. [DOI] [PubMed] [Google Scholar]
- 40. Gietz D., St Jean A., Woods R.A., Schiestl R.H.. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992; 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Davis N.G., Horecka J.L., Sprague G.F. Jr. Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J. Cell Biol. 1993; 122:53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Malyarchuk S., Castore R., Harrison L.. DNA repair of clustered lesions in mammalian cells: involvement of non-homologous end-joining. Nucleic Acids Res. 2008; 36:4872–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hoffman C.S., Winston F.. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987; 57:267–272. [DOI] [PubMed] [Google Scholar]
- 44. Anandhakumar J., Moustafa Y.W., Chowdhary S., Kainth A.S., Gross D.S.. Evidence for multiple mediator complexes in yeast independently recruited by activated heat shock factor. Mol. Cell. Biol. 2016; 36:1943–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wells W.A., Murray A.W.. Aberrantly segregating centromeres activate the spindle assembly checkpoint in budding yeast. J. Cell Biol. 1996; 133:75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hartl F.U., Pfanner N., Nicholson D.W., Neupert W.. Mitochondrial protein import. Biochim. Biophys. Acta. 1989; 988:1–45. [DOI] [PubMed] [Google Scholar]
- 47. Westermann B., Neupert W.. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast (Chichester, England). 2000; 16:1421–1427. [DOI] [PubMed] [Google Scholar]
- 48. King M.P., Attardi G.. Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol. 1996; 264:304–313. [DOI] [PubMed] [Google Scholar]
- 49. Kushwaha A.K., Grove A.. Mycobacterium smegmatis Ku binds DNA without free ends. Biochem. J. 2013; 456:275–282. [DOI] [PubMed] [Google Scholar]
- 50. Kushwaha A.K., Grove A.. C-terminal low-complexity sequence repeats of Mycobacterium smegmatis Ku modulate DNA binding. Biosci. Rep. 2013; 33:175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Giloni L., Takeshita M., Johnson F., Iden C., Grollman A.P.. Bleomycin-induced strand-scission of DNA. Mechanism of deoxyribose cleavage. J. Biol. Chem. 1981; 256:8608–8615. [PubMed] [Google Scholar]
- 52. Frank B.L., Worth L., Christner D.F., Kozarich J.W., Stubbe J., Kappen L.S., Goldberg I.H.. Isotope effects on the sequence-specific cleavage of DNA by neocarzinostatin: kinetic partitioning between 4΄- and 5΄-hydrogen abstraction at unique thymidine sites. J. Am. Chem. Soc. 1991; 113:2271–2275. [Google Scholar]
- 53. Sitlani A., Long E.C., Pyle A.M., Barton J.K.. DNA photocleavage by phenanthrenequinone diimine complexes of rhodium(III): shape-selective recognition and reaction. J. Am. Chem. Soc. 1992; 114:2303–2312. [Google Scholar]
- 54. Sugiyama H., Tsutsumi Y., Fujimoto K., Saito I.. Photoinduced deoxyribose C2΄ oxidation in DNA. Alkali-dependent cleavage of erythrose-containing sites via a retroaldol reaction. J. Am. Chem. Soc. 1993; 115:4443–4448. [Google Scholar]
- 55. Ling F., Makishima F., Morishima N., Shibata T.. A nuclear mutation defective in mitochondrial recombination in yeast. EMBO J. 1995; 14:4090–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Power S.D., Lochrie M.A., Sevarino K.A., Patterson T.E., Poyton R.O.. The nuclear-coded subunits of yeast cytochrome c oxidase. I. Fractionation of the holoenzyme into chemically pure polypeptides and the identification of two new subunits using solvent extraction and reversed phase high performance liquid chromatography. J. Biol. Chem. 1984; 259:6564–6570. [PubMed] [Google Scholar]
- 57. Dagsgaard C., Taylor L.E., O’Brien K.M., Poyton R.O.. Effects of anoxia and the mitochondrion on expression of aerobic nuclear COX genes in yeast: evidence for a signaling pathway from the mitochondrial genome to the nucleus. J. Biol. Chem. 2001; 276:7593–7601. [DOI] [PubMed] [Google Scholar]
- 58. Fangman W.L., Henly J.W., Churchill G., Brewer B.J.. Stable maintenance of a 35-base-pair yeast mitochondrial genome. Mol. Cell. Biol. 1989; 9:1917–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pastukh V.M., Gorodnya O.M., Gillespie M.N., Ruchko M.V.. Regulation of mitochondrial genome replication by hypoxia: the role of DNA oxidation in D-loop region. Free Rad. Biol. Med. 2016; 96:78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ling F., Niu R., Hatakeyama H., Goto Y., Shibata T., Yoshida M.. Reactive oxygen species stimulate mitochondrial allele segregation toward homoplasmy in human cells. Mol. Biol. Cell. 2016; 27:1684–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Asai T., Bates D.B., Kogoma T.. DNA replication triggered by double-stranded breaks in E. coli: dependence on homologous recombination functions. Cell. 1994; 78:1051–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.