Abstract
Many analyses for the detection of biological phenomena rely on a multiple sequence alignment as input. The results of such analyses are often further studied through parametric bootstrap procedures, using sequence simulators. One of the problems with conducting such simulation studies is that users currently have no means to decide which insertion and deletion (indel) parameters to choose, so that the resulting sequences mimic biological data. Here, we present SpartaABC, a web server that aims to solve this issue. SpartaABC implements an approximate-Bayesian-computation rejection algorithm to infer indel parameters from sequence data. It does so by extracting summary statistics from the input. It then performs numerous sequence simulations under randomly sampled indel parameters. By computing a distance between the summary statistics extracted from the input and each simulation, SpartaABC retains only parameters behind simulations close to the real data. As output, SpartaABC provides point estimates and approximate posterior distributions of the indel parameters. In addition, SpartaABC allows simulating sequences with the inferred indel parameters. To this end, the sequence simulators, Dawg 2.0 and INDELible were integrated. Using SpartaABC we demonstrate the differences in indel dynamics among three protein-coding genes across mammalian orthologs. SpartaABC is freely available for use at http://spartaabc.tau.ac.il/webserver.
INTRODUCTION
Sequence simulation is an extremely important component of phylogenetic studies and many sequence simulators have been previously developed (1–12). The tasks for which sequence simulations are used vary greatly and span a wide range of scientific questions. For example, Worobey et al. used sequence simulations to investigate the origins of influenza A virus within and between hosts (13). Shapiro et al. utilized sequence simulations in their study of early events of ecological differentiation of bacterial genomes (14). Gossmann and Schmid included sequence simulations as part of their analysis of post-duplication selective forces on genes in Arabidopsis thaliana (15). Sequence simulators are also often used in studies that aim to evaluate the performance of alignment and tree reconstruction algorithms (16–23). Finally, sequence simulations are an integral part of parametric bootstrap test procedures, which are used, for example, to test for the constancy of evolutionary rates (24), to study the fit of various evolutionary models to real sequences (25,26), to detect traits that impact the rate of evolution (27,28) and to compare competing tree topologies (29–31).
Sequence simulators provide in-silico generated datasets under different evolutionary scenarios. The complete evolutionary process relies on a substitution model (e.g. 32–38) as well as a model of insertion and deletion. The occurrence of indel events is defined relative to the substitution process and is controlled by the IR parameter—the indel-to-substitution rate ratio. The length of the indel is often modeled using a power–law distribution, controlled by its shape parameter ‘A’. This distribution is characterized by a reverse relationship between an indel size and its probability. Finally, the root length parameter RL controls the length of the sequence at the root of the tree (the start of the simulation). Although the root length is not a pure indel parameter, it strongly affects the resulting multiple sequence alignment (MSA). Until recently, no methodology was available for users to determine the values of these parameters in a way that best reflects the indel dynamics in their datasets of interest.
We recently developed the SpartaABC algorithm (Levy Karin, Shkedy et al. submitted for publication), an approximate Bayesian computation rejection algorithm to infer indel parameters from sequence data. SpartaABC focuses on the inference of the above-mentioned three indel parameters. To this end, SpartaABC extracts a vector of summary statistics from its input; it then performs repeated simulations using an integrated sequence simulator (8,12) under various indel parameters. From each such simulated dataset it extracts a vector of summary statistics and computes its distance from the vector extracted for the input using a weighted Euclidean distance. SpartaABC retains a subset of the simulations for which the distance from the input was small enough. The parameter sets from simulations with a small distance are used to estimate the indel parameters behind the input. Using a simulation study, the SpartaABC algorithm was shown to accurately infer indel parameter values under various conditions (Levy Karin, Shkedy et al. submitted for publication). Thus, sequences simulated using the SpartaABC inferred indel parameters resemble the input data in terms of their indel properties, much more so, than when sequence simulators are run with default parameters.
Here we use the SpartaABC algorithm as part of a broader web service, which provides the following: (i) MSA reconstruction (optional), (ii) tree reconstruction (optional), (iii) inference of indel dynamics and (iv) sequence simulation based on the inferred indel parameters (optional). Visual and textual outputs of these services are offered as downloadable files.
MATERIALS AND METHODS
Input
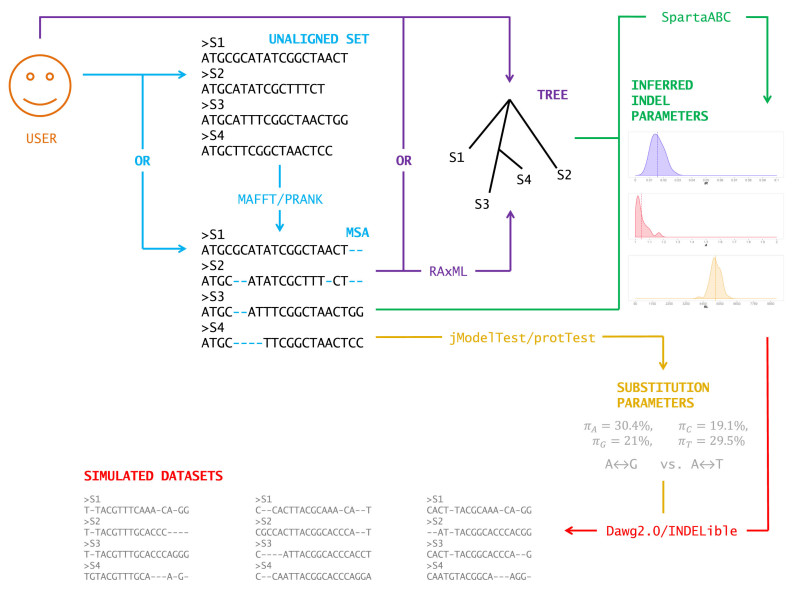
The SpartaABC web server requires sequence data (either nucleotide or protein) as input. The user can provide either an MSA or a set of unaligned sequences. If unaligned data are provided, the user will be asked to choose between the programs MAFFT (39,40) and PRANK (41) to align them. An optional input to the SpartaABC web server is a phylogenetic tree. If the user does not provide a phylogenetic tree, the maximum likelihood tree will be computed based on the MSA of the sequences, using RAxML (42). SpartaABC integrates two sequence simulators: Dawg 2.0 (12), which is the default, and INDELible (8). Finally, the user can indicate the number of simulated datasets to produce based on the indel parameters inferred from the input. An illustration of the computational stages performed by the SpartaABC web server is presented in Figure 1.
Figure 1.
An illustration of the computational stages performed by the SpartaABC web server.
Summary statistics
The summary statistics computed by SpartaABC are detailed in the OVERVIEW section of the web server. Among them are the average gap length, the total number of gaps and the MSA length. Based on the summary statistics extracted from the input MSA and each simulation, SpartaABC computes a weighted Euclidean distance. The weights used by SpartaABC are also available for download from the SOURCE & USAGE section of the web server. In addition, the extracted summary statistics values from the input MSA and each simulation in the SpartaABC run are available for download.
Indel parameters search space
Throughout its computation, SpartaABC proposes 100,000 indel parameter combinations by sampling values of each of the parameters from a prior uniform distribution. Specifically, the ‘A’ parameter value is sampled from a wide range: (1, 2]; the IR parameter value is sampled by default from the range: [0, 0.05], but this range can be extended by the user up to [0, 0.1]. Finally, the RL parameter range is determined empirically according to the input provided by the user. Let L denote the longest sequence in the user-provided input, then the search range of the RL parameter is [50, 1.1 × L].
Output
SpartaABC provides a step-by-step progress report and estimation of the expected run time. Upon completion of the SpartaABC computation, all examined indel parameter combinations and their distance from the input dataset are available to the user as a downloadable file. Out of these, the 50 parameter combinations with the smallest distance are used to approximate the posterior distributions of the indel parameters. These distributions are presented to the user in three plots, where the x-axis is the entire search range of each indel parameter and the y-axis is the density. An example for such plots is given in Figure 2. In addition, SpartaABC computes the posterior expectations based on the inferred posterior distributions to yield point estimates of the indel parameters. As its final step, the web server simulates datasets using the indel parameters point estimates, according to the number of replicates determined by the user. The substitution model and parameters used in the sequence simulation step are estimated and selected according to the AICc (43) criterion by jModelTest (44) or protTest (45), for DNA or protein input, respectively. The user can download a zipped file of these simulated datasets as well as the sequence simulator control file. Finally, the MSA and phylogenetic tree from which SpartaABC inferred the indel parameters are presented visually using Wasabi (46).
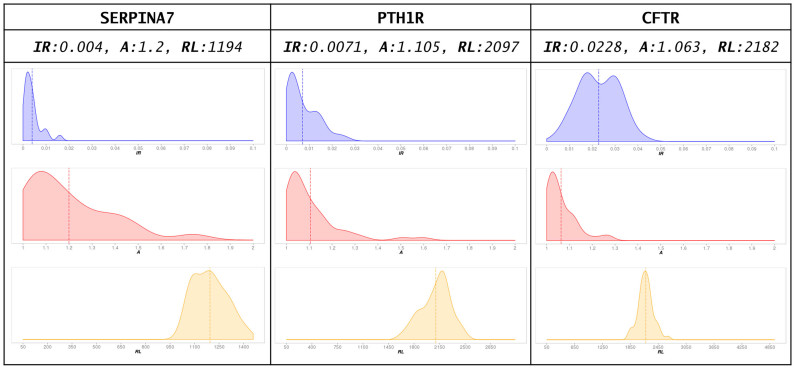
Figure 2.
SpartaABC analyses of three genes involved in human diseases across mammalian orthologs. The point estimates of each of the indel parameters are presented above the approximated posterior distribution plots. IR: indel to substitution rate ratio; A: the shape parameter controlling the power–law distribution describing indel lengths; RL: root length.
Implementation
The SpartaABC web server runs on a Linux cluster of 2.6 GHz AMD Opteron processors, equipped with 4 GB RAM per quad-core node. The server runs up-to-date versions of the supported multiple alignment and tree reconstruction programs. The SpartaABC algorithm was implemented in C++. We provide its source code, a precompiled version for UNIX systems, a short manual and a run example in the SOURCE & USAGE section of the web server. In addition, the web server contains a frequently asked questions page to provide additional information concerning the algorithm and methodology.
CASE STUDY
SERPINA7, PTH1R and CFTR are genes known to play a role in the human diseases: thyroxine-binding globulin deficiency, chondrodysplasia and cystic fibrosis, respectively (47–49). In order to examine their indel dynamics, we obtained their sets of unaligned coding sequences across >30 mammalian orthologous species from the OrthoMam database (50). These datasets are available for download at the GALLERY section of the web server. We then analyzed each of these sets using the SpartaABC web server. First, an MSA was computed for each unaligned set of sequences using the server's default MSA program, MAFFT (40). Second, a phylogenetic tree was reconstructed using RAxML (42). Third, the MSA and the tree were used to infer indel parameters. We found, that in spite of the fact that all three analyzed coding sequences are involved in human diseases and have roughly the same number of mammalian orthologs, they display substantially different indel dynamics (Figure 2). Specifically, the IR parameter inferred for SERPINA7 is 5-fold smaller than that inferred for CFTR, with PTH1R taking an IR value in between the other two. In addition, the inferred RL parameters corresponded to the different lengths of the genes. Finally, all genes displayed a tendency for longer indels as evident by their inferred ‘A’ parameter. All three inferred ‘A’ values were close to 1.0, yielding power low distributions where longer indels are more probable compared to power low distributions with a high ‘A’ value. From these results we conclude that even when examining orthologous genes within the same taxonomic class and similar biological contexts, it is important to characterize the indel dynamics of each gene individually in order to best mimic biological data.
In the following example, we demonstrate the utility of the SpartaABC web server to test specific evolutionary hypotheses using a parametric bootstrap procedure, in which the sequences are generated based on the indel parameters inferred from the data. Here, we focused on the comparison of the indel dynamics between the coding region of SERPINA7 (as analyzed above) to the entire SERPINA7 gene (exons and introns included). To this end, we obtained the full SERPINA7 gene sequences across 35 mammalian orthologs from the ENSEMBL database (51). Using SpartaABC web server, we found that over the whole gene, the IR parameter is much higher (0.0166) compared to that inferred in the coding sequence only (0.004), suggesting that indels are much more frequent when intronic regions are included in the analysis compared to examining only exonic regions. A much smaller difference was found in the inferred ‘A’ parameter (1.036 and 1.2 for the full and coding-only SERPINA7, respectively), suggesting that the main difference between the full and coding-only SERPINA7 is the frequency of indels, rather than their size. We hypothesized that such a difference may stem from selection against the introduction of indels in a specific region that resides within the exons of the analyzed gene. To statistically test this hypothesis, we first measured the longest stretch of consecutive columns without gap characters in the MSA of the full SERPINA7 gene. We found that this stretch was 174 columns in length, which reside within the second human exon of this gene (starting at position 3726 of the MSA). Using simulations which do not prefer one sequence position over the other for indel events, we could test how likely it is to observe a gap-free stretch of consecutive columns of such length. We thus compared the length of the SERPINA7 stretch to those computed from 100 simulated MSAs produced by the SpartaABC web server according to the SERPINA7 inferred indel parameters using Dawg 2.0 (12). In all 100 simulated datasets we found that the longest stretch without any gap characters did not exceed 85 columns in length, suggesting the SERPINA7 stretch is significantly longer than one could expect (empirical P-value < 0.01). The data (e.g., MSAs and trees) and the analyses associated with this example are provided in the GALLERY section of the web server. In conclusion, indel dynamics can vary along a specific gene and using sequence simulations it is possible to detect gene regions that deviate from the average indel dynamics inferred for the entire sequence.
FUNDING
BSF [2015247 to R.A.C., T.P.]; ISF [1092/13, 802/16 to T.P.]; Edmond J. Safra Center for Bioinformatics at Tel Aviv University Fellowship (to E.L.K.). Funding for open access charge: BSF [2015247 to R.A.C., T.P.]; ISF [1092/13, 802/16 to T.P.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Rambaut A., Grassly N.C.. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput. Appl. Biosci. 1997; 13:235–238. [DOI] [PubMed] [Google Scholar]
- 2. Stoye J., Evers D., Meyer F.. Rose: generating sequence families. Bioinformatics. 1998; 14:157–163. [DOI] [PubMed] [Google Scholar]
- 3. Cartwright R.A. DNA assembly with gaps (Dawg): simulating sequence evolution. Bioinformatics. 2005; 21:iii31–iii38. [DOI] [PubMed] [Google Scholar]
- 4. Gesell T., von Haeseler A.. In silico sequence evolution with site-specific interactions along phylogenetic trees. Bioinformatics. 2006; 22:716–722. [DOI] [PubMed] [Google Scholar]
- 5. Strope C.L., Scott S.D., Moriyama E.N.. indel-Seq-Gen: a new protein family simulator incorporating domains, motifs, and indels. Mol. Biol. Evol. 2007; 24:640–649. [DOI] [PubMed] [Google Scholar]
- 6. Hall B.G. Simulating DNA coding sequence evolution with EvolveAGene 3. Mol. Biol. Evol. 2008; 25:688–695. [DOI] [PubMed] [Google Scholar]
- 7. Shavit Grievink L., Penny D., Hendy M.D., Holland B.R.. LineageSpecificSeqgen: generating sequence data with lineage-specific variation in the proportion of variable sites. BMC Evol. Biol. 2008; 8:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fletcher W., Yang Z.. INDELible: a flexible simulator of biological sequence evolution. Mol. Biol. Evol. 2009; 26:1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sipos B., Massingham T., Jordan G.E., Goldman N.. PhyloSim - Monte Carlo simulation of sequence evolution in the R statistical computing environment. BMC Bioinformatics. 2011; 12:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koestler T., von Haeseler A., Ebersberger I.. REvolver: modeling sequence evolution under domain constraints. Mol. Biol. Evol. 2012; 29:2133–2145. [DOI] [PubMed] [Google Scholar]
- 11. Dalquen D.A., Anisimova M., Gonnet G.H., Dessimoz C.. ALF–a simulation framework for genome evolution. Mol. Biol. Evol. 2012; 29:1115–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cartwright R.A. Dawg 2.0. 2017; https://github.com/reedacartwright/dawg/tree/develop.
- 13. Worobey M., Han G.-Z., Rambaut A.. A synchronized global sweep of the internal genes of modern avian influenza virus. Nature. 2014; 508:254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shapiro B.J., Friedman J., Cordero O.X., Preheim S.P., Timberlake S.C., Szabó G., Polz M.F., Alm E.J.. Population genomics of early events in the ecological differentiation of bacteria. Science. 2012; 336:48–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gossmann T.I., Schmid K.J.. Selection-driven divergence after gene duplication in Arabidopsis thaliana. J. Mol. Evol. 2011; 73:153–165. [DOI] [PubMed] [Google Scholar]
- 16. Charleston M.A., Hendy M.D., Penny D.. The effects of sequence length, tree topology, and number of taxa on the performance of phylogenetic methods. J. Comput. Biol. 1994; 1:133–151. [DOI] [PubMed] [Google Scholar]
- 17. Izquierdo-Carrasco F., Smith S.A., Stamatakis A.. Algorithms, data structures, and numerics for likelihood-based phylogenetic inference of huge trees. BMC Bioinformatics. 2011; 12:470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blackburne B.P., Whelan S.. Measuring the distance between multiple sequence alignments. Bioinformatics. 2012; 28:495–502. [DOI] [PubMed] [Google Scholar]
- 19. Kück P., Mayer C., Wägele J.-W., Misof B.. Long branch effects distort maximum likelihood phylogenies in simulations despite selection of the correct model. PLoS One. 2012; 7:e36593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Löytynoja A., Vilella A.J., Goldman N.. Accurate extension of multiple sequence alignments using a phylogeny-aware graph algorithm. Bioinformatics. 2012; 28:1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thi Nguyen M.A., Gesell T., von Haeseler A.. ImOSM: intermittent evolution and robustness of phylogenetic methods. Mol. Biol. Evol. 2012; 29:663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ashkenazy H., Cohen O., Pupko T., Huchon D.. Indel reliability in indel-based phylogenetic inference. Genome Biol. Evol. 2014; 6:3199–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sela I., Ashkenazy H., Katoh K., Pupko T.. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 2015; 43:W7–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adell J.C., Dopazo J.. Monte Carlo simulation in phylogenies: an application to test the constancy of evolutionary rates. J. Mol. Evol. 1994; 38:305–309. [DOI] [PubMed] [Google Scholar]
- 25. Goldman N. Simple diagnostic statistical tests of models for DNA substitution. J. Mol. Evol. 1993; 37:650–661. [DOI] [PubMed] [Google Scholar]
- 26. Goldman N. Statistical tests of models of DNA substitution. J. Mol. Evol. 1993; 36:182–198. [DOI] [PubMed] [Google Scholar]
- 27. Mayrose I., Otto S.P.. A likelihood method for detecting trait-dependent shifts in the rate of molecular evolution. Mol. Biol. Evol. 2011; 28:759–770. [DOI] [PubMed] [Google Scholar]
- 28. Levy Karin E., Wicke S., Pupko T., Mayrose I.. An integrated model of phenotypic trait changes and site-specific sequence evolution. Syst. Biol. 2017; doi:10.1093/sysbio/syx032. [DOI] [PubMed] [Google Scholar]
- 29. Bull J.J., Cunningham C.W., Molineux I.J., Badget M.R., Hillis D.M.. Experimental molecular evolution of bacteriophage T7. Evolution (N. Y). 1993; 47:993–1007. [DOI] [PubMed] [Google Scholar]
- 30. Swofford D.L., Olsen G.J., Waddell P.J., Hillis D.M.. Hillis DM, Moritz C, Mable BK. Phylogenetic inference. Molecular Systematics. 1996; Sunderland: Sinauer Associates, Inc; 407–514. [Google Scholar]
- 31. Levy Karin E., Susko E., Pupko T.. Alignment errors strongly impact likelihood-based tests for comparing topologies. Mol. Biol. Evol. 2014; 31:3057–3067. [DOI] [PubMed] [Google Scholar]
- 32. Jukes T.H., Cantor C.R.. Munro HN, Allison JB. Evolution of protein molecules. Mammalian Protein Metabolism. 1969; NY: Academic Press; 21–132. [Google Scholar]
- 33. Tavare S. Some probabilistic and statistical problems on the analysis of DNA sequences. Lect. Math. Life Sci. 1986; 17:57–86. [Google Scholar]
- 34. Hasegawa M., Kishino H., Yano T.. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985; 22:160–174. [DOI] [PubMed] [Google Scholar]
- 35. Jones D.T., Taylor W.R., Thornton J.M.. The rapid generation of mutation data matrices from protein sequences. Bioinformatics. 1992; 8:275–282. [DOI] [PubMed] [Google Scholar]
- 36. Goldman N., Yang Z.. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 1994; 11:725–736. [DOI] [PubMed] [Google Scholar]
- 37. Whelan S., Goldman N.. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001; 18:691–699. [DOI] [PubMed] [Google Scholar]
- 38. Le S.Q., Gascuel O.. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008; 25:1307–1320. [DOI] [PubMed] [Google Scholar]
- 39. Katoh K., Misawa K., Kuma K., Miyata T.. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002; 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Katoh K., Standley D.M.. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013; 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Löytynoja A., Goldman N.. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science. 2008; 320:1632–1635. [DOI] [PubMed] [Google Scholar]
- 42. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014; 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sullivan J., Joyce P.. Model selection in phylogenetics. Annu. Rev. Ecol. Evol. Syst. 2005; 36:445–466. [Google Scholar]
- 44. Darriba D., Taboada G.L., Doallo R., Posada D.. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012; 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Darriba D., Taboada G.L., Doallo R., Posada D.. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011; 27:1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Veidenberg A., Medlar A., Löytynoja A.. Wasabi: an integrated platform for evolutionary sequence analysis and data visualization. Mol. Biol. Evol. 2016; 33:1126–1130. [DOI] [PubMed] [Google Scholar]
- 47. Mori Y., Seino S., Takeda K., Flink I.L., Murata Y., Bell G.I., Refetoff S.. A mutation causing reduced biological activity and stability of thyroxine-binding globulin probably as a result of abnormal glycosylation of the molecule. Mol. Endocrinol. 1989; 3:575–579. [DOI] [PubMed] [Google Scholar]
- 48. Rommens J.M., Iannuzzi M.C., Kerem B., Drumm M.L., Melmer G., Dean M., Rozmahel R., Cole J.L., Kennedy D., Hidaka N.. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989; 245:1059–1065. [DOI] [PubMed] [Google Scholar]
- 49. Schipani E., Kruse K., Jüppner H.. A constitutively active mutant PTH-PTHrP receptor in Jansen-type metaphyseal chondrodysplasia. Science. 1995; 268:98–100. [DOI] [PubMed] [Google Scholar]
- 50. Douzery E.J.P., Scornavacca C., Romiguier J., Belkhir K., Galtier N., Delsuc F., Ranwez V.. OrthoMaM v8: a database of orthologous exons and coding sequences for comparative genomics in mammals. Mol. Biol. Evol. 2014; 31:1923–1928. [DOI] [PubMed] [Google Scholar]
- 51. Aken B.L., Achuthan P., Akanni W., Amode M.R., Bernsdorff F., Bhai J., Billis K., Carvalho-Silva D., Cummins C., Clapham P. et al. . Ensembl 2017. Nucleic Acids Res. 2017; 45:D635–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]