Abstract
Background
Major depressive disorder is a debilitating illness, which is most commonly treated with antidepressant drugs. As the majority of patients do not respond on their first trial, there is great interest in identifying biological factors that indicate the most appropriate treatment for each patient. Studies suggest that microRNA represent excellent biomarkers to predict antidepressant response.
Methods
We investigated the expression of miR-1202, miR-135a, and miR-16 in peripheral blood from 2 cohorts of depressed patients who received 8 weeks of antidepressant therapy. Expression was quantified at baseline and after treatment, and its relationship to treatment response and depressive symptoms was assessed.
Results
In both cohorts, responders displayed lower baseline miR-1202 levels compared with nonresponders, which increased following treatment.
Conclusions
Ultimately, our results support the involvement of microRNA in antidepressant response and suggest that quantification of their levels in peripheral samples represents a valid approach to informing treatment decisions.
Keywords: microRNA, antidepressant response, major depressive disorder
Introduction
Major depressive disorder (MDD) is a prevalent and debilitating illness and a leading cause of premature death worldwide (Global Burden of Disease Study, 2015). The most commonly used treatments are antidepressant drugs. Unfortunately, the majority of patients do not respond on their first trial, and typically 30% to 40% of patients do not respond following several adequate trials (Trivedi et al., 2006). As such, there is great interest in identifying biological factors that can indicate the most appropriate treatment for each patient. Increasing evidence suggests that microRNAs (miRNA) may be good biomarkers for treatment response (Dwivedi, 2016). MiRNA can be easily quantified in peripheral tissues, and several have been shown to mediate and predict antidepressant effects (Belzeaux et al., 2016). MiR-16 was demonstrated to be involved in the molecular mechanisms underlying the therapeutic effects of antidepressants (Baudry et al., 2010; Launay et al., 2011). MiR-1202 has been found to be downregulated in the brains of depressed individuals and to predict response to the antidepressant citalopram (Lopez et al., 2014, 2017). Finally, miR-135a was shown to mediate antidepressant actions in humans and animal models of depression (Issler et al., 2014). In this study, we investigated these miRNAs in 2 cohorts of depressed patients undergoing antidepressant therapy and assessed their relationships to presence and improvement of depressive symptoms.
Methods
Cohort 1
This cohort was comprised of 55 patients with MDD. Patients were treated with either escitalopram (27 patients, selective serotonin reuptake inhibitor, 10, 15 or 20 mg) or desvenlafaxine (28 patients, serotonin-norepinephrine reuptake inhibitor [SNRI], 50 or 100 mg). Subjects were assessed before treatment (BT) and after 8 weeks (AT) using the Hamilton Depression Rating Scale, 21 items (HAM-D). Whole blood was collected at both timepoints in PAXgene Blood RNA Tubes (PreAnalytix) and total RNA was extracted using the miRNeasy Mini Kit (Qiagen). MiRNA were quantified using Taqman assays. This study was approved by the local institutional review board and written informed consent was obtained from all subjects.
Cohort 2
This cohort consisted of 124 MDD patients who were enrolled in a clinical trial (www.ClinicalTrials.gov 11984A NCT00635219; 11918A NCT00599911; 13267A NCT01140906) and treated with duloxetine (SNRI, 60 mg) for up to 8 weeks. Whole blood was collected before and after treatment in PAXgene Blood RNA Tubes (PreAnalytix) and total RNA was extracted using the PAXgene Blood miRNA Kit (Qiagen, Canada). Depression scores were assessed using the Montgomery-Asberg Depression Rating Scale (MADRS). MiRNA were quantified using Firefly BioWorks miRNA multiplex assay.
Response to Treatment
HAM-D (Cohort 1) and MADRS (Cohort 2) scores were used to classify patients as either treatment responders (RES) or nonresponders (NRES) based on a 50% reduction in depressive scores after treatment. These scales have undergone rigorous studies and are accepted as valid standards of symptom outcome in MDD. Importantly, these 2 scales correlate well with each other and are comparable when assessing symptoms in depressed patients (Jiang and Ahmed, 2009).
Results
Cohort 1
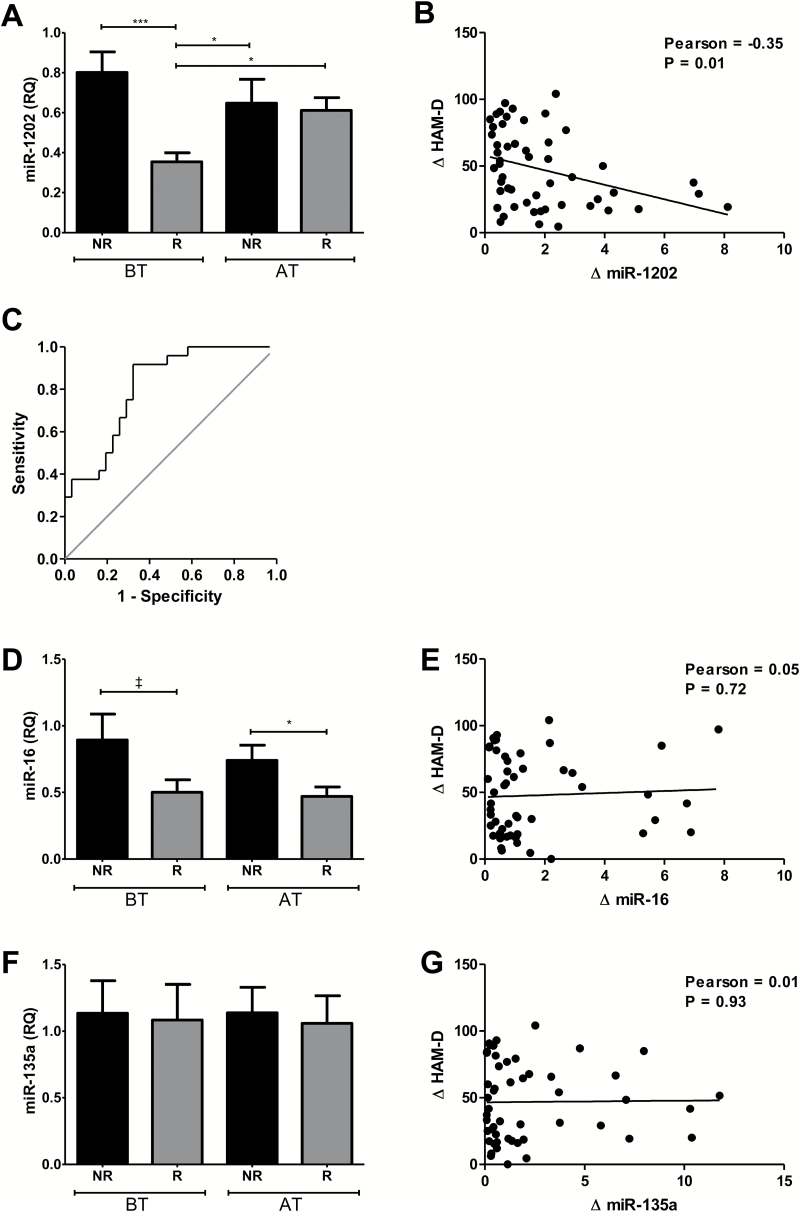
Of the 55 patients in this cohort, 31 were classified as responders (citalopram: 15, desvenlafaxine: 16) and 24 were classified as nonresponders (citalopram: 12, desvenlafaxine: 12). Both groups were clinically depressed according to baseline HAM-D scores (RES: 29±5, NRES: 32±6). MiR-1202 displayed reduced baseline expression in responders, which was increased after treatment, where it displayed similar levels to nonresponders (Figure 1A) and healthy controls (Supplemental Figure 1). Furthermore, we observed a significant correlation between depression symptoms and ratios of miR-1202 levels across time (Figure 1B). We performed a Receiver-Operating Characteristic (ROC) curve analysis and found that baseline miR-1202 levels had a predictive value for antidepressant nonresponse following treatment (AUC=0.812, 95% CI=0.70–0.92, P=.00008) (Figure 1C). Using the Youden Index, miR-1202 levels predicted nonresponse with a sensitivity of 91.7% and specificity of 67.7%. MiR-16 was decreased in responders but unchanged after treatment (Figure 1D-E). Mir-135a levels were unchanged and did not differ between groups (Figure 1F-G).
Figure 1.
Assessment of miRNA in Cohort 1. Levels of miR-1202 (A), miR-16 (D), and miR-135a (F) were quantified by Taqman miRNA assays with RNU6B as the endogenous control using the relative quantitation (RQ) method. Assessments were performed in 24 nonresponders (NR, black), and 31 responders (R, grey) at baseline (BT) and after 8 weeks (AT). Changes in HAM-D scores were calculated using the ratio of scores at AT over BT and are plotted against ratios at BT over AT for miR-1202 (B), miR-16 (E), and miR-135a (G). A ROC curve analysis was performed using baseline miR-1202 levels to predict nonresponse (C). *P<.05; ***P <.0001; ‡ 0.1< P >.05.
Cohort 2
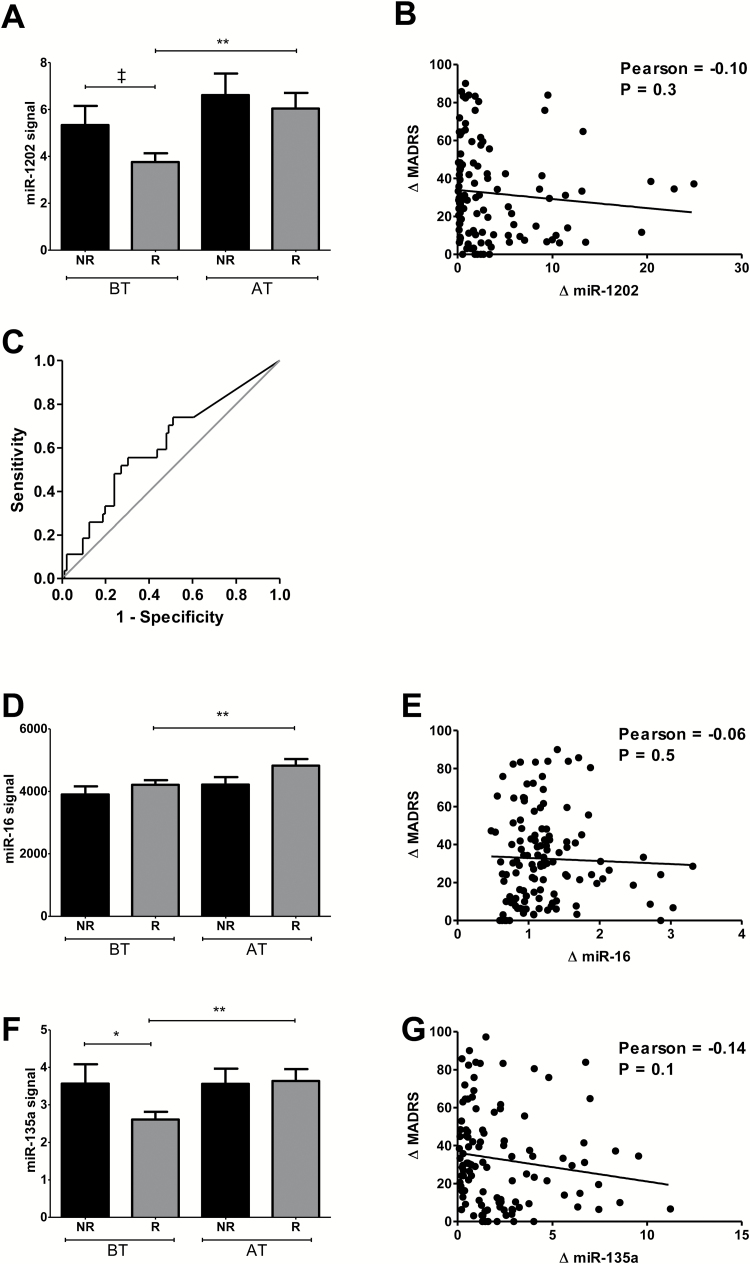
This cohort included 97 responders and 27 nonresponders. Both groups were clinically depressed at baseline (RES: 31±4, NRES: 32±4). Consistent with Cohort 1, at baseline, miR-1202 displayed a trend for decreased levels in responders relative to nonresponders (P=.058), which increased and displayed similar levels to nonresponders following treatment (P=.67) (Figure 2A). We performed a ROC curve analysis using baseline miR-1202 levels and found a trend for significance (AUC=0.61, 95% CI=0.49–0.74, P=.07) (Figure 2C). Although there were no differences in baseline miR-16, it was increased in responders after treatment (Figure 2D). MiR-135a showed a similar pattern of expression as miR-1202, with a decrease at baseline and increase in responders after treatment (Figure 2F). There were no significant correlations between changes in MADRS scores and miRNA levels for any of the miRNA (Figure 2B,E,G).
Figure 2.
Assessment of miRNA in Cohort 2. Levels of miR-1202 (A), miR-16 (D), and miR-135a (F) were quantified using Firefly BioWorks multiplex miRNA assay and normalized to the geometric means of miR-29b-3p, miR-19b-3p, Let-7i-5p, and Let-7b-5p. Assessments were performed in 27 nonresponders (black) and 97 responders (grey) at baseline (BT) and after treatment with duloxetine (AT). Changes in MADRS scores were calculated using the ratio of scores at AT over BT and are plotted against ratios at BT over AT for miR-1202 (B), miR-16 (E), and miR-135a (G). A ROC curve analysis was performed using baseline miR-1202 levels to predict nonresponse (C). *P<.05; **P<.01; ‡ 0.1<P>.05.
Discussion
Overall, we identified lower baseline expression of the 3 miRNAs in treatment responders. Our most consistent findings were with miR-1202, which displayed decreased baseline levels in responders in both cohorts and increased levels as a function of treatment response. As these findings were found for both antidepressant classes (selective serotonin reuptake inhibitors and SNRIs), this could indicate that lower levels may represent a biomarker for clinical response to antidepressant monotherapy in general. Indeed, previous studies by our group found that miR-1202 levels were upregulated following chronic treatment of neural progenitor cells using either citalopram or imipramine, but unchanged by treatment with nonserotonergic drugs (Lopez et al., 2014). Glutamate receptor 4 (GRM4) was also identified as being an important target of miR-1202, and a recent study identified a variant in the 3’ UTR of GRM4 that was predicted to disrupt miR-1202 binding and was associated with MDD (Dadkhah et al., 2017). The expression of a number of glutamate receptors, including GRM4, have been found to be dysregulated in depressed suicide completers (Gray et al., 2015), and they have been shown to be altered by antidepressant treatment (Cruceanu et al., 2016). It is thus reasonable to speculate that at least some of our findings with miR-1202 are related to its relationship with the glutamate system.
Unlike miR-1202, the results with miR-16 and miR-135a displayed variability between cohorts, suggesting that their effects may be influenced by variables not investigated in this study. Indeed, a number of studies have investigated the relationship between miR-16 and both depression- and anxiety-related phenotypes and found it to be both significantly upregulated or downregulated depending upon the phenotype and tissue being examined (Bai et al., 2012; Honda et al., 2013; Song et al., 2015; Zurawek et al., 2016b). This has also been observed with miR-135a (Mannironi et al., 2013; Zurawek et al., 2016a). Accordingly, the baseline differences we observed between our 2 cohorts may reflect underlying differences in anxiety or stress-related variables between the 2 populations. One mechanism for these effects could be through their actions on the serotonin system, as both miR-16 and miR-135a have been shown to regulate the expression of the serotonin transporter (SERT) (Baudry et al., 2010; Launay et al., 2011; Issler et al., 2014). This may also partially explain some of the drug-specific effects we observed in the current study.
Ultimately, these results support an involvement of miR-1202, miR-16, and miR-135a in antidepressant response and suggest that quantification of their levels in peripheral samples may represent a valid approach to informing treatment decisions.
Statement of Interest
S.H.K. has received research funding or honoraria from the following sources: Allergan, AstraZeneca, BMS, Brain Canada, Canadian Institutes for Health Research (CIHR), Eli Lilly, Janssen, Lundbeck, Lundbeck Institute, OMHF, Ontario Brain Institute, Ontario Research Fund (ORF), Pfizer, Servier, St. Jude Medical, Sunovion and Xian-Janssen. G.T. holds a Canada Research Chair (Tier 1), Fonds de Recherche du Québec - Santé Chercheur National salary award, and a NARSAD Distinguished Investigator Award. G.T. is supported by grants from the CIHR (FDN148374, MOP93775, MOP11260, MOP119429, and MOP119430), from the US National Institutes of Health (1R01DA033684), by the FRQS through the Quebec Network on Suicide, Mood Disorders and Related Disorders, and through an investigator-initiated research grant from Pfizer.
Supplementary Material
Acknowledgments
This research was funded by the Canadian Institutes of Health Research and with the support of the Canadian Biomarker Integration Network in Depression (CAN-BIND), an Integrated Discovery Program, with funding from the Ontario Brain Institute, an independent nonprofit corporation, funded partially by the Ontario government. Additional funding for CAN-BIND was provided by the CIHR, Brain Canada, Lundbeck, Bristol-Myers Squibb, Pfizer, and Servier.
References
- Bai M, Zhu X, Zhang Y, Zhang S, Zhang L, Xue L, Yi J, Yao S, Zhang X. (2012) Abnormal hippocampal BDNF and miR-16 expression is associated with depression-like behaviors induced by stress during early life. PLoS One 7:e46921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry A, Mouillet-Richard S, Schneider B, Launay JM, Kellermann O. (2010) miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science 329:1537–1541. [DOI] [PubMed] [Google Scholar]
- Belzeaux R, Lin CW, Ding Y, Bergon A, Ibrahim EC, Turecki G, Tseng G, Sibille E. (2016) Predisposition to treatment response in major depressive episode: a peripheral blood gene coexpression network analysis. J Psychiatr Res 81:119–126. [DOI] [PubMed] [Google Scholar]
- Cruceanu C, Lopez JP, Tsai WT, Turecki G. (2016) Dysregulation of the glutamatergic receptors after antidepressant treatment in human neural progenitor cells. Mol Psychiatry doi: 10.1038/mp.2016.138. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Dadkhah T, Rahimi-Aliabadi S, Jamshidi J, Ghaedi H, Taghavi S, Shokraeian P, Akhavan-Niaki H, Tafakhori A, Ohadi M, Darvish H. (2017) A genetic variant in miRNA binding site of glutamate receptor 4, metabotropic (GRM4) is associated with increased risk of major depressive disorder. J Affect Disord 208:218–222. [DOI] [PubMed] [Google Scholar]
- Dwivedi Y. (2016) Pathogenetic and therapeutic applications of microRNAs in major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry 64:341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Burden of Disease Study C (2015) Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 386:743–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray AL, Hyde TM, Deep-Soboslay A, Kleinman JE, Sodhi MS. (2015) Sex differences in glutamate receptor gene expression in major depression and suicide. Mol Psychiatry 20:1057–1068. [DOI] [PubMed] [Google Scholar]
- Honda M, Kuwano Y, Katsuura-Kamano S, Kamezaki Y, Fujita K, Akaike Y, Kano S, Nishida K, Masuda K, Rokutan K. (2013) Chronic academic stress increases a group of microRNAs in peripheral blood. PLoS One 8:e75960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issler O, Haramati S, Paul ED, Maeno H, Navon I, Zwang R, Gil S, Mayberg HS, Dunlop BW, Menke A, Awatramani R, Binder EB, Deneris ES, Lowry CA, Chen A. (2014) MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron 83:344–360. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Ahmed S. (2009) An analysis of correlations among four outcome scales employed in clinical trials of patients with major depressive disorder. Ann Gen Psychiatry 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launay JM, Mouillet-Richard S, Baudry A, Pietri M, Kellermann O. (2011) Raphe-mediated signals control the hippocampal response to SRI antidepressants via miR-16. Transl Psychiatry 1:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JP, Pereira F, Richard-Devantoy S, Berlim M, Chachamovich E, Fiori LM, Niola P, Turecki G, Jollant F. (2017) Co-variation of peripheral levels of miR-1202 and brain activity and connectivity during antidepressant treatment. Neuropsychopharmacology doi: 10.1038/npp.2017.9. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JP, Lim R, Cruceanu C, Crapper L, Fasano C, Labonte B, Maussion G, Yang JP, Yerko V, Vigneault E, El Mestikawy S, Mechawar N, Pavlidis P, Turecki G. (2014) miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat Med 20:764–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannironi C, Camon J, De Vito F, Biundo A, De Stefano ME, Persiconi I, Bozzoni I, Fragapane P, Mele A, Presutti C. (2013) Acute stress alters amygdala microRNA miR-135a and miR-124 expression: inferences for corticosteroid dependent stress response. PLoS One 8:e73385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MF, Dong JZ, Wang YW, He J, Ju X, Zhang L, Zhang YH, Shi JF, Lv YY. (2015) CSF miR-16 is decreased in major depression patients and its neutralization in rats induces depression-like behaviors via a serotonin transmitter system. J Affect Disord 178:25–31. [DOI] [PubMed] [Google Scholar]
- Trivedi MH, Rush AJ, Wisniewski SR, Nierenberg AA, Warden D, Ritz L, Norquist G, Howland RH, Lebowitz B, McGrath PJ, Shores-Wilson K, Biggs MM, Balasubramani GK, Fava M, Team SDS (2006) Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am J Psychiatry 163:28–40. [DOI] [PubMed] [Google Scholar]
- Zurawek D, Kusmider M, Faron-Gorecka A, Gruca P, Pabian P, Solich J, Kolasa M, Papp M, Dziedzicka-Wasylewska M. (2016a) Reciprocal MicroRNA expression in mesocortical circuit and its interplay with serotonin transporter define resilient rats in the chronic mild stress. Mol Neurobiol 2016. Sep 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawek D, Kusmider M, Faron-Gorecka A, Gruca P, Pabian P, Kolasa M, Solich J, Szafran-Pilch K, Papp M, Dziedzicka-Wasylewska M. (2016b) Time-dependent miR-16 serum fluctuations together with reciprocal changes in the expression level of miR-16 in mesocortical circuit contribute to stress resilient phenotype in chronic mild stress - an animal model of depression. Eur Neuropsychopharmacol 26:23–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.