Abstract
Metformin is widely prescribed as a first-choice antihyperglycemic drug for treatment of type 2 diabetes mellitus, and recent epidemiological studies showed its utility also in cancer therapy. Although it is in use since the 1970s, its molecular target, either for antihyperglycemic or antineoplastic action, remains elusive. However, the body of the research on metformin effect oscillates around mitochondrial metabolism, including the function of oxidative phosphorylation (OXPHOS) apparatus. In this study, we focused on direct inhibitory mechanism of biguanides (metformin and phenformin) on OXPHOS complexes and its functional impact, using the model of isolated brown adipose tissue mitochondria. We demonstrate that biguanides nonspecifically target the activities of all respiratory chain dehydrogenases (mitochondrial NADH, succinate, and glycerophosphate dehydrogenases), but only at very high concentrations (10−2–10−1 M) that highly exceed cellular concentrations observed during the treatment. In addition, these concentrations of biguanides also trigger burst of reactive oxygen species production which, in combination with pleiotropic OXPHOS inhibition, can be toxic for the organism. We conclude that the beneficial effect of biguanides should probably be associated with subtler mechanism, different from the generalized inhibition of the respiratory chain.
1. Introduction
Metformin (dimethyl biguanide) is the most widely used frontline drug for treatment of type II diabetes mellitus [1, 2]. At the whole-body level, it effectively decreases blood glucose and insulin levels during hyperglycemia [3]. Several possible mechanisms of its action have been suggested, including inhibition of adenylate cyclase [4], interference with mitochondrial dynamics [5], alterations in gut microbiota composition [6], or inhibition of mitochondrial respiratory chain [7, 8]. Some of the proposed mechanisms oscillate around AMP-activated protein kinase (AMPK) activation, which in itself was also suggested as a direct target for biguanides [5, 9]. However, precise molecular mechanism of its action remains questionable [10, 11].
Metformin utility was also explored in the model of heart failure where epidemiologic evidence suggests its protective effect [12, 13]. Nevertheless, at the molecular level, a direct effect on mitochondria is observed in some cases [14], but not in others [15].
In addition to its antihyperglycemic effect, a broad range of epidemiologic studies showed that chronic metformin treatment is associated with a reduced risk of cancer [16–18]. As well as in the case of diabetes, the explicit mechanism of its antineoplastic action is not yet clear [11]. Metformin was proposed to act either indirectly by decreasing levels of insulin [19] or directly by suppression of mitochondrial-dependent biosynthetic pathways [20, 21]. One of the most studied possible molecular targets for biguanides action is their inhibitory action on respiratory chain complex I (NADH dehydrogenase, NDH), first described in liver tissue [8, 22]. Since then, it was confirmed for various tissues and cellular models and crucially also for cancer cells [11, 23, 24]. Hypothetical model proposes that NDH inhibition leads to decreased respiration and consequently to activation of AMPK, the key player in cellular energy homeostasis [25]. The exact site of biguanide binding to NDH is ambiguous; it was found to influence reactivity of its flavin cofactor, but it also inhibits ubiquinone reduction in a noncompetitive manner [23]. In addition to NDH, metformin was shown to inhibit other enzymes of the mitochondrial oxidative phosphorylation apparatus, including mitochondrial glycerophosphate dehydrogenase (mGPDH) and ATP synthase [7, 10, 23]. Interestingly, the viability of cancer cells lacking mitochondrial DNA (rho0 cells) is also affected by the drug, making its putative action on respiratory chain complexes rather questionable [26].
Respiratory chain is also a predominant source of reactive oxygen species (ROS), and targeting individual complexes may modulate ROS production and thus influence various pathological processes and/or signaling in different metabolic pathways [27]. Using specific substrates and inhibitors of respiratory chain enzymes, it is possible to localize the site of ROS generation or conversely the binding site of the inhibitor. In isolated mitochondria, the electron leak occurs mainly under the conditions of high NAD(P)H pool reduction [28, 29] or high ubiquinone pool reduction (high proton motive force) [29–31]. Essentially, all of the dehydrogenases in the respiratory chain were under certain conditions demonstrated to allow electron leak and ROS production. Flavin site of complex I (IF) was identified as a site of superoxide production using NADH-linked substrates [32, 33]. Under the conditions of high flux from succinate oxidation and high proton motive force, the electrons can backflow to NDH and escape at the level of Q site (IQ) towards the molecular oxygen [30, 31]. SDH itself was shown to produce significant amounts of ROS, especially when succinate levels are low (submillimolar). Under these conditions, flavin site (IIF) is not fully occupied by the substrate and is more accessible to oxygen allowing superoxide generation [34]. Mitochondrial mGPDH was also shown to act as a potent ROS producer [35] even in mitochondria from tissues with low amount of the enzyme [36]. ROS production from mGPDH can reach the levels of ROS from complex III when inhibited with antimycin A (the most potent source of ROS in mitochondria) [37]. Since mGPDH-dependent ROS production increases linearly with increasing GP concentration, the most plausible site of electron leak is the Q site or the semiquinone formed here [27].
Given the conflicting reports regarding the molecular target of biguanides in mitochondria, in this study, we addressed the direct impact of biguanides metformin and phenformin on NDH, SDH, and mGPDH, mitochondrial glycerophosphate dehydrogenases, which feed electrons into respiratory chain and were proposed to be a target of these drugs. We used mitochondria isolated from brown adipose tissue (BAT), which are advantageous for such study as they contain comparable amounts of these dehydrogenases. Furthermore, uncoupling protein 1 (UCP1) dissipates mitochondrial membrane potential (Δψm) in the isolated BAT mitochondria so that the cationic biguanides are much less concentrated in mitochondrial matrix, and therefore, the determined concentrations of these drugs are very close to the actual ones required for inhibition. We compared the effects of biguanides on dehydrogenases with their typical inhibitors (rotenone, atpenin A5, and iGP-1) using activity assays, mitochondrial oxygen consumption, and ROS production.
2. Materials and Methods
2.1. Isolation of Mitochondria from Brown Adipose Tissue
We used interscapular brown adipose tissue (BAT) of four-week-old Wistar rats kept at 22°C under 12 h/12 h light/dark cycle on a standard diet and water supply ad libitum. All animal works were approved by the institutional ethics committee and were in accordance with the EU Directive 2010/63/EU for animal experiments. Mitochondria were isolated in STE medium (250 mM sucrose, 10 mM Tris–HCl, 1 mM EDTA, pH 7.4) and supplemented with BSA (10 mg/mL) by differential centrifugation [38]. Quality of isolation was routinely checked by oxygraph, and fresh mitochondria were used for measurements of oxygen consumption and hydrogen peroxide production. Subsequently, frozen-thawed mitochondria were used for determination of enzymatic activities; as in such preparations, the integrity of mitochondrial membrane is disrupted and NADH has therefore access to the oxidation site of NDH. Also, freezing/thawing ensures that the membrane potential is not maintained.
2.2. Enzyme Activity Assays
Activities of mitochondrial dehydrogenases were determined spectrophotometrically as NADH oxidoreductases (NDH, monitored at 340 nm, ε340 = 6.22 mM−1·cm−1) or CoQ1 oxidoreductases (SDH and mGPDH, monitored at 275 nm, ε275 = 13.6 mM−1·cm−1). The assay medium contained 50 mM KCl, 20 mM Tris–HCl, 1 mM EDTA, 1 mg/mL BSA, 2 mM KCN, pH 7.4, and 50 μM CoQ1. The reaction was started by adding 100 μM NADH, 25 mM glycerophosphate (GP), or 25 mM succinate, respectively, and after 5–10 minutes, changes of absorbance were monitored at 30°C. Enzyme activities were expressed as pmol/s/mg protein.
2.3. Determination of Mitochondrial Membrane Potential
The changes in mitochondrial membrane potential (Δψm) were measured with TPP+-selective electrode as described in [39, 40]. Isolated BAT mitochondria (0.4 mg/mL) were resuspended in KCl medium and subsequently supplemented with 10 mM GP, 0.3% BSA, 1 mM GDP, and 1 μM carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP). The electrode was calibrated by stepwise addition of TPP+ (1–6 μM) before each measurement, and Δψm changes were plotted as pTPP, that is, the negative common logarithm of TPP+ concentration.
2.4. Fluorometric Detection of Hydrogen Peroxide Production
Hydrogen peroxide production was determined fluorometrically by measuring oxidation of Amplex UltraRed (Thermo Fisher) essentially as before [27]. Fluorescence of the Amplex UltraRed oxidation product was measured at 37°C using Tecan Infinite M200 multiwell fluorometer. Excitation/emission wavelengths were 544 nm (bandwidth 15 nm)/590 nm (bandwidth 30 nm). The assay was performed with 10 μg of mitochondrial protein in KCl-based medium (120 mM KCl, 3 mM HEPES, 5 mM KH2PO4, 3 mM MgSO4, 1 mM EGTA, 3 mg/mL BSA, pH 7.2) supplemented either with 10 mM pyruvate plus 2 mM malate, 10 mM succinate, or 10 mM GP. Amplex UltraRed was used at the final concentration of 50 μM with horseradish peroxidase (HRP) at 1 U/mL. Fluorescence signal from the well containing all substrates and inhibitors, but not mitochondria, was subtracted as a background for every experimental condition used. Background, caused mostly by autoxidation of the dye or nonenzymatic effect of inhibitors on apparent ROS production, varied between conditions from 0 to 1 pmol/s/mg and was uniform across respective titration points. Signal was calibrated using H2O2 at the final concentration of 0–5 μM, and H2O2 stock concentration was routinely checked by measuring its absorption at 240 nm.
2.5. Western Blotting
BAT and liver homogenates were denatured at 65°C for 15 min in a sample lysis buffer (2% (v/v) 2-mercaptoethanol, 4% (w/v) SDS, 50 mM Tris–HCl, pH 7.0, 10% (v/v) glycerol, and 0.017% (w/v) Coomassie Brilliant Blue R-250), and Tricine SDS-PAGE [41] was performed on 10% (w/v) polyacrylamide slab gels at room temperature. The gels were blotted onto a PVDF membrane (Immobilon P, Merck Millipore) by semidry electrotransfer at 0.8 mA/cm2 for 1 h. Membranes were blocked in 5% nonfat dried milk dissolved in TBS (150 mM NaCl, 10 mM Tris–HCl, pH 7.5) for 1 h at room temperature and incubated for 2 h with the following primary antibodies: antibody to NADH dehydrogenase (NDUFA9 subunit of complex I)—Abcam ab14713, succinate dehydrogenase complex (subunit A (SDHA) of complex II)—Abcam ab14715, Core2 subunit of complex III—Abcam ab14745, actin—Millipore MAB1501, rabbit polyclonal antibody to porin (1 : 1000) was a kind gift from Professor Vito de Pinto (Dipartimento di Scienze Chimiche, Catania, Italy), and rabbit polyclonal antibody to mGPDH was custom prepared [42]. Membranes were then incubated for 1 h with corresponding secondary fluorescent antibodies, IRDye 680- or 800-conjugated donkey anti-mouse IgG (Thermo Fischer) or donkey anti-rabbit IgG (LI-COR Biosciences), respectively. The fluorescence was detected using ODYSSEY infra-red imaging system (LI-COR Biosciences), and the signal was quantified using Aida 3.21 Image Analyzer software (RayTest).
2.6. Polarographic Detection of Oxygen Consumption
Oxygen consumption was measured at 30°C as described before [35] using Oxygraph-2k (Oroboros, Austria). Measurements were performed in 2 mL of KCl medium (80 mM KCl, 10 mM Tris–HCl, 3 mM MgCl2, 1 mM EDTA, 5 mM K-Pi, pH 7.4) using 15–60 μg protein/mL of freshly isolated mitochondria. For measurements, 10 mM pyruvate plus 2 mM malate, 10 mM succinate, or 10 mM GP, respectively, were used. The oxygen consumption was expressed in pmol oxygen/s/mg protein.
3. Results and Discussion
The direct mechanistic knowledge on how biguanides (metformin, phenformin) influence mitochondrial function is not yet clear. While their inhibitory effect on NADH dehydrogenase (complex I, NDH) received most attention [8, 23, 24], it was also reported that biguanides can inhibit other dehydrogenases in the mitochondrial respiratory chain, namely, succinate dehydrogenase (SDH) [10] and mitochondrial glycerophosphate dehydrogenase (mGPDH) [7]. As those dehydrogenases substantially differ in their architecture of substrate sites, coenzyme Q (CoQ), binding sites and pathways of the electron transfer from substrate to CoQ, such as lack of selectivity, is rather surprising. Therefore, we focused on the action of biguanides on NDH, SDH, and mGPDH and compared it with canonical specific inhibitors of ubiquinone binding site of complex I (rotenone) [31], ubiquinone site of SDH (atpenin A5) [43], and the novel specific inhibitor of mGPDH (iGP-1) [44].
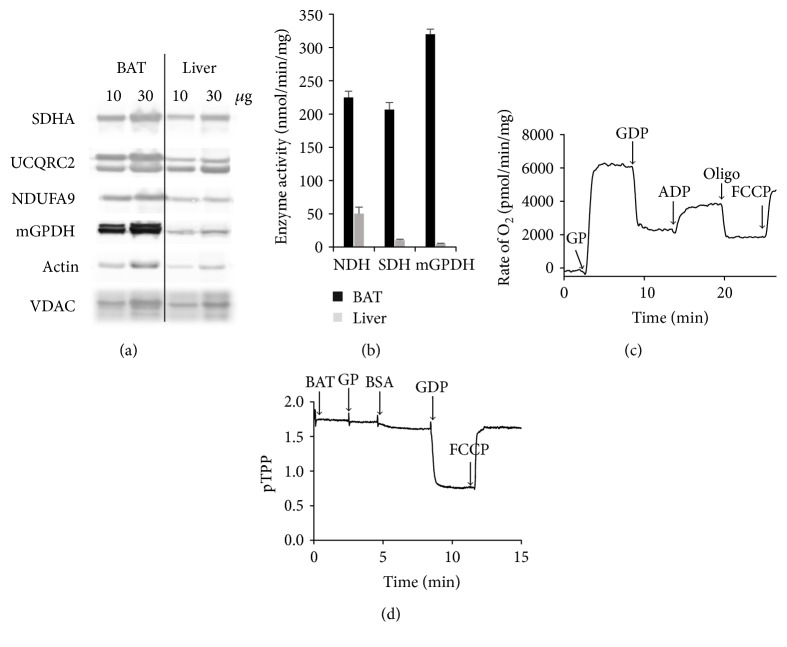
As a model, we chose isolated mitochondria from brown adipose tissue (BAT), as they have several advantages for such study. First, compared to the liver, where the inhibitory action of biguanides on mGPDH was originally described [7], they contain near equimolar levels of all three dehydrogenases studied. This can be documented both at the level of protein quantity (Figure 1(a)) and enzyme activity (NDH, SDH, and mGPDH, Figure 1(b)). The differences in response between dehydrogenases in BAT cannot therefore be ascribed to the varying content of enzymes studied. It is well established [36] and also obvious from Figure 1(b) that mGPDH activity in the liver is particularly low, and this can render the accurate measurement of inhibitory effects on GQR quite difficult. Another advantage of BAT mitochondria is the presence of UCP1 protein [45] which under native conditions allows the flow of protons back to the matrix (Figure 1(c)) and thus effectively discharges the mitochondrial membrane potential to the same level as prototypic uncoupler FCCP as can be seen by direct Δψm determination on TPP+-selective electrode (Figure 1(d)). This means that studied ROS production is independent of OXPHOS coupling. On the other hand, mitochondria can easily be coupled by the inhibitory action of guanosine diphosphate (GDP) on the UCP1 and contribution of electron backflow can then be established. Finally, lack of coupling together with physiologically low levels of mitochondrial FoF1 ATP synthase makes the action of biguanides on mitochondrial dehydrogenases rather independent of ATP synthase activity. This is quite important as FoF1 ATP synthase was also shown to be inhibited by biguanides [23] and thus may blur the picture in other model systems.
Figure 1.
Characterization of brown adipose tissue (BAT) mitochondria. (a) SDS-PAGE and Western blot analysis of mitochondrial respiratory chain complexes in BAT and liver homogenates by polyclonal antibodies against mGPDH and VDAC and monoclonal antibodies against representative subunits of NDH (subunit NDUFA9), SDH (subunit SDHA), complex III (subunit UCQRC2), and actin. Two protein concentrations were loaded as indicated, representative image of three biological replicates. (b) Enzyme activities of mitochondrial dehydrogenases in mitochondria isolated from the BAT and liver. Activities were determined as rates of CoQ1 reduction using respective substrates. Results are means ± SEM from three independent measurements. (c) Representative quality control curve of O2 consumption and (d) representative trace of Δψm measurement in isolated BAT mitochondria. The following compounds were added: 10 mM glycerophosphate (GP), 0.3% BSA, 1 mM GDP, 1 mM ADP, 1 μM oligomycin (oligo), and 1 μM carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP).
3.1. Effect of Biguanides on Enzymatic Activities
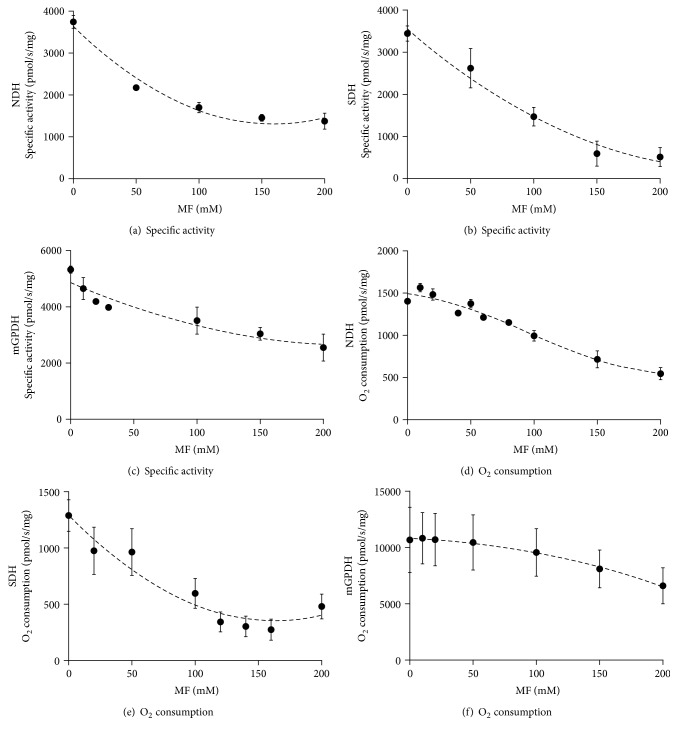
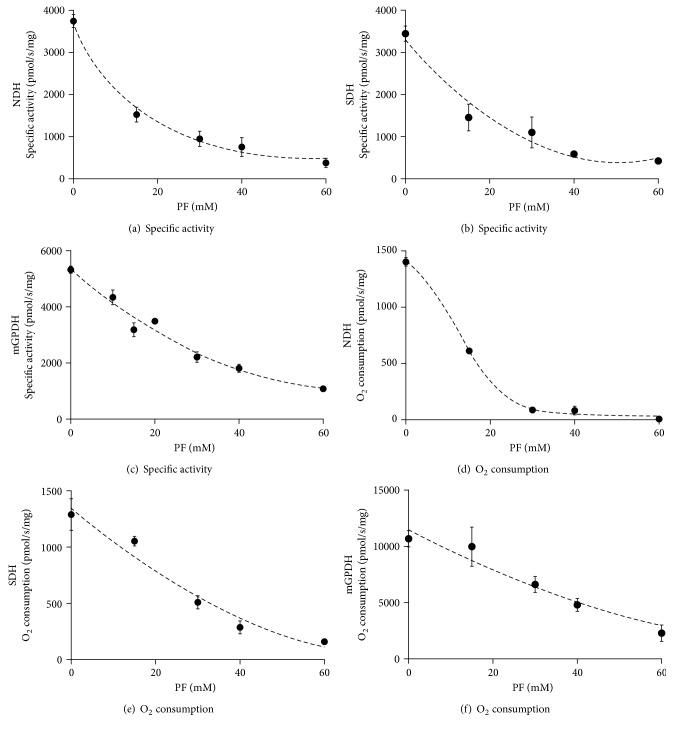
As a first step, we estimated direct impact of metformin on activity of each complex measured as individual dehydrogenase activity (substrate : coenzyme Q) in frozen-thawed mitochondria or in the context of electron transport chain in intact mitochondria measured as oxygen consumption using respective substrates (Figure 2). Metformin inhibited all enzymes studied, but only at very high concentrations (IC50 varied from 80 to 180 mM, Figures 2(a), 2(b), and 2(c)). Out of those activities, NDH was the most sensitive to the inhibitory action of metformin (Figure 2(a)). Similarly, at the same concentrations, metformin also inhibited respiration with pyruvate and malate used as substrates for NDH (Figure 2(d)), succinate for SDH (Figure 2(e)), and glycerophosphate (GP) for mGPDH (Figure 2(f)). Another biguanide, phenformin, behaved analogously to metformin, but its inhibitory effect was more efficient; the IC50 varied between 10 and 25 mM for both isolated enzyme activities and respiration of all three dehydrogenases (Figure 3). Because of the limited permeability of the biguanides, we waited to reach plateau (approximately 5 min) before proceeding to the next addition. It is to be stressed that IC50 values of observed inhibitory effects in uncoupled mitochondria (oxygraphy) were the same or slightly higher than corresponding IC50 values in frozen-thawed mitochondria (spectrophotometry), thus conferring comparable concentration of biguanides on both sides of the inner mitochondrial membrane. Determined IC50 in our case is rather high in comparison with that previously reported [7, 33]. This can be most likely attributed to the uncoupled state of BAT mitochondria which prevents accumulation of biguanides in mitochondria (see below).
Figure 2.
Respiratory chain dehydrogenase sensitivity to metformin titration. Specific enzyme activities (a, b, c) or oxygen consumption (d, e, f) of BAT mitochondria was titrated with 0–200 mM metformin using 100 μM NADH (a) or 10 mM pyruvate plus 2 mM malate (d), 10 mM succinate (b, e), and 25 mM (c) or 10 mM glycerophosphate (f). Individual points represent means ± SEM of at least three independent measurements.
Figure 3.
Respiratory chain dehydrogenases sensitivity to phenformin titration. Specific enzyme activities (a, b, c) or oxygen consumption (d, e, f) of BAT mitochondria was titrated with 0–60 mM phenformin using 100 μM NADH (a) or 10 mM pyruvate plus 2 mM malate (d), 10 mM succinate (b, e), and 25 mM (c) or 10 mM glycerophosphate (f). Individual points represent means ± SEM of at least three independent measurements.
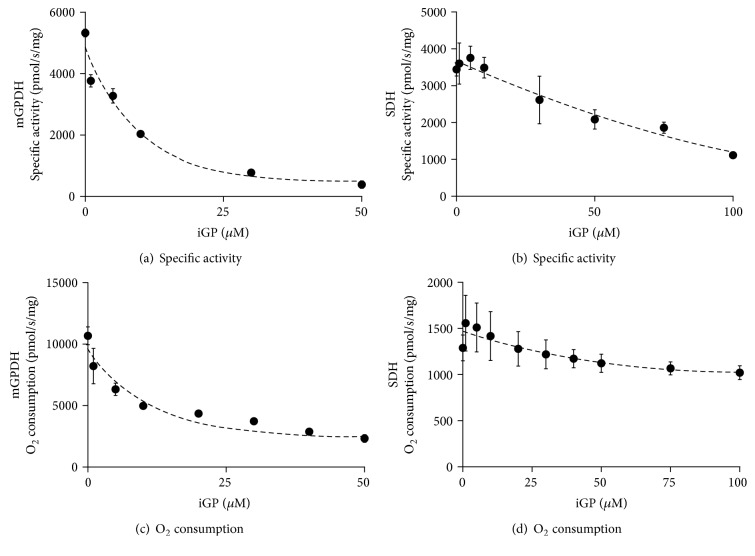
Subsequently, we focused on the action of canonical inhibitors on individual complexes. As expected, rotenone completely inhibited complex I enzyme activity as well as pyruvate plus malate-supported oxygen consumption at 500 nM. Similarly, complete inhibition of SDH by atpenin A5 using succinate as a substrate was achieved at even lower concentrations (20 nM) and activity of mGPDH was abolished at 50 μM of iGP-1. Interestingly, we also observed some nonspecific actions of iGP-1 on SDH (Figures 4(b) and 4(d)), but the IC50 was significantly higher (~80 μM) than that in the case of mGPDH (Figures 4(a) and 4(c)).
Figure 4.
Comparison of iGP-1 effect on respiratory chain dehydrogenases mGPDH and SDH. Specific enzyme activities (a, b) or oxygen consumption (c, d) of BAT mitochondria was titrated with 0–100 μM iGP-1 using 25 mM (a) or 10 mM glycerophosphate (c) and 10 mM succinate (b, d). Individual points represent means ± SEM of at least three independent measurements.
Given the extremely high IC50 we identified for phenformin and metformin, it is to be questioned whether their effects on respiratory chain enzymes are pharmacologically relevant and can affect mitochondrial function in tumors. As already mentioned, isolated BAT mitochondria with inherently low levels of mitochondrial membrane potential represent a model free of the effect of metformin partitioning due to its charge. Biguanides carry a positive charge, which means that they do accumulate in cells and mitochondria in dependence on membrane potential across both cellular and mitochondrial membranes. Thus, the concentration could be 100 to 300 times higher inside energized mitochondria (compared to concentration in cytosol) with the difference in mitochondrial membrane potential between 120 and 150 mV [23]. Since the reported values for metformin concentration inside tumors are in 1–10 μM range [21], the actual concentration in respiring mitochondria could reach mM values, which is likely not sufficient for the inhibitory action on mitochondrial oxidative phosphorylation. Indeed, in this regard, it is important to note that two recent reports demonstrated higher tumor antiproliferative efficiency for mitochondrially targeted (through positive charge of conjugated TPP moiety) analogs of metformin [46, 47]. However, metformin and its conjugated analogs accumulate in the mitochondrial matrix, where they can act on enzyme complexes facing this compartment, namely, NDH and SDH. The effect on mGPDH is more questionable, since this enzyme is located on the outer face of the inner membrane, facing the mitochondrial intermembrane space and does not span to the matrix [48]. One of the reports using mitochondrially targeted metformin analogs [46] reported also higher potency of that compound on GP-dependent respiration, which would be in line with rather generalized nonspecific mode of action for example through the influencing of mitochondrial membrane properties.
3.2. Reactive Oxygen Species Production
Action of pharmacologically active compounds on suppression of cancer cell proliferation may not only be achieved by decreased flow of electrons through respiratory chain and associated impairment of aerobic ATP production. Another successful antiproliferative strategy may be to increase in reactive oxygen species (ROS) production and subsequent induction of apoptosis [49, 50]. It was also proposed for biguanides that their primary antiproliferative effect is manifested through increase in ROS production [21, 23], which also holds true for the mitochondria-targeted analogs [51]. In this case, the mitochondrial ROS also interfered with redox signaling events. Mitochondrial respiratory chain contains numerous sites which may leak electrons to molecular oxygen and produce superoxide [27, 34, 35, 52–55]. The major superoxide-producing sites in mitochondrial respiratory chain were proposed to be NDH [53, 56], complex III [53, 57], mGPDH [27, 35], and, under certain circumstances, also SDH [34].
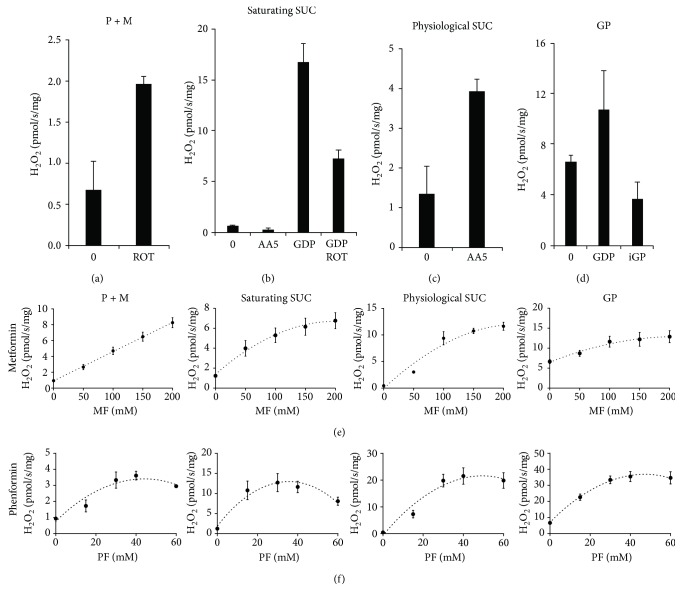
To distinguish sites in mitochondrial respiratory chain, where biguanides may induce electron leak, we followed the rates of ROS production (detected as H2O2 upon conversion by superoxide dismutase) in fresh mitochondria from BAT, where the respiratory chain is not reduced due to uncoupling by UCP1. First, using the established inhibitors of individual dehydrogenases and their respective substrates, we established ROS production pattern characteristic for each site in BAT mitochondria (Figure 5). As expected, also in BAT mitochondria, rotenone increased the rate of H2O2 production with NADH-linked substrates (pyruvate and malate), indicative of ROS production from IF site (Figure 5(a)).
Figure 5.
Effect of biguanides on reactive oxygen species (ROS) production compared to typical inhibitors. The rate of H2O2 generation was estimated by Amplex red assay. H2O2 production with (a) 10 mM pyruvate and 2 mM malate (P + M) with or without 1 μM rotenone (ROT), (b) 10 mM succinate (saturating SUC) with or without 0.5 μM atpenin A5 (AA5) or 1 μM rotenone, (c) 0.4 mM succinate (physiological SUC) with or without 0.5 μM atpenin A5, and (d) 10 mM glycerophosphate (GP) with or without 100 μM iGP-1 (iGP) were indicated; BAT mitochondria were coupled with 1 mM guanosine diphosphate (GDP). (e) Titration of H2O2 production by biguanides metformin (MF, 0–200 mM) or (f) phenformin (PF, 0–60 mM) using the same substrate concentrations as in respective experiments in (a–d). Each point is the mean ± SEM of at least three independent measurements.
Using high concentrations of succinate (10 mM), very low levels of ROS were produced in uncoupled state, which is in agreement with the previously published data [29] and this leak occurs at the Q site of SDH (Figure 5(b)). After GDP coupling, ROS were produced by the electron backflow at the IQ site of NDH (Figure 5(b)). Low levels of succinate led to pronounced increase in ROS production from flavin of SDH (inhibition of the Q site by atpenin A5, Figure 5(c)).
GP-dependent ROS production was quite high even when respiratory chain was not reduced and did not increase further after GDP addition, again in agreement with previously reported data [37, 58], and it was inhibited by mGPDH inhibitor iGP-1 (Figure 5(d)).
Compared to typical inhibitors, both metformin and phenformin induced dose-dependent increase in ROS production, but it seemed rather indiscriminative, as its pattern was highly similar for all substrates tested (Figures 5(e) and 5(f)). It suggests that these compounds do not bind directly to either flavin or ubiquinone-binding site of either of the dehydrogenases studied and that biguanides can affect mitochondria by different mechanisms, for example, they can influence membrane phospholipid environment.
4. Conclusions
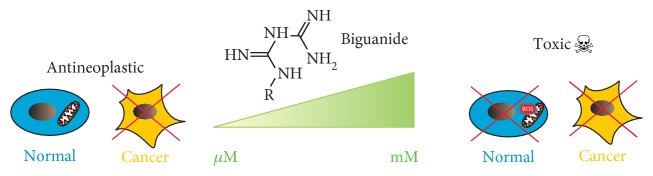
Our data demonstrate that the inhibitory effect of biguanides on OXPHOS enzymes is rather pleiotropic including the previously reported inhibition of NADH and mGPDH dehydrogenases. Drug sensitivity of respiratory chain complexes in brown adipose tissue was comparable, with IC50 higher than 80 mM in case of metformin or ranging from 20 to 30 mM in case of the more potent phenformin. Moreover, these biguanide concentrations induce nonspecific increase of mitochondrial reactive oxygen species production. Our data suggest that biguanides do not bind to any specific sites on respiratory chain dehydrogenases and require high concentrations to be effective. Under these conditions, their effect on dehydrogenases remains therapeutically questionable. Indeed, phenformin use had to be discontinued due to induction of lactic acidosis in treated patients. As summarized in Figure 6, we propose that pharmacologically relevant concentrations of biguanides confer their antineoplastic effect through yet unidentified target in mitochondrial metabolism, different from the inhibition of individual dehydrogenases.
Figure 6.
Proposed scheme of biguanide action. While at micromolar concentrations, biguanides confer antineoplastic action; at millimolar ones, they inhibit mitochondrial respiratory chain complexes and are toxic for all the cells.
Acknowledgments
This work was supported by the Grant Agency of the Czech Republic (16-12726S) and the Ministry of Education, Youth and Sports of the Czech Republic (ERC CZ: LL1204). Institutional support was provided through the Project RVO:67985823.
Abbreviations
- BAT:
Brown adipose tissue
- NDH:
NADH dehydrogenase
- SDH:
Succinate dehydrogenase
- mGPDH:
Mitochondrial glycerophosphate dehydrogenase
- OXPHOS:
Oxidative phosphorylation
- GDP:
Guanosine diphosphate
- ROS:
Reactive oxygen species
- MF:
Metformin
- PF:
Phenformin
- UCP1:
Uncoupling protein 1
- IQ:
Coenzyme Q binding site of respiratory chain complex I
- IF:
Flavin site of respiratory chain complex I
- IIF:
Flavin site of respiratory chain complex II.
Conflicts of Interest
All authors declare that there is no conflict of interests.
References
- 1.Bosi E. Metformin—the gold standard in type 2 diabetes: what does the evidence tell us? Diabetes, Obesity & Metabolism. 2009;11(Supplement 2):3–8. doi: 10.1111/j.1463-1326.2008.01031.x. [DOI] [PubMed] [Google Scholar]
- 2.Campbell R. K., White J. R., Jr., Saulie B. A. Metformin: a new oral biguanide. Clinical Therapeutics. 1996;18(3):360–371. doi: 10.1016/S0149-2918(96)80017-8. [DOI] [PubMed] [Google Scholar]
- 3.Knowler W. C., Barrett-Connor E., Fowler S. E., et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. The New England Journal of Medicine. 2002;346(6):393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller R. A., Chu Q., Xie J., Foretz M., Viollet B., Birnbaum M. J. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature. 2013;494(7436):256–260. doi: 10.1038/nature11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q., Zhang M., Torres G., et al. Metformin suppresses diabetes-accelerated atherosclerosis via the inhibition of Drp1-mediated mitochondrial fission. Diabetes. 2017;66(1):193–205. doi: 10.2337/db16-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forslund K., Hildebrand F., Nielsen T., et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528(7581):262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madiraju A. K., Erion D. M., Rahimi Y., et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature. 2014;510(7506):542–546. doi: 10.1038/nature13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Owen M. R., Doran E., Halestrap A. P. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. The Biochemical Journal. 2000;348(3):607–614. doi: 10.1042/bj3480607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou G., Myers R., Li Y., et al. Role of AMP-activated protein kinase in mechanism of metformin action. The Journal of Clinical Investigation. 2001;108(8):1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drahota Z., Palenickova E., Endlicher R., et al. Biguanides inhibit complex I, II and IV of rat liver mitochondria and modify their functional properties. Physiological Research. 2014;63(1):1–11. doi: 10.33549/physiolres.932600. [DOI] [PubMed] [Google Scholar]
- 11.Foretz M., Guigas B., Bertrand L., Pollak M., Viollet B. Metformin: from mechanisms of action to therapies. Cell Metabolism. 2014;20(6):953–966. doi: 10.1016/j.cmet.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 12.Eurich D. T., Majumdar S. R., McAlister F. A., Tsuyuki R. T., Johnson J. A. Improved clinical outcomes associated with metformin in patients with diabetes and heart failure. Diabetes Care. 2005;28(10):2345–2351. doi: 10.2337/diacare.28.10.2345. [DOI] [PubMed] [Google Scholar]
- 13.Masoudi F. A., Inzucchi S. E., Wang Y., Havranek E. P., Foody J. M., Krumholz H. M. Thiazolidinediones, metformin, and outcomes in older patients with diabetes and heart failure: an observational study. Circulation. 2005;111(5):583–590. doi: 10.1161/01.CIR.0000154542.13412.B1. [DOI] [PubMed] [Google Scholar]
- 14.Sun D., Yang F. Metformin improves cardiac function in mice with heart failure after myocardial infarction by regulating mitochondrial energy metabolism. Biochemical and Biophysical Research Communications. 2017;486(2):329–335. doi: 10.1016/j.bbrc.2017.03.036. [DOI] [PubMed] [Google Scholar]
- 15.Benes J., Kazdova L., Drahota Z., et al. Effect of metformin therapy on cardiac function and survival in a volume-overload model of heart failure in rats. Clinical Science. 2011;121(1):29–41. doi: 10.1042/CS20100527. [DOI] [PubMed] [Google Scholar]
- 16.Decensi A., Puntoni M., Goodwin P., et al. Metformin and cancer risk in diabetic patients: a systematic review and meta-analysis. Cancer Prevention Research. 2010;3(11):1451–1461. doi: 10.1158/1940-6207.CAPR-10-0157. [DOI] [PubMed] [Google Scholar]
- 17.Libby G., Donnelly L. A., Donnan P. T., Alessi D. R., Morris A. D., Evans J. M. New users of metformin are at low risk of incident cancer: a cohort study among people with type 2 diabetes. Diabetes Care. 2009;32(9):1620–1625. doi: 10.2337/dc08-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollak M. Potential applications for biguanides in oncology. The Journal of Clinical Investigation. 2013;123(9):3693–3700. doi: 10.1172/JCI67232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollak M. The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nature Reviews Cancer. 2012;12(3):159–169. doi: 10.1038/nrc3215. [DOI] [PubMed] [Google Scholar]
- 20.Griss T., Vincent E. E., Egnatchik R., et al. Metformin antagonizes cancer cell proliferation by suppressing mitochondrial-dependent biosynthesis. PLoS Biology. 2015;13(12, article e1002309) doi: 10.1371/journal.pbio.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X., Romero I. L., Litchfield L. M., Lengyel E., Locasale J. W. Metformin targets central carbon metabolism and reveals mitochondrial requirements in human cancers. Cell Metabolism. 2016;24(5):728–739. doi: 10.1016/j.cmet.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Mir M. Y., Nogueira V., Fontaine E., Averet N., Rigoulet M., Leverve X. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. The Journal of Biological Chemistry. 2000;275(1):223–228. doi: 10.1074/jbc.275.1.223. [DOI] [PubMed] [Google Scholar]
- 23.Bridges H. R., Jones A. J., Pollak M. N., Hirst J. Effects of metformin and other biguanides on oxidative phosphorylation in mitochondria. The Biochemical Journal. 2014;462(3):475–487. doi: 10.1042/BJ20140620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wheaton W. W., Weinberg S. E., Hamanaka R. B., et al. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. eLife. 2014;3, article e02242 doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viollet B., Guigas B., Sanz Garcia N., Leclerc J., Foretz M., Andreelli F. Cellular and molecular mechanisms of metformin: an overview. Clinical Science. 2012;122(6):253–270. doi: 10.1042/CS20110386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X., Chhipa R. R., Pooya S., et al. Discrete mechanisms of mTOR and cell cycle regulation by AMPK agonists independent of AMPK. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(4):E435–E444. doi: 10.1073/pnas.1311121111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mracek T., Holzerova E., Drahota Z., et al. ROS generation and multiple forms of mammalian mitochondrial glycerol-3-phosphate dehydrogenase. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2014;1837(1):98–111. doi: 10.1016/j.bbabio.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Kudin A. P., Bimpong-Buta N. Y., Vielhaber S., Elger C. E., Kunz W. S. Characterization of superoxide-producing sites in isolated brain mitochondria. The Journal of Biological Chemistry. 2004;279(6):4127–4135. doi: 10.1016/j.biopha.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 29.Quinlan C. L., Treberg J. R., Perevoshchikova I. V., Orr A. L., Brand M. D. Native rates of superoxide production from multiple sites in isolated mitochondria measured using endogenous reporters. Free Radical Biology and Medicine. 2012;53(9):1807–1817. doi: 10.1016/j.freeradbiomed.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korshunov S. S., Skulachev V. P., Starkov A. A. High protonic potential actuates a mechanism of production of reactive oxygen species in mitochondria. FEBS Letters. 1997;416(1):15–18. doi: 10.1016/S0014-5793(97)01159-9. [DOI] [PubMed] [Google Scholar]
- 31.Lambert A. J., Brand M. D. Superoxide production by NADH: ubiquinone oxidoreductase (complex I) depends on the pH gradient across the mitochondrial inner membrane. The Biochemical Journal. 2004;382(Part 2):511–517. doi: 10.1042/BJ20040485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrero A., Barja G. Sites and mechanisms responsible for the low rate of free radical production of heart mitochondria in the long-lived pigeon. Mechanisms of Ageing and Development. 1997;98(2):95–111. doi: 10.1016/S0047-6374(97)00076-6. [DOI] [PubMed] [Google Scholar]
- 33.Hirst J., King M. S., Pryde K. R. The production of reactive oxygen species by complex I. Biochemical Society Transactions. 2008;36(Part 5):976–980. doi: 10.1042/BST0360976. [DOI] [PubMed] [Google Scholar]
- 34.Quinlan C. L., Orr A. L., Perevoshchikova I. V., Treberg J. R., Ackrell B. A., Brand M. D. Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. The Journal of Biological Chemistry. 2012;287(32):27255–27264. doi: 10.1074/jbc.M112.374629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drahota Z., Chowdhury S. K., Floryk D., et al. Glycerophosphate-dependent hydrogen peroxide production by brown adipose tissue mitochondria and its activation by ferricyanide. Journal of Bioenergetics and Biomembranes. 2002;34(2):105–113. doi: 10.1023/A:1015123908918. [DOI] [PubMed] [Google Scholar]
- 36.Mracek T., Pecinova A., Vrbacky M., Drahota Z., Houstek J. High efficiency of ROS production by glycerophosphate dehydrogenase in mammalian mitochondria. Archives of Biochemistry and Biophysics. 2009;481(1):30–36. doi: 10.1016/j.abb.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 37.Vrbacky M., Drahota Z., Mracek T., et al. Respiratory chain components involved in the glycerophosphate dehydrogenase-dependent ROS production by brown adipose tissue mitochondria. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2007;1767(7):989–997. doi: 10.1016/j.bbabio.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 38.Cannon B., Lindberg O. Mitochondria from brown adipose tissue: isolation and properties. Methods in Enzymology. 1979;55:65–78. doi: 10.1016/0076-6879(79)55010-1. [DOI] [PubMed] [Google Scholar]
- 39.Labajova A., Vojtiskova A., Krivakova P., Kofranek J., Drahota Z., Houstek J. Evaluation of mitochondrial membrane potential using a computerized device with a tetraphenylphosphonium-selective electrode. Analytical Biochemistry. 2006;353(1):37–42. doi: 10.1016/j.ab.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 40.Pecinova A., Drahota Z., Nuskova H., Pecina P., Houstek J. Evaluation of basic mitochondrial functions using rat tissue homogenates. Mitochondrion. 2011;11(5):722–728. doi: 10.1016/j.mito.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 41.Schagger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Analytical Biochemistry. 1987;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 42.Mracek T., Jesina P., Krivakova P., et al. Time-course of hormonal induction of mitochondrial glycerophosphate dehydrogenase biogenesis in rat liver. Biochimica et Biophysica Acta (BBA) - General Subjects. 2005;1726(2):217–223. doi: 10.1016/j.bbagen.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 43.Miyadera H., Shiomi K., Ui H., et al. Atpenins, potent and specific inhibitors of mitochondrial complex II (succinate-ubiquinone oxidoreductase) Proceedings of the National Academy of Sciences of the United States of America. 2003;100(2):473–477. doi: 10.1073/pnas.0237315100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orr A. L., Ashok D., Sarantos M. R., et al. Novel inhibitors of mitochondrial sn-glycerol 3-phosphate dehydrogenase. PLoS One. 2014;9(2, article e89938) doi: 10.1371/journal.pone.0089938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Houstek J., Cannon B., Lindberg O. Glycerol-3-phosphate shuttle and its function in intermediary metabolism of hamster brown-adipose tissue. European Journal of Biochemistry. 1975;54(1):11–18. doi: 10.1111/j.1432-1033.1975.tb04107.x. [DOI] [PubMed] [Google Scholar]
- 46.Boukalova S., Stursa J., Werner L., et al. Mitochondrial targeting of metformin enhances its activity against pancreatic cancer. Molecular Cancer Therapeutics. 2016;15(12):2875–2886. doi: 10.1158/1535-7163.MCT-15-1021. [DOI] [PubMed] [Google Scholar]
- 47.Cheng G., Zielonka J., Ouari O., et al. Mitochondria-targeted analogues of metformin exhibit enhanced antiproliferative and radiosensitizing effects in pancreatic cancer cells. Cancer Research. 2016;76(13):3904–3915. doi: 10.1158/0008-5472.CAN-15-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mracek T., Drahota Z., Houstek J. The function and the role of the mitochondrial glycerol-3-phosphate dehydrogenase in mammalian tissues. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2013;1827(3):401–410. doi: 10.1016/j.bbabio.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 49.Kluckova K., Sticha M., Cerny J., et al. Ubiquinone-binding site mutagenesis reveals the role of mitochondrial complex II in cell death initiation. Cell Death & Disease. 2015;6, article e1749 doi: 10.1038/cddis.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rohlena J., Dong L. F., Kluckova K., et al. Mitochondrially targeted alpha-tocopheryl succinate is antiangiogenic: potential benefit against tumor angiogenesis but caution against wound healing. Antioxidants & Redox Signaling. 2011;15(12):2923–2935. doi: 10.1007/s12013-017-0796-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalyanaraman B., Cheng G., Hardy M., et al. Modified metformin as a more potent anticancer drug: mitochondrial inhibition, redox signaling, antiproliferative effects and future EPR studies. Cell Biochemistry and Biophysics. 2017 doi: 10.1007/s12013-017-0796-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cadenas E., Davies K. J. Mitochondrial free radical generation, oxidative stress, and aging. Free Radical Biology and Medicine. 2000;29(3-4):222–230. doi: 10.1016/S0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 53.Chen Q., Vazquez E. J., Moghaddas S., Hoppel C. L., Lesnefsky E. J. Production of reactive oxygen species by mitochondria: central role of complex III. The Journal of Biological Chemistry. 2003;278(38):36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 54.Schonfeld P., Wojtczak L. Brown adipose tissue mitochondria oxidizing fatty acids generate high levels of reactive oxygen species irrespective of the uncoupling protein-1 activity state. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2012;1817(3):410–418. doi: 10.1016/j.bbabio.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 55.Tretter L., Adam-Vizi V. Generation of reactive oxygen species in the reaction catalyzed by alpha-ketoglutarate dehydrogenase. The Journal of Neuroscience. 2004;24(36):7771–7778. doi: 10.1523/JNEUROSCI.1842-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kussmaul L., Hirst J. The mechanism of superoxide production by NADH:ubiquinone oxidoreductase (complex I) from bovine heart mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(20):7607–7612. doi: 10.1073/pnas.0510977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quinlan C. L., Gerencser A. A., Treberg J. R., Brand M. D. The mechanism of superoxide production by the antimycin-inhibited mitochondrial Q-cycle. The Journal of Biological Chemistry. 2011;286(36):31361–31372. doi: 10.1074/jbc.M111.267898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shabalina I. G., Vrbacky M., Pecinova A., et al. ROS production in brown adipose tissue mitochondria: the question of UCP1-dependence. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2014;1837(12):2017–2030. doi: 10.1074/jbc.M111.267898. [DOI] [PubMed] [Google Scholar]