Abstract
Data produced with short-read sequencing technologies result in ambiguous haplotyping and a limited capacity to investigate the full repertoire of biologically relevant forms of genetic variation. The notion of haplotype-resolved sequencing data has recently gained traction to reduce this unwanted ambiguity and enable exploration of other forms of genetic variation; beyond studies of just nucleotide polymorphisms, such as compound heterozygosity and structural variations. Here we describe Droplet Barcode Sequencing, a novel approach for creating linked-read sequencing libraries by uniquely barcoding the information within single DNA molecules in emulsion droplets, without the aid of specialty reagents or microfluidic devices. Barcode generation and template amplification is performed simultaneously in a single enzymatic reaction, greatly simplifying the workflow and minimizing assay costs compared to alternative approaches. The method has been applied to phase multiple loci targeting all exons of the highly variable Human Leukocyte Antigen A (HLA-A) gene, with DNA from eight individuals present in the same assay. Barcode-based clustering of sequencing reads confirmed analysis of over 2000 independently assayed template molecules, with an average of 753 reads in support of called polymorphisms. Our results show unequivocal characterization of all alleles present, validated by correspondence against confirmed HLA database entries and haplotyping results from previous studies.
INTRODUCTION
The progression of most DNA sequencing technologies are currently being geared toward constructing long sequences of phased information. Increasing the length of sequence blocks, without compromising on accuracy, is essential for improving current genotyping capabilities. Long sequence blocks can be obtained by long read sequencing platforms, by statistical inference of short reads, or by linking short reads experimentally. Single molecule long read sequencing platforms, as commercialized by Pacific Biosciences (1) and Oxford Nanopore Technologies (2), have been established as viable solutions to obtain long blocks of phased sequence information, but their uses are limited by relatively high error rates and low-throughput (3–5). With computational approaches unable to resolve de novo variants (6), many research groups are looking toward linked-read methods (7–9) to obtain accurate haplotype-resolved genomes (10). These approaches typically involve more elaborate library preparation procedures but benefit from higher sequencing accuracy and throughput of short-read massively parallel sequencing platforms. The recently established technology from 10× Genomics offers an appealing tool for linked-read haplotyping on a genome-wide scale, but for biological questions with loci-specific sequencing needs, such a system is not cost efficient. Specific loci of interest may be covered to some extent by this technology but considering the costs associated with instrumentation and consumable kits it is a waste of resources not to use a targeted approach when applicable. We recently described a method (11) for targeted phasing of multiple amplicons from single DNA molecules. In this study we describe Droplet Barcode Sequencing (DB-Seq), an improved approach for targeted linked-read haplotyping, featuring a much-simplified workflow with cheaper reagents and increased phasing capacity. The method is independent of specialty microfluidics equipment or reagent kits, making it a cost efficient alternative to achieve long sequence data blocks while utilizing the superior accuracy and throughput of readily available short-read sequencing technologies.
Combining the use of picoliter-scale emulsion droplets with beads to localize clonal populations of barcoded oligonucleotides to confined spaces has been a reoccurring theme for many new methods (9,11–13). While useful in many aspects, barcoded beads are typically laborious and costly to produce. As described by Klein et al. (12), the generation of barcoded gel beads requires a microfluidic device to form droplets which, after gel polymerization, undergoes multiple extension cycles to build up encapsulated oligonucleotide sequences combinatorially in 384 well plates. The scale of this bead production approach is limited to 384∧2 (∼1.5*10∧5) different barcode combinations, and it requires synthesis of 768 barcode oligonucleotides. Another related approach by Macosko et al. (13) outlines the production of barcoded polymer beads by reverse-direction phosphoramidite synthesis, performing twelve split-and-pool cycles to yield a barcoded library complexion of 4∧12 (∼1.7*10∧7). The previous DNA phasing method from our lab (11) employed shake-emulsion droplets with magnetic beads to produce millions of uniquely barcoded beads by clonal polymerase chain reaction (PCR) amplification of degenerated barcoding molecules, enabling a theoretical complexity of 4∧15 (∼1.1*10∧9) barcodes from the synthesis of a single oligonucleotide. Regardless of which production method is used, the need for barcoded beads results in assays for which specialty reagents must be generated before proceeding with the intended assay reactions. A method described by Lan et al. (8) has circumvented the need for barcoded beads by microfluidic manipulation of droplets in several steps. Many high-throughput methods that rely on precise mixing of picoliter-scale reagents, or a succession of incompatible enzymatic reactions, have instigated a need for intricate microfluidic devices, which require niched expertise to develop (8,14,15). With our DB-Seq, users are relieved of these dependencies as emulsion droplets are formed without beads by simple shaking, and a single enzymatic reaction is used to both generate target amplicons from single template molecules and to link them to a unique population of droplet barcodes that is generated at the same time.
We have applied our method to perform long-range phasing of the Human Leukocyte Antigen A (HLA-A) gene. The HLA gene family is the most polymorphic loci in the human genome, it encodes for the major histocompatibility complexes which function to mediate interactions between cells and antigens (16). It has been the focus of many research studies of diseases relating to the immune system and histocompatibility screenings in a quest to characterize variant alleles (17). Due to the large number of nucleotide polymorphisms spanning over thousands of bases, having long stretches of accurately phased sequence information is highly advantageous when needing to distinguish alleles (18). Before the advent of sequencing-based typing, serological typing and then PCR-based methods were used to investigate allelic diversity within the HLA loci, albeit with very limited throughput, accuracy and capacity for allele discrimination (19). DNA sequencing has since then become the preferred approach, but with most systems generating incomplete and often ambiguous phasing of alleles there remains a demand for technologies able to produce full-length phase blocks. Single molecule long read sequencers have been used for HLA typing (18,20), and while these technologies are able to generate full length haplotype data, sequencing platforms have yet to be widely implemented. DB-Seq enables users of massively parallel sequencing platforms to unequivocally link regions with polymorphic positions from the same molecule of origin, across entire genes. To showcase the method we used DB-Seq to phase all eight exons of the HLA-A gene from related individuals, two families consisting of three trios (mother/father/offspring); to derive the alleles for each individual and compare the results with previously characterized HLA-typing results from whole genome sequencing data.
MATERIALS AND METHODS
Samples
Extracted human genomic DNA from eight individuals (NA12877, NA12878, NA12882, NA10860, NA11992, NA11993, NA12891 and NA12892; hereafter named according to sample IDs 01, 02, 03, 04, 05, 06, 07 and 08, respectively) were obtained from the Coriell Institute (NJ, USA). These individuals make up three sets of family trios, two of which belong to the same extended family (Supplementary Table S1).
HLA-A long range PCR
A 50 µl PCR mix was prepared with 1× PrimeStar GXL Buffer (Takara Bio, Shiga, Japan), 1.25 U PrimeStar GXL DNA Polymerase (Takara Bio), 200 µM dNTPs (Invitrogen, CA, USA), 50 ng of gDNA (Coriell Institute), 200 nM of reverse primer (LR.R) and 200 nM of an indexed forward primer (LR.F01–LR.F.08; Supplementary Table S2). Each reaction was cycled in a Mastercycler Pro S (Eppendorf, Hamburg, Germany), starting with 1 min at 98°C, followed by 15 cycles at 98°C for 15 s, 60°C for 30 s, 68°C for 6 min and ended with 10 min at 68°C. Following PCR the samples were purified by polyethylene glycol precipitation on carboxylic-acid beads (21) using a Magnatrix™ 1200 Biomagnetic Workstation (NorDiag ASA, Oslo, Norway). Sample concentrations were determined by Qubit 3.0 (Life Technologies (Thermo Fisher Scientific), MA, USA) before diluting to 1 pM for use in emulsion reactions.
Emulsion reactions
Assay reactions consist of an aqueous phase and an oil phase enabling the formation of picoliter scale droplets (reaction compartments) when mixed (Supplementary Note 1 and Supplementary Figure S7). The aqueous constituent of each assay reaction consisted of PCR reagents to a total volume of 50 μl, containing 1× Ex Taq Buffer (Takara), 500 μM dNTPs (Invitrogen, Carlsbad, CA, USA), 1 M Betaine (Sigma Aldrich, MO, USA), 3% Dimethyl Sulfoxide (DMSO) (Thermo Scientific), 2.5 U Ex Taq HS (Takara), 40 fM HLA-A long range amplicon and a cocktail of synthetic oligonucleotides (see composition in Supplementary Table S3). The same protocol was used for all emulsion reactions, except for one multiplex reaction; to which an equimolar pool of HLA-A long range amplicons from eight individuals were added instead; at a final concentration of 75 fM. The aqueous phase was added on top of 100 μl HFE-7500 oil with 5%(w/v) 008-Flourosurfactant (Ran Biotechnologies, MA, USA), and the two phases were emulsified by shaking for 8 min at 15 Hz in a Qubit™ (Life Technologies) tube, using a Tissuelyser instrument (Qiagen, MD, USA). After shaking, the entire reaction volume was transferred to a thin-walled PCR tube and 85 μl mineral oil (Sigma) was added on top to prevent evaporation. The reaction was then cycled in a Mastercycler Pro S (Eppendorf) using the protocol outlined in Supplementary Table S4.
Emulsion breakage
Following emPCR the mineral oil on top was carefully removed and discarded. A 15 μl volume of Ethylenediaminetetraacetic acid (EDTA, 100 mM) (Invitrogen) was added and the entire emulsion reaction was transferred to a 0.5 ml DNA LoBind tube (Eppendorf). A volume of 200 μl 1H,1H,2H,2H-Perfluoro-1-octanol (Sigma) was then added to the reaction volume, followed by vortexing for 10 s at maximum speed. The mixture was then centrifuged for 2 min at 25 000 × g. With complete separation of aqueous and oil/perfluoro-octanol phases achieved (evaluated visually by the absence of a non-transparent emulsion phase), the aqueous phase (on top) could then carefully be withdrawn and transferred to a fresh reaction tube for downstream processing.
Sample enrichment
The library underwent a size selection procedure to remove excess amplification primers and non-coupled barcoding amplicons. This was done by polyethylene glycol precipitation on carboxylic-acid beads using a Magnatrix™ 1200 Biomagnetic Workstation (NorDiag ASA) as described by Lundin et al. (21). An enrichment was then performed to fish out the coupled amplicons (barcoded loci-specific products) from the excess of non-barcoded loci-specific amplicons. The sample was incubated with 20 µl Dynabeads MyOne Streptavidin T1 (Life Technologies) beads under rotation for 1 h at RT, after which the supernatant was discarded and the beads were washed meticulously; starting with Elution Buffer (Qiagen) and followed by four washes with NaOH (125 mM) before neutralization with Elution Buffer (Qiagen). To release the enriched bead-bound products a second round of PCR was performed. A 50 µl PCR reaction was prepared with 1× Ex Taq Buffer (Takara), 500 uM dNTPs (Invitrogen), 3% DMSO (Thermo Fisher Scientific), 1.25 U Ex Taq HS (Takara) and 200 nM of each of H1 and H3 oligonucleotides (Supplementary Table S2). The PCR protocol started with 2 min at 94°C, followed by 10 cycles at 94°C for 1 min, 50°C for 1 min, 72°C for 2 min and ended with 5 min at 72°C. Following PCR the beads were discarded and primers were removed from the supernatant by polyethylene glycol precipitation on carboxylic-acid beads as previously described.
Sequencing library preparation
Indexing qPCR reactions were then performed to prepare the samples for sequencing, consisting of 1× Ex Taq SYBR Ready Mix (Takara), 3% DMSO (Thermo Scientific), 200 nM of each of i5-H1 and i7΄(Index)-H3 primers (Supplementary Table S2). Reactions were cycled on a CFX96 instrument (Bio-Rad, CA, USA) instrument according to manufacturer's recommendations and taken out individually during the beginning of the exponential amplification stage to avoid over-amplification. Samples were then purified to remove excess primers before the concentrations were determined by Qubit 3.0 (Life Technologies) and samples were diluted to 2 nM as recommended by the preparation protocol for sequencing by Illumina. Samples were sequenced using a MiSeq v3 600 cycle kit, loading a 8 pM library and covering 305 bases each with read 1 and read 2.
Data analysis
Scripts used for barcode clustering, allele identification (and grouping) and classification of alleles by matching to the IPD-IMGT/HLA database are available online (https://github.com/elhb/DBS_Analysis), and each step is described briefly below. An overview of the bioinformatic solution applied to the data generated in this study is presented in Supplementary Figure S3. Picard tools (https://broadinstitute.github.io/picard/) and pysam (https://github.com/pysam-developers/pysam) were extensively used in the scripts to facilitate data handling. All data from this study has been uploaded to the Sequence Read Archive (SRA), accession number SRP100498.
Barcode clustering and mapping
The process of identifying handles and the barcode sequence in each read, and then grouping reads into barcode clusters was based on scripts used by Borgstrom et al (11). The barcode clustering was performed using CD-HIT-454 (22). Reads where a target amplicon sequence could be identified between known handles (H2 and H3; Supplementary Figure S2) were then mapped to a gene reference sequence extracted from hg19 chr6:29907000-29917000 using bowtie2 (23).
Allele identification
The identification of alleles was based on all clusters with ≥20 reads, according to the procedure detailed in Supplementary Figure S4. To identify positions different from the reference genome we defined two conditions for positions in any given barcode cluster; (i) ‘non-reference base calls’ defined as positions where ≥80% of the reads support one base that is not the reference base and (ii) ‘mixed base calls’ defined as positions where ≥20% and ≤80% of the reads do not correspond to the reference base. Barcode clusters displaying mixed base calls in a position, where at least two other barcode clusters display a non-reference base call, were excluded from further analysis. All barcode clusters with full coverage of the target amplicon regions (defined as having ≥5 base calls with a phred-scaled base calling quality value ≥20 for each position) were used as seeds for candidate alleles. Allele representation strings (defined as a concatenated sequence of non-reference positions) were first produced for each of these seed barcode clusters, and candidate alleles were then generated by grouping allele representation strings with an edit distance of 1. Barcode clusters without full coverage (at least one position with insufficient read support) were then used as added support for the identified candidate alleles. In this process, support clusters featuring non-reference bases that matched none or multiple candidate alleles, in more than one position, were removed from the dataset.
Allele classification
Consensus sequences based on data from both seed and support clusters were built for each of the target regions (Supplementary Figures S3 and S4). To classify these consensus sequences according to the HLA gene nomenclature (http://hla.alleles.org/), sequences were matched against a set of reference gene sequences from the IPD-IMGT/HLA database (http://www.ebi.ac.uk/ipd/imgt/hla/). Only sequences with perfect match to all targeted regions and in the expected order were considered as potential HLA-A allele names.
METHOD DESCRIPTION
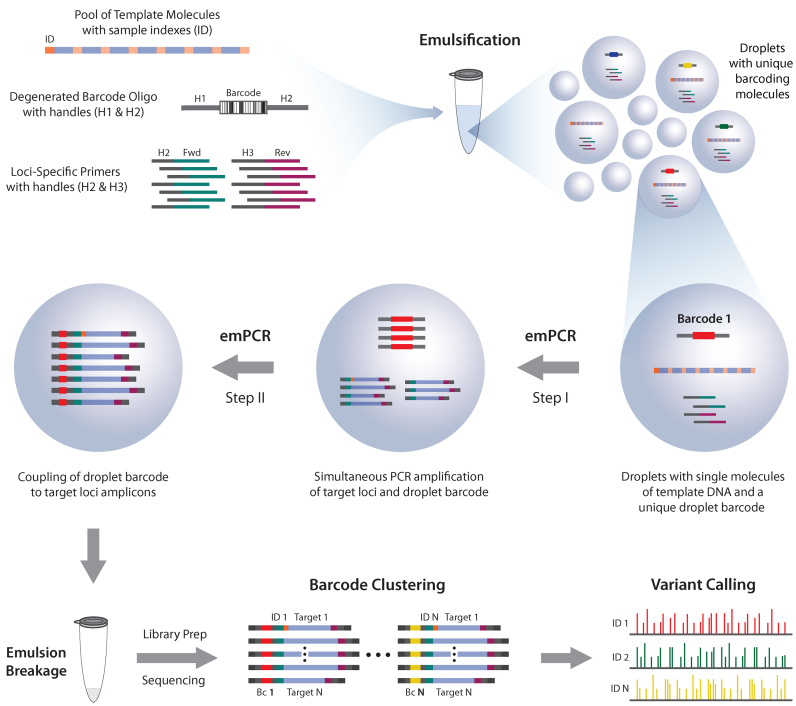
Droplet-based barcoding has recently been presented to investigate single DNA molecules and the biological content of single cells in a high-throughput manner. While all methods rely on barcoded beads or intricate microfluidic devices to automate a chain of enzymatic reactions, we here demonstrate a technique that relies on emulsion droplets formed by controlled shaking and ordinary PCR. The method simultaneously generates populations of unique droplet barcodes and clonally amplified target regions, and couples the two components together (Figure 1). Inside a subset of active droplets, barcode populations are formed from single copies of a barcoding oligonucleotide featuring a stretch of 20 semi-degenerated bases (H1-(BDVH)5-H2, sequence degeneracy of 3∧20 (∼3.5*10∧9); Supplementary Table S3). Combined with amplification of a single DNA template molecule the reaction enables the biological information within the template to be exclusively coupled to a unique droplet barcode. Coupling is achieved by asymmetric amplification of both component molecules, initially yielding populations of ssDNA products that share a complimentary handle sequence at the 3΄ end. As the concentration of ssDNA substrates increases they will hybridize to each other and couple the barcode information to each template amplicon (see Supplementary Figure S1 for a detailed assay overview). Dilution of reagents ensures that a sufficient number of droplets contain only one copy of both barcode and template components (Supplementary Notes 1 and 2), while droplets without barcode and/or template components will not yield coupled amplicons. Following emulsion PCR, downstream processing of the samples ensures that amplicons formed in droplets missing either the barcoding oligonucleotide or the template molecule will be removed from the library. Post sequencing, the read pairs are clustered based on the droplet barcode and sample origin is identified by a sample-specific ID tag incorporated into the forward primer used to prepare DNA templates. The preparation of DNA templates is done by means of an indexed long range PCR, enabling analysis of molecules from multiple sources. Sequence variations are called within each barcode cluster and then combined to form a phased consensus sequence for each DNA molecule (Supplementary Figures S3 and S4).
Figure 1.
Technological assay overview. DNA template molecules and degenerated barcoding oligonucleotides are encapsulated into droplets to contain one or zero of each molecule. Primers are added to enable amplification of each component in parallel until a critical concentration is reached, facilitating interaction between the two clonally amplified PCR products. A barcode sequence exclusive to each droplet is thereby coupled to each target loci from the original template molecule. After emulsion breakage the target products are enriched and prepared for sequencing. Read pairs then undergo barcode-based clustering and prevalent variations are called to produce a set of alleles for each sample ID (as determined by an ID-tag at the 5΄ end of template molecules and target 1 amplicons).
RESULTS
To validate the phasing capacity of DB-Seq we amplified the whole HLA-A gene through long range PCR and generated libraries with template DNA from eight different individuals separately, as well as one assay reaction with an equimolar mix of template molecules from all eight individuals. The basis of phasing in this technique is the process of post-sequencing barcode-based clustering, where the reads with any given barcode are defined as sharing a mutually exclusive droplet barcode as well as the expected read structure with known handle sequences (Supplementary Figure S2). Each barcode cluster used in the analysis is required to consist of at least 20 reads. These clusters may or may not include sequence representation from all seven amplicons, but only clusters containing all the targets may be used as a seed for the identification of candidate alleles (Supplementary Figure S4). Once candidate alleles have been identified from seed clusters, remaining clusters are used to add support. Sequencing reads from the eight separate singleplex reactions and the eight-plex reaction yielded from 788 to 1544 and 2057 barcode clusters, respectively (Table 1 and Supplementary Table S5). In the eight-plex reaction, seven alleles (A-G; Table 1) were identified with an average support of 753 reads per barcode cluster and 6.78% of the total read population per individual. As shown in Supplementary Figure S3, additional alleles (H-M) were identified but these were supported only by a single barcode cluster and featured a low proportion of the total read population (0.02–0.04% per allele), which is why they were discarded. These alleles did not match any entries in the IPD/IMGT-HLA database, suggesting that they may stem from droplets containing either multiple or chimeric template molecules. With a relatively low sequencing depth of 200 K reads per template molecule, two phased alleles could be identified for each sample in the eight-plex reaction and be classified to the same degree of specificity as for the singleplex reactions. In addition, each allele is supported by a similar proportion of the read population; indicating that amplification bias is not an issue. When classifying the alleles according to entries in the IPD/IMGT-HLA database, some alleles exhibited a perfect match to more than one database entry. Variations of these database entries are in the fourth digit of the HLA nomenclature, corresponding to polymorphisms in non-coding regions of the HLA-A gene that are not covered by the targeted loci in this study. Despite this we see that for all experimentally derived alleles the classification is more specific than that of previously published HLA-A haplotyping data (24,25) based on whole genome, whole exome and targeted sequencing data of the same individuals.
Table 1. Experimentally derived haplotypes for HLA-A.
| Reaction sample | Reads† [% trc] | Identified alleles | Barcode clusters | % trc | Found in sample ID | IPD-IMGT/HLA CORRESPONDENCE | External references |
|---|---|---|---|---|---|---|---|
| 8-Plex ID 01-08 | 1.74 M [54.2%] | A | 667 | 14.37% | 01, 03, 04, 05, 08 | A*02:01:01:01 | A*02:01 |
| B | 362 | 13.38% | 02, 03, 08 | A*11:01:01:01 | A*11:01 | ||
| C | 409 | 10.23% | 02, 05, 07 | A*01:01:01:01 | A*01:01 | ||
| D | 215 | 5.64% | 04, 06 | A*29:02:01:01, A*29:02:01:02 | A*29:02 | ||
| E | 134 | 3.98% | 01 | A*03:01:01:01 | No Data | ||
| F | 142 | 3.68% | 07 | A*24:02:01:01 | A*24:02 | ||
| G | 122 | 2.74% | 06 | A*26:01:01:01, A*26:01:01:03N | A*26:01 | ||
| ID 01 | 862 K [53.1%] | E | 744 | 34.81% | 01 | A*03:01:01:01 | No Data |
| A | 486 | 18.27% | A*02:01:01:01 | ||||
| ID 02 | 865 K [60.9%] | C | 563 | 33.17% | 02 | A*01:01:01:01 | A*01:01 |
| B | 419 | 27.44% | A*11:01:01:01 | A*11:01 | |||
| ID 03 | 1.05 M [72.9%] | B | 783 | 42.37% | 03 | A*11:01:01:01 | No Data |
| A | 730 | 30.93% | A*02:01:01:01 | ||||
| ID 04 | 833 K [57.3%] | D | 369 | 32.65% | 04 | A*29:02:01:01, A*29:02:01:02 | A*29:02 |
| A | 423 | 27.11% | A*02:01:01:01 | A*02:01 | |||
| ID 05 | 1.06 M [63.5%] | C | 552 | 37.74% | 05 | A*01:01:01:01 | A*01:01 |
| A | 524 | 32.13% | A*02:01:01:01 | A*02:01 | |||
| ID 06 | 1.05 M [64.1%] | D | 460 | 30.9% | 06 | A*29:02:01:01, A*29:02:01:02 | A*29:02 |
| G | 433 | 27.57% | A*26:01:01:01, A*26:01:01:03N | A*26:01 | |||
| ID 07 | 857 K [63.6%] | F | 718 | 33.13% | 07 | A*24:02:01:01 | A*24:02 |
| C | 787 | 30.50% | A*01:01:01:01 | A*01:01 | |||
| ID 08 | 1.02 M [69.0%] | B | 880 | 44.36% | 08 | A*11:01:01:01 | A*11:01 |
| A | 667 | 24.58% | A*02:01:01:01 | A*02:01 |
Results for the eight-plex and all singleplex reactions, featuring correspondence to the IPD-IMGT/HLA database and two independent sources (24,25) of haplotyping data. † Reads counts of barcode clusters used for classification of alleles. % trc corresponds to percentage of total read counts. Only matching database entries that have been confirmed and published are included. The table details the data analysis output of all alleles with support from at least two independent barcode clusters.
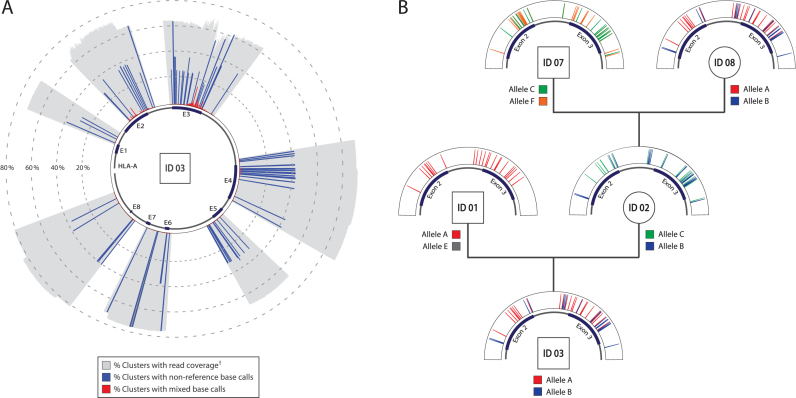
In this proof of concept study, seven targeted loci were designed to cover all eight exons of the HLA-A gene and surrounding intronic regions. Figure 2A depicts the sequencing coverage of these targeted loci, and all positions identified as non-reference base calls or mixed base calls within sample ID 03. The sequencing coverage is a measure of what proportion of barcode clusters met the criteria for being used for base calling of each position. The blue lines represent all positions at which non-reference base calls (polymorphisms) have been identified for both alleles. A few of the identified polymorphisms make up 100% of clusters supporting that base call, meaning that the two alleles present within the sample share the same non-reference base calls for those positions. In most cases, non-reference base calls constitute around half of the reported coverage; portraying variations that are only present in one of the two alleles and indicating that the read support for the two alleles is roughly the same. Mixed base calls that do not overlap with non-reference base calls can arise either from emulsion reaction polymerase errors or sequencing errors. In contrast, mixed base calls that stem from droplets with multiple template molecules (i.e. non-clonal barcode clusters) are expected to overlap with non-reference base calls. From Figure 2A we see that most positions with a large proportion of mixed base calls do not overlap with the called non-reference variants, supporting the notion that these errors do not stem from non-clonal amplification of target molecules. Error prone positions yielding mixed base calls are predominantly localized to small but problematic GC-rich regions of exon 2 and 3, and the same pattern is observed in all samples (Supplementary Figure S5). This was confirmed by a dip in sequencing quality within these regions, as well as an observation that amplicon formation was negligible when alternative polymerases were used in the emulsion PCR (data not shown).
Figure 2.
Assay haplotyping results visualized. (A) Data from all clusters with ≥20 reads, for the singleplex reaction carried out with sample ID 03. † Base calling of each position supported by ≥5 reads with a quality score of ≥20. (B) Pedigree visualization of allele heredity for extended family of individuals and a more focused view (targets covering exons 2 and 3) of non-reference base calls for each allele identified within these samples.
Figure 2B depicts a more detailed distribution of non-reference base calls of the two alleles in each sample, within two trios constituting an extended family of individuals. The resulting pedigree is meant to visualize allele heredity within the family and as expected we clearly see that one allele per parent ends up in the offspring for each trio (see also Supplementary Figure S6 for results visualizing non-reference base calls across all targets regions, for all samples). Noteworthy for sample ID 01 is that he has inherited the same allele as his unrelated partner's mother (allele A), and that allele E appears to be identical to the reference for all positions within two targets covering exons 2 and 3. By pure coincidence it seems that sample ID 03 possesses the same set of alleles as his grandmother (sample ID 08), which is supported by haplotyping results based on all loci as shown in Table 1. It is also worth noting that, unlike what is typical for computational phasing approaches, our assay does not require linkage information from trios to resolve variant alleles, nor was this genetic relation between samples in any way used in the process of identifying alleles within our datasets. Sample trios in general, and these samples in particular as their genomes previously have been extensively sequenced and characterized, were only chosen as they provide a convenient way of illustrating the data and knowing whether the results are reasonable.
DISCUSSION
Our results show that DB-Seq is a method that in a high-throughput manner enables barcoding of single DNA molecules and long range phasing of all exonic single base variations present in a highly polymorphic genomic loci. Since the method features single molecule resolution it is independent of sample pool complexity, and given that each individual sample is assigned a unique ID-tag, both alleles from any individual can be correctly phased and its sample origin can be demultiplexed. Being able to resolve single molecules is a requirement for applications where templates share significant homology but contain a few polymorphic positions with biological relevance. The analysis of such template molecules is likely to be error prone with other methods that rely on the encapsulation of many template molecules in each droplet, especially if one considers using samples from multiple sources in the same assay. The data suggest that DB-Seq is a robust method for phasing long single DNA molecules, potentially for hundreds of samples at once, as supported by the fact that each identified allele is supported by roughly the same number of reads and barcode clusters.
Prior to the emulsion reaction, we opted to perform an enrichment of the target gene by means of long range PCR. This was done to circumvent the problem of a ‘double random’ Poisson distribution, which would arise from trying to encapsulate single copies of both barcoding oligonucleotides and DNA template molecules. A potential drawback of using long range PCR as an enrichment step prior to library preparation is that universal base substitutions (polymerase errors) or chimeric products can arise and lead to identification of false positive alleles. We addressed this issue by running a minimal number of long range PCR cycles, enabled by the fact that only 2 amol of template is required for the assay reactions. Our results show that we do not identify any alleles with more than a single barcode cluster in support that could potentially be of chimeric origin. While this aspect of long range PCR limits the phasing length capacity of our assay to ∼30 kb, the length of template molecules does not impact the efficiency of the assay or the subsequent sequencing accuracy. This is in contrast to long read sequencing technologies where shorter fragments are over-represented in the dataset and sequencing quality decreases with size. Sequencing of the HLA-A gene with Oxford Nanopore Technologies has been previously reported to yield consensus sequences matching that of short-read sequencing platforms when a very high coverage is used to compensate for low accuracy (20). A higher coverage (and thus improved accuracy) can be obtained through an added library preparation step where multiple copies of an amplicon can be concatenated by means of rolling circle amplification (26). These studies suggest that nanopore sequencing has the potential to be used in clinical settings. However, given the wide availability of instrumentation, established bioinformatic tools and the superior sequencing accuracy of short-read sequencing technologies; the alternative of linking reads from short-read sequencing platforms remains highly relevant. To study loci beyond the range of long range PCR one could envision omitting this step and instead reoptimizing the process of diluting template fragments, to end up with droplets with multiple genomic fragments but a single copy of the loci of interest. However, without a sample-specific ID-tag such an approach would not enable multiplex assay reactions to be performed.
The advantage of DB-Seq as a non-commercial linked-read approach is that minimal resources can be spent on assay reagents, and it does not require speciality instrumentation, reagent kits or microfluidic devices. In laboratories where microfluidic devices for droplet generation are available, our method for simultaneous barcode generation and target barcoding would likely benefit from such devices; as it is easier to evaluate the necessary dilution requirements with a monodisperse population of droplets (Supplementary Note 1). However, this study shows that such devices are not a necessity for high-throughput assays enabled by emulsion droplet technologies. Having a robust and simple-to-execute targeted phasing approach as an alternative to genome-wide phasing technologies offers researchers the choice of flexibility to tailor costs after the research question of their interest. In this study the HLA-A gene was used as a model system to showcase our assay's usefulness in clinical studies, but DB-Seq is relevant for a broad range of applications where linking sequence information from multiple loci is of interest, for instance studies of compound heterozygosity, structural variations of alleles and as exemplified by studies of the HLA loci; to investigate the relationship between genetic variation and disease susceptibility. It is also tempting to speculate that the same type of platform-independent method could be used to obtain linked reads across an entire genome, potentially through randomly primed amplification of single molecules, but at a fraction of the cost of commercial systems.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Sanja Vickovic for valuable input relating to the illustrations, and the National Genomics Infrastructure (NGI) for providing infrastructure and sequencing support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Stiftelsen Olle Engkvist Byggmästare [2015/347]; Stockholms Läns Landsting [LS2016-0764]; Knut and Alice Wallenberg Foundation [2011.0113]. Funding for open access charge: Royal Institute of Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1. Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G., Peluso P., Rank D., Baybayan P., Bettman B. et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009; 323:133–138. [DOI] [PubMed] [Google Scholar]
- 2. Clarke J., Wu H.C., Jayasinghe L., Patel A., Reid S., Bayley H.. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 2009; 4:265–270. [DOI] [PubMed] [Google Scholar]
- 3. Koren S., Schatz M.C., Walenz B.P., Martin J., Howard J.T., Ganapathy G., Wang Z., Rasko D.A., McCombie W.R., Jarvis E.D. et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat. Biotechnol. 2012; 30:693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Laver T., Harrison J., O’Neill P.A., Moore K., Farbos A., Paszkiewicz K., Studholme D.J.. Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol. Detect. Quantif. 2015; 3:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quail M.A., Smith M., Coupland P., Otto T.D., Harris S.R., Connor T.R., Bertoni A., Swerdlow H.P., Gu Y.. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012; 13:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Browning S.R., Browning B.L.. Haplotype phasing: existing methods and new developments. Nat. Rev. Genet. 2011; 12:703–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amini S., Pushkarev D., Christiansen L., Kostem E., Royce T., Turk C., Pignatelli N., Adey A., Kitzman J.O., Vijayan K. et al. Haplotype-resolved whole-genome sequencing by contiguity-preserving transposition and combinatorial indexing. Nat. Genet. 2014; 46:1343–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lan F., Haliburton J.R., Yuan A., Abate A.R.. Droplet barcoding for massively parallel single-molecule deep sequencing. Nat. Commun. 2016; 7:11784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zheng G.X., Lau B.T., Schnall-Levin M., Jarosz M., Bell J.M., Hindson C.M., Kyriazopoulou-Panagiotopoulou S., Masquelier D.A., Merrill L., Terry J.M. et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat. Biotechnol. 2016; 34:303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Snyder M.W., Adey A., Kitzman J.O., Shendure J.. Haplotype-resolved genome sequencing: experimental methods and applications. Nat. Rev. Genet. 2015; 16:344–358. [DOI] [PubMed] [Google Scholar]
- 11. Borgstrom E., Redin D., Lundin S., Berglund E., Andersson A.F., Ahmadian A.. Phasing of single DNA molecules by massively parallel barcoding. Nat. Commun. 2015; 6:7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klein A.M., Mazutis L., Akartuna I., Tallapragada N., Veres A., Li V., Peshkin L., Weitz D.A., Kirschner M.W.. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015; 161:1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015; 161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Streets A.M., Zhang X., Cao C., Pang Y., Wu X., Xiong L., Yang L., Fu Y., Zhao L., Tang F. et al. Microfluidic single-cell whole-transcriptome sequencing. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:7048–7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marcus J.S., Anderson W.F., Quake S.R.. Microfluidic single-cell mRNA isolation and analysis. Anal. Chem. 2006; 78:3084–3089. [DOI] [PubMed] [Google Scholar]
- 16. Erlich H.A., Opelz G., Hansen J.. HLA DNA typing and transplantation. Immunity. 2001; 14:347–356. [DOI] [PubMed] [Google Scholar]
- 17. Robinson J., Halliwell J.A., Hayhurst J.D., Flicek P., Parham P., Marsh S.G.. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015; 43:D423–D431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang C.J., Chen P.L., Yang W.S., Chao K.M.. A fault-tolerant method for HLA typing with PacBio data. BMC Bioinformatics. 2014; 15:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Middelton D. HLA typing from serology to sequencing era. Iran J. Allergy Asthma Immunol. 2005; 4:53–66. [PubMed] [Google Scholar]
- 20. Ammar R., Paton T.A., Torti D., Shlien A., Bader G.D.. Long read nanopore sequencing for detection of HLA and CYP2D6 variants and haplotypes. F1000Res. 2015; 4:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lundin S., Stranneheim H., Pettersson E., Klevebring D., Lundeberg J.. Increased throughput by parallelization of library preparation for massive sequencing. PLoS One. 2010; 5:e10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Niu B., Fu L., Sun S., Li W.. Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinformatics. 2010; 11:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Major E., Rigo K., Hague T., Berces A., Juhos S.. HLA typing from 1000 genomes whole genome and whole exome illumina data. PLoS One. 2013; 8:e78410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Erlich R.L., Jia X., Anderson S., Banks E., Gao X., Carrington M., Gupta N., DePristo M.A., Henn M.R., Lennon N.J. et al. Next-generation sequencing for HLA typing of class I loci. BMC Genomics. 2011; 12:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li C., Chng K.R., Boey E.J., Ng A.H., Wilm A., Nagarajan N.. INC-Seq: accurate single molecule reads using nanopore sequencing. Gigascience. 2016; 5:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.