Abstract
Alternative translation initiation mechanisms such as leaky scanning and reinitiation potentiate the polycistronic nature of human transcripts. By allowing for reprogrammed translation, these mechanisms can mediate biological responses to stimuli. We combined proteomics with ribosome profiling and mRNA sequencing to identify the biological targets of translation control triggered by the eukaryotic translation initiation factor 1 (eIF1), a protein implicated in the stringency of start codon selection. We quantified expression changes of over 4000 proteins and 10 000 actively translated transcripts, leading to the identification of 245 transcripts undergoing translational control mediated by upstream open reading frames (uORFs) upon eIF1 deprivation. Here, the stringency of start codon selection and preference for an optimal nucleotide context were largely diminished leading to translational upregulation of uORFs with suboptimal start. Interestingly, genes affected by eIF1 deprivation were implicated in energy production and sensing of metabolic stress.
INTRODUCTION
Qualitative and quantitative characterization of gene expression is indispensable to understand dynamic phenotypes of eukaryotic cells. Through technological advances in high-throughput sequencing and proteomics, it is now possible to follow gene expression from transcription to protein turnover (1–5). One of the remaining fundamental challenges in modern biology includes the unraveling of the full diversity of proteoforms (i.e. the different molecular forms of proteins) (6,7) expressed from single genes. An increasing line of evidence suggests that mRNA translation may both be a rapid means of gene expression control (8–10) as well as a major source of proteoforms (11–14). However, genes undergoing translational control (8,15) and regulation of proteoform expression (16–18) remain poorly investigated.
Alternative translation initiation mechanisms allow to select between multiple start codons and open reading frames (ORFs) within a single mRNA molecule. Here, the scanning ribosomes may omit less efficient upstream start codons (e.g. non-AUG start codons and start codons embedded in a suboptimal nucleotide context) to initiate translation downstream in a process referred to as leaky scanning (8,19). Reinitiation, another alternative translation initiation mechanism (8,19,20), may occur when post-termination ribosomes are retained on the mRNA molecule after completing translation of an upstream ORF (uORF) and reused to support translation of a proximal downstream ORF. A particular role in alternative translation was postulated for short ORFs situated in the mRNA 5΄ leaders (uORFs) or upstream and partially overlapping the main protein-coding sequence (CDS) (upstream-overlapping ORFs or u-oORFs). Due to the directionality of ribosomal scanning, these short ORFs may regulate protein translation (21,22) or even impact on the selection of alternative translation sites giving rise to alternative protein N-termini and thus N-terminal proteoforms (16–18). The importance of u(-o)ORFs was supported by sequencing of ribosome associated mRNA regions (ribosome profiling, or ribo-seq) (5,23) which provided evidence for the ubiquitous translation from non-AUG start sites situated outside annotated protein-coding regions. Prevalence of regulatory features in 5΄ leaders was further highlighted by translation complex profile sequencing (TCP-seq), a ribo-seq derived method, which specifically tracks the footprints of small ribosomal subunits during the scanning process (4). uORFs were characterized in a variety of organisms and conditions (9,10,24–26), and their impact on the translation efficiency of proteins was found to be conserved among orthologous genes (24,25). Considering the directionality of scanning, ribosome profiling experiments revealed that ribosomes distribute asymmetrically across ORFs, as they readily accumulate at translation initiation and termination sites (5), an effect which may be enlarged due to pretreatment with translation elongation inhibitors (5,27), overall warranting caution when interpreting uORF expression levels. Importantly however, further studies reveled that ribosome footprints of 5΄ leaders generally resemble those of coding sequences, suggesting genuine translation of these regions (23).
Translation initiation is a determining control step in translation (28). In consequence, translational control is mainly facilitated by eukaryotic translation initiation factors (eIFs) which may readily respond to (extra)cellular conditions by changing the global rates of protein synthesis at the ribosome. To reduce the high energy cost of protein production, translational control through reinitiation can be triggered by eIF2α phosphorylation in response to nutrient deprivation and accumulation of unfolded proteins (15). On the other hand, eIF1 was shown to orchestrate leaky scanning by stabilizing ‘open’, scanning-competent conformation of the ribosome (29) and thereby regulate translation initiation rates at suboptimal translation initiation start sites (30,31). Besides, eIF1 protein levels and its phosphorylation have been linked to reprogrammed translation of uORFs (32,33) and responses to stress stimuli, including arsenite (33); glucose or oxygen deprivation (10). Although eIF1 plays a central role in translation initiation (34), a genome-wide assessment of its role in translational regulation is lacking. By combining tailored proteomic strategies with ribosome profiling and mRNA sequencing we here identified the biological targets of the translation control exerted by eIF1.
MATERIALS AND METHODS
Cell culture
The human colon cancer cell line HCT116 was kindly provided by the Johns Hopkins Sidney Kimmel Comprehensive Cancer Center (Baltimore, USA). The HAP1 wild type and CRISPR/Cas9 engineered knockout cell lines were obtained from Horizon Genomics GmbH, Vienna. In particular, a single eIF1B knockout clone and two eIF1 knockout clones were acquired (i.e. an eIF1-14bp deletion knock out (eIF1KO cl. 1) and eIF1-265bp insertion knock out (eIF1KO cl. 2)). For details, see Supporting File 1: Supporting Methods.
Knockdown experiments
Cells were transfected with either control si-RNA (si-Ctrl, ON-TARGETplus Non-targeting Control siRNAs: D-001810-01-05), si-RNAs targeting eIF1 (si-eIF1, SMARTpool: M-015804-01-0005) or si-RNAs targeting eIF1B (si-eIF1B, SMARTpool: M-019996-00-0005, Dharmacon, GE Healthcare Life Sciences). For details, see Supporting File 1: Supporting Methods.
Label-free shotgun proteomics
For label-free shotgun proteome analyses of HAP1 cells, three biological replicate samples of WT cells, eIF1B knockout and both eIF1 knockout clones were prepared. For the label-free shotgun proteome analyses of HCT116 cells, two biological replicate samples of si-Ctrl cells and si-eIF1 were prepared. The same sample input material was used for preparation of the ribo-seq samples (See ribosome profiling). Cells were lysed in 4 M Gu.HCl, 50 mM NH4HCO3 pH 7.9, sonicated and centrifuged for 30 min at 3500 g (4°C), followed by a precipitation of proteins from the supernatant with 4× volumes of –20°C acetone for 2 h at –20°C. Precipitated proteins were digested overnight at 37°C using mass spectrometry grade trypsin/Lys-C. Solid phase extraction of peptides was performed using C18 reversed phase sorbent containing 100μL pipette tips (Piece C18 tips—Thermo Scientific) according to the manufacturer's instructions. For details, see Supporting File 1: Supporting Methods.
Ribosome profiling
For ribosome profiling, cells were incubated with either 50 μM lactimidomycin (LTM) (35,36) or 100 μg/ml cycloheximide (CHX) (Sigma, USA). Cells were lysed in ice-cold lysis buffer (10 mM Tris–HCl, pH 7.4, 5 mM MgCl2, 100 mM KCl, 1% Triton X-100, 2 mM dithiothreitol (DTT), 100 μg/ml CHX, 1 × complete and EDTA-free protease inhibitor cocktail (Roche)) (37) and passed through QIAshredder spin columns (Qiagen). The flow-through was clarified and subjected to RNase I (Thermo Fisher Scientific Inc.) digestion. Subsequent steps were performed as described previously (38) with minor adjustments. The resulting ribosome profiling libraries were sequenced on a NextSeq 500 instrument (Illumina) to yield 75 bp single-end reads. For details, see Supporting File 1: Supporting Methods.
Ribosome profiling data analysis
CHX and LTM ribo-seq data were analysed in parallel using the PROTEOFORMER pipeline (39). Reads were initially mapped onto small nuclear RNA, tRNA and rRNA sequences to remove contaminant sequences. The remaining reads were then mapped onto the human GRCh38 reference genome (Ensembl annotation bundle 82) using STAR 2.4.0i allowing only unique mapping with a maximum of two mismatches. The TIS calling algorithm was applied with default PROTEOFORMER settings, followed by ORF delineation, as described previously (7,39). Protein sequence database of in-silico translated Ensembl annotated CDS sequences (‘aTIS database’) was generated from PROTEOFORMER output disregarding any single nucleotide polymorphisms (SNPs) detected. For differential expression analysis, the most probable protein-coding transcript per gene was selected and only uniquely mapped CHX reads were counted in the annotated CDS and u(-o)ORF regions. To generate the custom protein library (‘custom database’) that contains both ribo-seq predicted and Ensembl annotated ORFs, ribo-seq data was mapped to the genome as described above, except that multiple mapping reads (up to 16 genomic loci) were allowed and SNP detection was enabled. For details, see Supporting File 1: Supporting Methods.
mRNA sequencing and data analysis
RNA was isolated with TRIzol reagent (Invitrogen, Thermo Fisher Scientific Inc.) according to manufacturer's instructions. RNA quality was assessed by Agilent Bioanalyzer RNA 600 Nano Kit and RIN values >9 were accepted. Random fragmentation, cDNA synthesis and library generation were performed according to TruSeq Stranded Total RNA Sample Preparation protocol (Illumina). Libraries were subjected to sequencing, mapped onto the human reference genome and the resulting unique reads were counted across annotated protein-coding transcripts. For details, see Supporting File 1: Supporting Methods.
Differential expression analysis at the transcript level
Differential expression analysis was performed according to Andreev et al. (9). mRNAs, CDSs and uORFs exceeding the minimal read count thresholds were considered. Translation efficiencies (TE) were calculated dividing the CDS or uORF read counts by their corresponding mRNA read counts. Fold changes in TE, mRNA, CDS and uORF expression between si-eIF1 and si-Ctrl condition were subjected to log 2 and Z-score transformation, followed by differential expression analysis at a 1% significance level (absolute Z-score value ≥ 2.58). Enrichment analysis (40) of annotation terms was performed with a corrected P-value threshold of 0.02. For details, see Supporting File 1: Supporting Methods.
LC–MS/MS analysis of label-free proteomes
Samples were analyzed by LC-MS/MS using an UltiMate 3000 RSLC nano HPLC (Dionex) in-line connected to a Q-Exactive HF mass spectrometer (Thermo Fisher Scientific Inc.). Spectra identification was performed with MaxQuant (version 1.5.3.30) using the Andromeda search engine (41) with false discovery rate (FDR) set at 1% on peptide and protein level. Spectra were searched against the ‘aTIS database’ or ‘custom database’. Proteins were quantified by the MaxLFQ algorithm integrated in the MaxQuant software (42). For details, see Supporting File 1: Supporting Methods.
Data analysis of label-free shotgun proteomics
Data analysis was performed with the Perseus software (43) (version 1.5.3.0). Following ‘aTIS database’ search, a multiple-sample ANOVA test was applied with S0 parameter set to 0.1 and P-value threshold of 0.01 (for HAP1 experiment) or 0.05 (for HCT116 experiment) enabling for a differential protein expression analysis. Enrichment analysis (40) of annotation terms was performed with a corrected P-value threshold of 0.02. In case of the HCT116 label-free shotgun proteomics data, the peptide identifications obtained from the ‘custom database’ search were visualized on the human GRCh38 reference genome (Ensembl annotation bundle 82) as a BED track (Supporting File 2). For details, see Supporting File 1: Supporting Methods.
For calculation of sequence conservation and RNA secondary structure analysis, RT-qPCR, cell viability and Western blot assays as well as ATP and mitochondrial membrane potential measurement, see Supporting File 1: Supporting Methods. For oligonucleotide sequences, see Supplementary Table S4.
RESULTS
Integrative OMICS to map the translational landscape
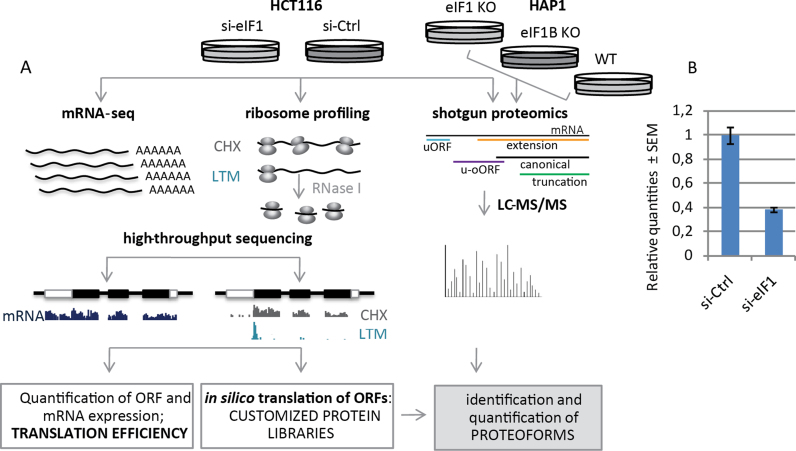
First, to obtain a comprehensive view of the cellular response upon reduced eIF1 levels, mRNA-seq, ribo-seq and label-free steady-state proteome analyses were performed upon siRNA mediated knockdown of eIF1 (si-eIF1) in the near-diploid and chromosomally stable HCT116 cell line (44) and compared to control (si-Ctrl) conditions (Figure 1A). qPCR experiments indicated a 62% eIF1 knockdown efficiency at the transcript level (Figure 1B), whereas shotgun proteomics data indicated a ∼50% knockdown efficiency at the protein level. In ribo-seq experiments we made use of lactimidomycin (LTM) (45) and cycloheximide (CHX) as translation inhibitors, enabling the study of translation initiation and elongation respectively. Subsequently, the PROTEOFORMER (39) pipeline was applied to map ribo-seq reads to the human genome, identify translation initiation sites (TIS) and assess translation efficiencies of specific ORFs across the genomic sequence.
Figure 1.
Experimental setup. (A) Integrative OMICS analysis of si-RNA mediated knockdown of eIF1 in HCT116 cells and shotgun proteomics data of eIF1 and eIF1B knockout HAP1 cell lines. (B) qPCR experiments indicated a 62% eIF1 knockdown efficiency at the transcript level, whereas shotgun proteomics experiments (A) indicated a 50% knockdown efficiency at the protein level.
To gain additional insights into the biological role of eIF1 and its paralog gene eIF1B, we complemented the studies performed upon eIF1 knockdown with label-free shotgun proteomics data from eIF1 knockout (eIF1KO) and eIF1B knockout (eIF1BKO) HAP1 cell lines (Figure 1A).
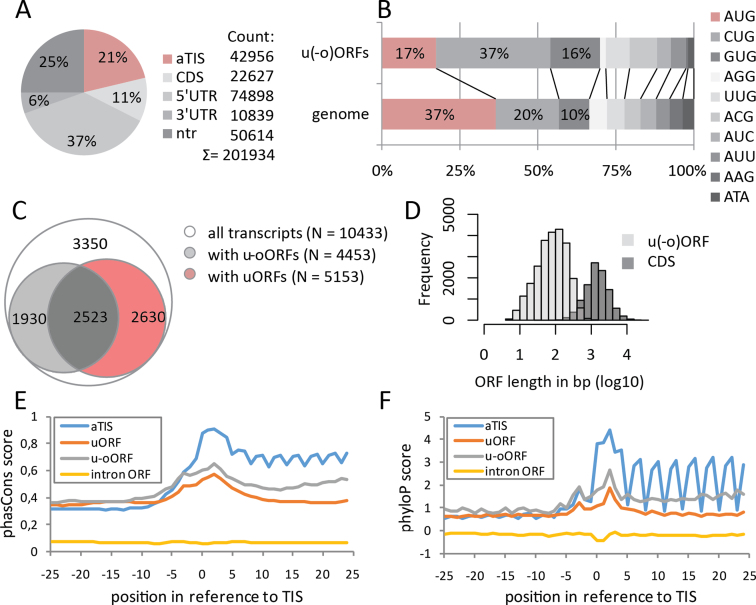
Overall, using ribo-seq in HCT116 cells we identified potential TIS at 201 934 unique genomic positions (Figure 2A). Of these, only 37% represented AUG codons (Figure 2B), an observation in line with previous reports (5). Given the plethora of potential translation start sites and annotated splice variants, an integrative OMICS analysis was facilitated by rationalized filtering of transcripts without any evidence of transcript-specific translation or poorly expressed transcripts (46), leading to the selection of a single (most) representative transcript per translated gene (see Supporting File 1: Supporting Methods). Using this approach in HCT116 cells, we quantified the expression changes of 4197 proteins and 10 433 transcripts with actively translated CDS. Additionally, in 7083 (68%) of these transcripts, translational activity in the 5΄ leader sequence was detected, pointing to a total of 15 894 uORFs and 8554 u-oORFs (Figure 2C). As expected, ORFs located in 5΄ leader sequences were generally 10- to 20-fold shorter, with a median length of 84 nucleotides (nt) for the u(-o)ORFs compared to 1440 nt for CDSs (Figure 2D). Next, we compared evolutionary conservation patterns of annotated, upstream and intronic AUG-initiated ORFs (47), which revealed that u(-o)ORFs have overall intermediate nucleotide conservation scores, nonetheless with clearly elevated scores around the start codon (47) (phastCons and phyloP analysis, Figure 2E and F) and –3 nucleotide position (phyloP analysis, Figure 2F), corresponding to the Kozak consensus sequence hallmarks (48). Despite the increased nucleotide conservation around upstream start sites (uTIS), u(-o)ORF sequence conservation was much lower as compared to CDS, underscoring their potential regulatory rather than peptide- or protein-coding roles (47).
Figure 2.
Overview of identified TIS and TIS-delineated ORFs. (A) Initiation sites were called at 201 934 unique genomic positions corresponding to aTIS, CDS, 5΄UTR, 3΄UTR regions or non-translated transcripts (ntr). (B) Percentage of TIS codon usage in the entire genome compared to upstream (uTIS) codon usage in 10433 selected transcripts. Frequencies of at least 10% were indicated. (C) Transcripts with (at least one) upstream (uORF) and/or upstream-overlapping ORFs (u-oORF) in their 5΄ leader sequences. (D) Comparison of observed u(-o)ORF and CDS length distribution. Nucleotide conservation contexts of AUG TIS (–25 to +25) were analyzed for several ORF categories using phastCons (E) and phyloP (F) scores. Each category comprises at least 500 AUG-initiated ORFs.
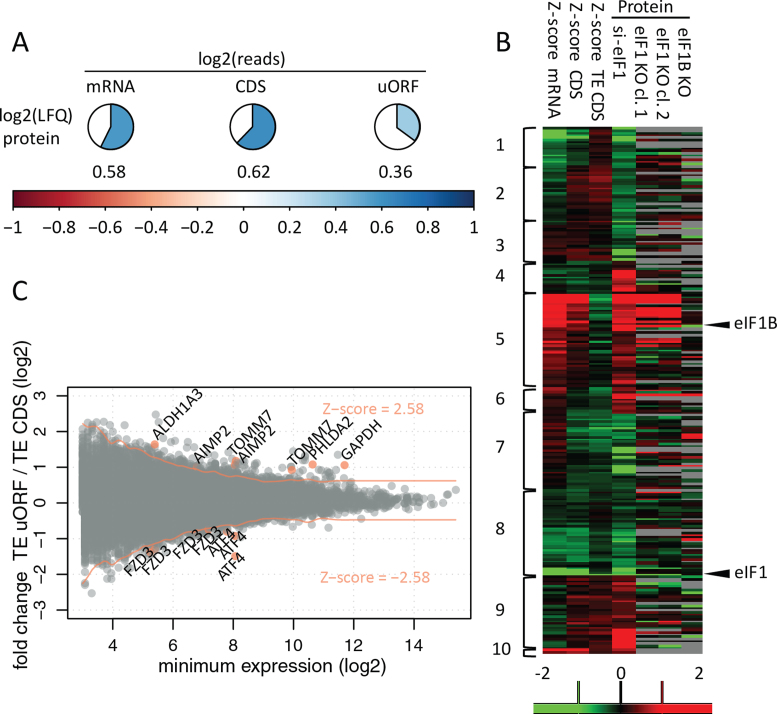
We then compared the steady-state levels of translation, mRNA and protein (Figure 3A). In line with previous reports in human and mouse (1,49,50), we detected a moderate to good correlation of protein and mRNA levels (Pearson coefficient r = 0.58). Protein synthesis rate was believed to largely explain the remaining variability (1,3,49), especially in the case of non-perturbed systems, and indeed, we have observed a slightly improved correlation of ribo-seq readout of translation to protein levels (r = 0.62), in line with previous reports (1,12). Our results further hint to the importance of downstream processes such as protein turnover in establishing a proteome level equilibrium. Moreover, inherent limitations of the applied technologies, namely the fact that ribo-seq captures a ‘snap-shot’ of translation while proteomics captures steady-state protein abundance, increase the difficulty in accounting for the potential delay in the manifestation of the translational response at the protein level (51), as may be the case in our study, since translatome and proteome samples were collected at the same time-point after knockdown. Of note however, translation of uORFs was only weakly correlated (r = 0.36) to protein changes (Figure 3A), in line with the postulated regulatory role of translationally active 5΄ leaders.
Figure 3.
Correlation and reproducibility at different OMICS levels. (A) Values represent average Pearson correlation coefficients for quantitative OMICS experiments performed in the si-Ctrl setup of HCT116 cells. LFQ; label free quantification of protein expression. (B) Identification of differentially expressed genes (P = 0.01) based on adjusted significance threshold in relation to data distribution obtained by Z-score transformation (Z-score cut-offs values of –2.58 and 2.58 corresponding to P = 0.01 are indicated with red lines). Three hundred thirty differentially regulated uORF/CDS pairs (from 245 transcripts) were identified at a 0.01 confidence level. Gene names were indicated for selected examples, also visualized in Figure 4, Supplementary Figures S2 and S3. (C). Significantly regulated proteins in the HCT116 shotgun proteomics experiment were clustered alongside their corresponding mRNA, CDS and TE CDS Z-score fold changes. Additionally, HAP1 LFQ data—not used for clustering—was visualized. Columns with protein data represent log2 fold change between average LFQ intensities when comparing knockdown/knockout versus control conditions. The scale ranges from red (upregulation in knockdown/knockout) to green (downregulation in knockdown/knockout) Clusters depict different modes of protein regulation: cluster 1—transcript downregulation; cluster 2—transcript downregulation not reflected by CDS translation; clusters 3 and 4—downregulated and upregulated proteins deviating from the expected transcript levels and translational regulation (31 proteins), respectively; cluster 5—transcript upregulation; cluster 6—transcript upregulation not reflected by CDS translation; cluster 7—translational downregulation; cluster 8—mixed (transcript and translational) downregulation; cluster 9—translational upregulation; cluster 10—mixed (transcript and translational) upregulation.
An OMICS perspective on eIF1 translational control
To study expression changes in response to eIF1 deprivation at the level of transcription, and CDS and uORF translation, we calculated ratios (fold changes) of normalized read counts between the eIF1 knockdown (si-eIF1) and control (si-Ctrl) conditions. Translation efficiency (TE) of the CDS or uORF was estimated by dividing normalized ribo-seq reads with normalized mRNA read count data of the corresponding transcript. Fold changes in TE were further calculated between both conditions. Finally, to determine the regulatory effect of uORFs on their downstream CDS, a ratio between uORF TE fold change and CDS TE fold change was calculated. In order to identify ORFs and transcripts significantly affected by differential eIF1 expression, we applied a Z-scoring strategy with adjustment for expression, as described by Andreev et al. (9). Using a threshold P-value of 0.01 corresponding to an absolute Z-score value ≥ 2.58 we detected significant deviations in response to eIF1 knockdown for 159 mRNAs, 125 CDSs and 291 u(-o)ORFs (Supplementary Figure S1, Supplementary Table S1). Additionally, 121 CDSs and 313 u(-o)ORFs (i.e. 81 u-oORFs and 232 uORFs) were affected at the TE level.
Protein expression data revealed 238 significantly regulated proteins identified by an ANOVA test with a threshold P-value of 0.05 (Supplementary Table S1). A systematic comparison of affected genes highlighted that the majority of changes in protein expression resulted from differential transcription or transcript stability (41%), translation (28%) or a combination thereof (18%) (Figure 3B). Interestingly, 31 proteins (13%) displayed significant changes in steady-state expression without notable changes in transcript or translation levels at the time-point of sampling, suggesting potential effects on protein turnover and/or posttranslational protein modification, although the exact mechanism and the role of eIF1 in modulating expression of these proteins remains to be determined (Figure 3B).
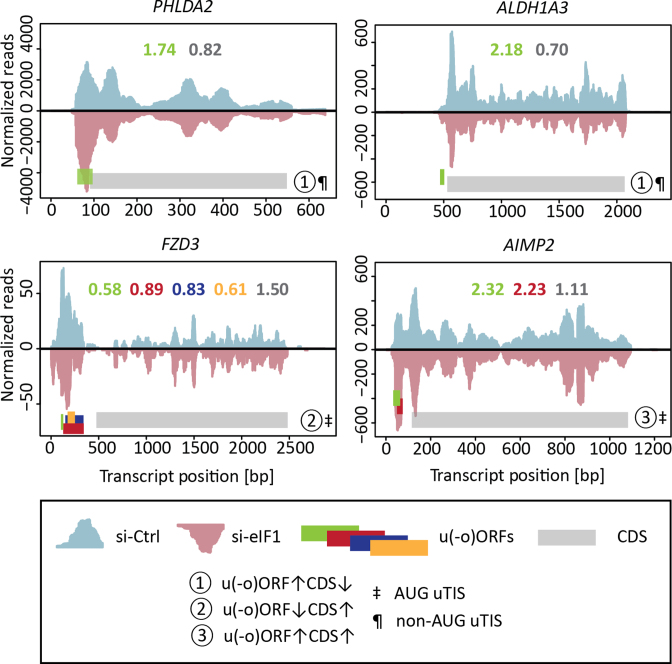
Finally, to explore the role of translated 5΄ leader sequences in gene expression changes observed upon eIF1 knockdown, we looked for evidence of differential regulation when comparing TE of transcript specific u(-o)ORF/CDS pairs. We confirmed significant differences in TE for 330 u(-o)ORF/CDS pairs, comprising 68 u-oORFs/CDS and 262 uORFs/CDS pairs, originating from 245 unique affected transcripts (Figure 3C). Genes regulated by their 5΄ leaders represented a broad spectrum of expression levels (Figure 3C). Although clustering of their corresponding expression values revealed the predominant effect of translation on expression of genes with regulatory u(-o)ORFs, in some cases translational regulation and transcript changes simultaneously contributed to the fine-tuning of gene expression (Supplementary Figure S2). Ribosome footprint distribution of individual affected genes pointed to different modes of regulation imposed by u(-o)ORFs (Figure 4, Supplementary Figure S3), including u(-o)ORF initiated at non-AUG uTIS with a possible inhibitory effect on CDS expression and AUG-initiated u(-o)ORF potentially enhancing CDS expression, among others.
Figure 4.
Ribosome density profiles of transcripts with actively translated u(-o)ORFs. Full length ribosome CHX reads of si-Ctrl and si-eIF1 samples were visualized across mRNA sequences as positive (blue) and negative (red) values, respectively. Only the 5΄ proximal part of the 3΄UTR sequence was included. Both uniquely and non-uniquely mapped reads were used for visualisation. CDS and u(-o)ORFs were marked using grey or green/red/blue/orange bars, respectively. Direction of expression changes (si-eIF1 versus si-Ctrl) and start codon of the most 5΄ proximal u(-o)ORF were indicated (see legend panel contained within figure). Additionally, profiles were quantified and si-eIF1/si-Ctrl fold change in uniquely mapped ribo-seq reads for individual ORFs were presented as numbers using a corresponding color scheme. Potential regulatory u(-o)ORFs were visualized for PHLDA2, ALDH1A3, FZD3 and AIMP2. These u(-o)ORFs contributed to significant Z-score TE uORF/TE CDS changes.
uORF features associated with eiF1 mediated translational control
Our analysis confirmed that uORFs are implicated in translational control exerted by eIF1, thereby contributing to regulation of protein expression. Therefore, we explored if certain intrinsic features of uORFs may determine their potential to enhance or repress the expression of downstream CDS.
uORF start codon
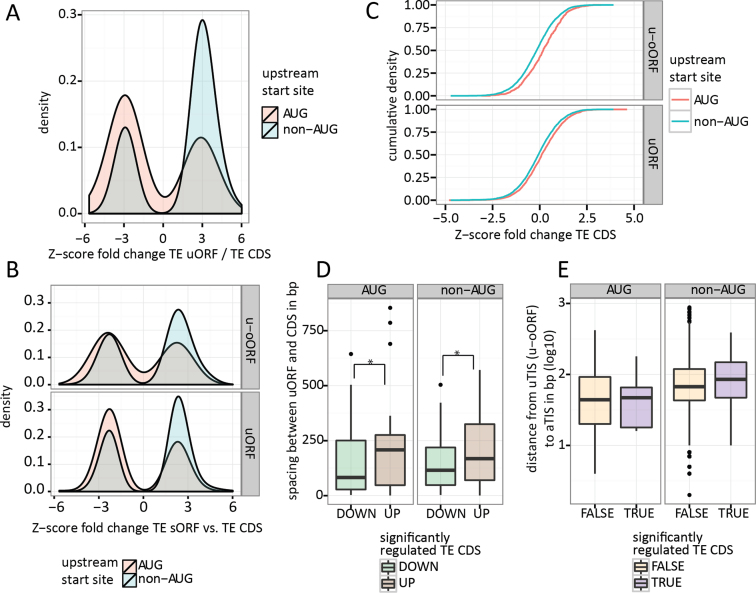
Studies reported by the Atkins group demonstrate that eIF1 levels may orchestrate the stringency of start codon selection (30,32). More specifically, overexpression of eIF1 resulted in a preference for AUG initiation. In light of these findings, we hypothesised that decreased eIF1 levels should lead to more flexibility in ribosomes initiating translation at non-AUG codons. Whereas the vast majority (99.6%) of annotated CDS detected in our study have AUG initiation sites, u(-o)ORFs display a broad spectrum of TIS codons (Figure 2B). In consequence, eIF1 knockdown, by increasing initiation (rates) at near-cognate start sites, is expected to impact the rate of leaky scanning and reinitiation, leading to altered incidences of both uTIS and aTIS initiation. Indeed, we observed a highly significant dependence between uTIS codon identity and the regulation of uORF/CDS pairs (Kruskal–Wallis test: P = 4.1e–14). This relationship was further confirmed on a subset of transcripts with a single uORF (P = 0.0014). uORFs with non-AUG start codons had frequently upregulated uORF/CDS TE ratio (Z-score TE uORF/CDS ≥ 2.58), whereas AUG-initiated uORFs were typically downregulated compared to their CDS (Z-score TE uORF/CDS ≤ –2.58; Figure 5A; χ2 test: P = 0.00035). Increased expression of non-AUG uORFs and decreased expression of AUG uORFs was also apparent when considering uORFs and u-oORFs separately (Figure 5B).
Figure 5.
Translational response to eIF1 deficiency is in part mediated by uORFs and depends on the identity of the uTIS codon and distance to CDS. (A) Lower eIF1 levels allow for translational upregulation of non-AUG uORFs and downregulation of AUG uORFs. Only significantly regulated uORF/CDS pairs were presented. (B) The relationship between uORF start codons and the expected directionality of translational regulation is maintained for both upstream and upstream-overlapping ORFs. (C) The effect of u(o-)ORF start site on downstream CDS translation. (D) Spacing between uORF stop codon and aTIS impacts CDS translation irrespective of the uTIS codon (AUG versus non-AUG). (E) Distance between u-oORF start site and aTIS has no observable impact on CDS translation. * Mann–Whitney test: P-value < 0.05.
When considering the impact of the uTIS codon identity on TE of the downstream CDS (Figure 5C), AUG uTIS codons more often associated with downregulation of uORF expression and enhanced expression of the corresponding CDS, while non-AUG-initiated uORFs displayed increased expression, thereby acting as CDS repressors. These results demonstrate that by relying on the principle of leaky scanning, eIF1 steers the stringency of start codon selection, thereby exerting translational control on protein-coding genes at a genome-wide scale. The direction (and likely also the degree) of eIF1-induced regulation is dependent on the cellular availability of the translation factor and on the nature of upstream and downstream start codons.
uORF location in reference to CDS
Sufficient spacing between the uORF and CDS is necessary to allow the occurrence of translation reinitiation in eukaryotes (52). Kozak et al. (52) illustrated how a 79 nt spacing stimulated efficient reinitiation in mammalian cells by enabling the 40S ribosome subunit to reload with translation initiation factors and initiator methionine tRNA, while resuming the scanning of mRNA. In line, our data shows that upregulated TE CDS coincided with higher distance between the uORF stop codon and the aTIS compared to a lower distance observed when TE CDS was repressed (Mann–Whitney test: P = 0.00091, Figure 5D). These results confirm that the relationship between repressiveness of uORFs and their distance to CDSs (24) are relevant parameters upon eIF1 deprivation and suggest that more distant uORFs may act as enhancers of CDS translation (via the reinitiation mechanism) whilst more proximal uORFs may act as repressors of CDS translation (reflecting perturbed rates of leaky scanning upon eIF1 deprivation). Interestingly, shorter uORFs were typically present in front of translationally upregulated CDSs compared to longer uORFs associated with downregulated CDS (Supplementary Figure S4A, Mann-Whitney test P = 0.0012 at Z-score TE CDS significance levels of 0.05). On the contrary to uORFs, and in line with previous reports (25), we did not observe any relationship between the u-oORF TIS/aTIS distance or the overlap with CDS and the significance of the fold change in TE CDS (Figure 5E, Supplementary Figure S4B).
Number of uORFs in the 5΄ leader
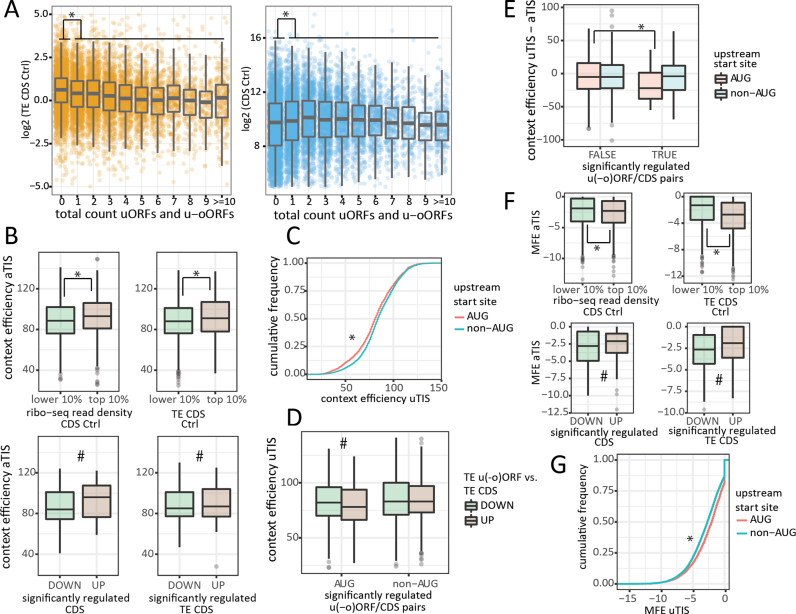
Next we investigated if the number of u(-o)ORFs is relevant for CDS expression levels. Higher ribo-seq coverage of CDSs and, by extension, higher translational signals, coincided with an increasing number of detected u(-o)ORFs (Figure 6A; Kruskal–Wallis test shows a significant relationship between log2 CDS ribo-seq read counts and u(-o)ORF count, P = 6.34e–07; Mann–Whitney test shows decreased CDS read counts in transcripts with no u(-o)ORFs, P = 7.73e–09). Despite this potential bias due to sequencing coverage, translation efficiency was dependent on the number of u(-o)ORFs (Kruskal Wallis: P < 2.2e–16), and TE rates were generally higher for CDSs without u(-o)ORFs (Mann–Whitney: P < 2.2e–16 Figure 6A). Interestingly, CDSs with one or more u(-o)ORFs were found to be significantly more repressed upon eIF1 knockdown (lower Z-score of TE CDS fold change; Mann–Whitney: P = 0.0007).
Figure 6.
5΄ leader, uORF and uTIS features relevant for translational regulation. (A) Translation efficiency of CDS is decreased in the presence of uORF(s) (left panel) despite the fact that highly expressed CDS tend to have more uORFs (right panel). (B) Higher aTIS context efficiencies can be expected for well expressed (upper left panel) and efficiently translated (upper right panel) CDSs at the steady-state level (si-Ctrl conditions). Increased CDS expression and translation efficiency upon eIF1 knockdown is also correlated with improved aTIS context efficiency (lower right and left panel, respectively). (C) Cumulative frequency plot demonstrating that non-AUG uTIS are embedded in more optimal nucleotide context compared to AUG uTIS. (D) Relaxed stringency of start codon selection by eIF1 allowed for significant regulation of u(-o) ORFs with suboptimal AUG uTIS context. (E) The impact of differences in AUG uTIS and aTIS context scores on regulation of single u(-o)ORF/CDS pairs. (F) Higher secondary structure (lower minimum free energy (MFE) values) around aTIS was observed for well expressed (upper left panel) and efficiently translated (upper right panel) CDS (si-Ctrl conditions). Conversely, less RNA structure (higher MFE) correlated with increased CDS expression and translation efficiency upon eIF1 knockdown (lower right and left panel, respectively). (G) non-AUG uTIS were embedded in more structured context compared to AUG uTIS. * Mann–Whitney test: P-value < 0.05; # Mann–Whitney test: P-value < 0.05 when Z-score significance threshold of 0.05 was considered.
Nucleotide context of TIS
Next, we sought to determine whether the primary nucleotide context surrounding uTIS codons was implicated in steering eIF1 regulation of u(-o)ORFs. Therefore, we first retrieved the sequence context of all aTIS and uTIS called by our PROTEOFORMER pipeline. As described by Noderer et al. (53), we assigned the entire nucleotide context with a single numeric score based on TIS context efficiencies experimentally determined in mammalian cells using FACS-seq. The best scoring context in the aTIS set (GCGAGTXXXGC, efficiency = 149) corresponded to the translation efficiency enhancing motif described by Noderer et al., whereas the Kozak consensus sequence GCC(A/G)CCXXXG (48) was assigned with an efficiency score of 122. Of note however, these were not the most frequent motifs observed in our aTIS dataset, instead GGGAAGXXXGC (score = 131) was most frequently observed (0.18%). Overall, better aTIS context scores were associated with higher baseline levels of expression and high translation efficiency of the corresponding CDS in the control sample (Mann–Whitney P ≤ 4.1e–05 for aTIS scores in 10% highest and 10% lowest expressed/translated genes; Figure 6B upper panel). Upon eIF1 downregulation, we also observed a positive relationship between aTIS context efficiency and both CDS and TE CDS fold change (Mann–Whitney: P = 0.014 and 0.0077 at 0.05 Z-score significance threshold, Figure 6B lower panel). However, aTIS context was clearly improved in the absence of u(-o)ORFs (Mann–Whitney: P = 7.3e–15), showing that 5΄ leader sequences cannot be disregarded when studying the impact of aTIS context on gene expression changes upon eIF1 deprivation. Using the scoring system established for aTIS, we assigned efficiencies to uTIS context sequences. Although uTIS identified in our dataset were clearly enriched in non-AUG codons, the nucleotide context score of these near-cognate start sites was generally much higher compared to AUG uTIS (Figure 6C, Mann–Whitney P < 2.2e–16) (54,55). Interestingly, the preference for an optimal consensus sequence was perturbed upon eIF1 knockdown, resulting in decreased uAUG context scores in the group of upregulated u(-o)ORF/CDS pairs (Figure 6D, Mann–Whitney P = 0.032 for Z-score TE uORF/CDS threshold of 0.05). When considering transcripts with a single u(-o)ORF, a decreased quality of the uTIS context sequence in relation to aTIS was especially apparent for regulated AUG uTIS compared to the non-regulated AUG uTIS group (Figure 6E, Mann–Whitney P = 0.024). To further asses the relationship between uTIS context and the direction of u(-o)ORF regulation we turned to a more simplified metric. The quality of context sequence was assumed to be ‘strong’ (indicated by –3 purine and +4 guanine relative to uTIS) or otherwise ‘weak’ (21). Using this metric, we observed that upregulated AUG uTIS had a significantly weaker context compared to downregulated AUG uTIS (Fisher's exact test P = 0.081 and P = 0.026 for Z-score TE uORF/CDS threshold 0.01 and 0.05, respectively). In contrast, such relationship was not detected for non-AUG uTIS, results corroborated by sequence logo analysis (http://weblogo.berkeley.edu, (56)) presented in Supplementary Figure S5). These results allow us to conclude that eIF1 knockdown perturbed translation initiation rates at u(-o)ORFs when their start codon was embedded in a suboptimal nucleotide context sequence. Although translation initiation rates at uTIS with a poor context was affected upon eIF1 knockdown, uTIS start codon identity (AUG versus non-AUG) seemed to be a stronger determinant of eIF1-driven start site selection.
RNA secondary structure of TIS regions
RNA secondary structure has a pivotal role in translation initiation. On one hand, RNA structures upstream of TIS, may impede the ability of ribosomes to bind and scan mRNA, consequently reducing the efficiency of initiation (8). On the other hand, start site recognition, especially in the case of suboptimal TIS, may be enhanced by a stable proximal downstream secondary structure, which temporary arrests scanning ribosomes and supresses leaky scanning (57). To determine RNA secondary structure, we calculated minimum free energy of aTIS and uTIS regions (±10 bp) (MFE, kcal/mol) using the ViennaRNA package (58). Overall, increased secondary structure at aTIS regions (low MFE values) corresponded to higher baseline expression and translation efficiency of the corresponding CDS in the control conditions (Figure 6F upper panel; Mann–Whitney P ≤ 4.1e–05). Conversely, upon eIF1 downregulation, we observed a positive relationship between aTIS MFE and TE CDS fold change (Mann–Whitney P = 0.093 and P = 3.83e–05 for Z-score TE CDS significance threshold of 0.01 and 0.05, respectively, Figure 6F lower panel), overall indicating more efficient translation initiation in the absence of secondary structures. Although eIF1 promotes ribosomal scanning, there is no evidence of eIF1 being required for translation initiation of structured mRNAs. In fact, Pestova et al. have shown that eIF1 is dispensable for ribosomal movement on 5΄ leaders containing secondary structures as long as helicases and ATP are present (59). Interestingly, we observed a significantly higher secondary structure at non-AUG compared to AUG upstream start site regions (Figure 6G, Mann–Whitney P < 2.2e–16), reminiscent of significantly better context efficiency at non-AUG uTIS compared to AUG uTIS (Figure 6C).
As convincingly shown by Chew et al. (24), individual RNA sequence features are not necessarily independent, complicating the interpretation of correlations observed in ribo-seq datasets (24). For example, in our dataset the number of u(-o)ORF in a transcript weakly correlated with aTIS and uTIS MFE (r = 0.122 P = 6.6e–35 and r = 0.081 P = 1.0e–10, respectively, Supplementary Figure S6), meaning that more u(-o)ORFs were found when RNA structure around TIS was relaxed. This observation may likely be explained by the fact that relaxed structure is correlated with increased AU base pair content and AU-rich 5΄ leader sequences are generally enriched in initiation codons (24).
Impact of eIF1 knockdown on cell metabolism and energy status
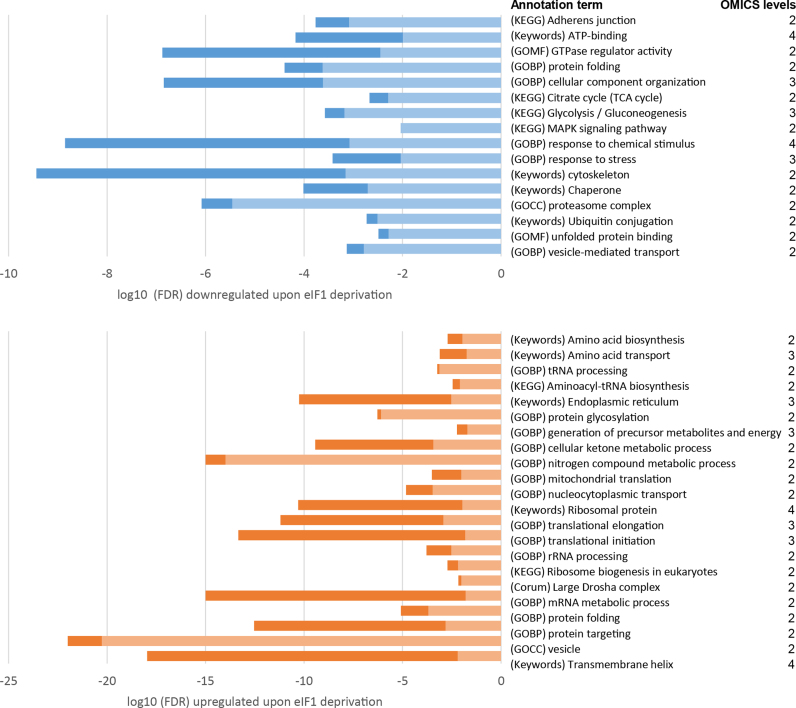
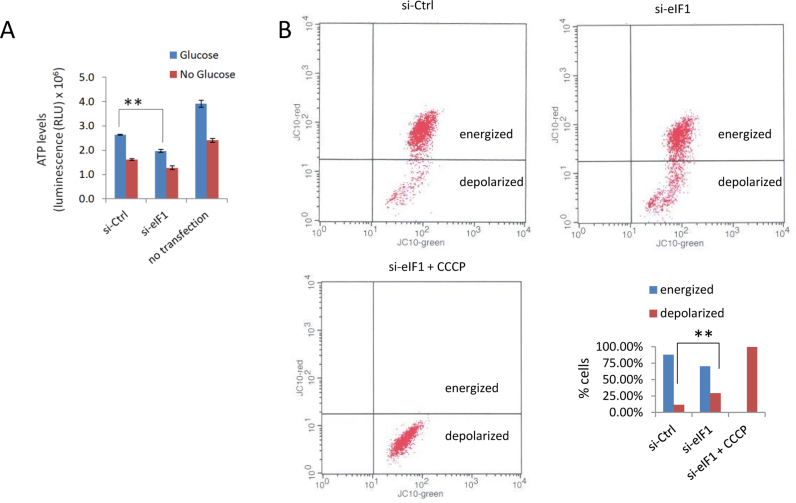
To investigate the consequences of eIF1 deficiency at the cellular level, we analysed changes in protein expression upon si-eIF1 treatment in HCT116 cells. Additionally, we validated our findings with label-free shotgun proteomics data from two independent eIF1 knockout (eIF1KO) HAP1 cell lines, which have shown overall good agreement with si-eIF1 results (Figure 3C, clusters 5 and 8). Combined annotation enrichment analysis of ribo-seq and proteomics data (Figure 7, Supplementary Table S2) revealed a decreased translation efficiency and, concomitantly, lowered expression of genes involved in glycolysis/gluconeogenesis (TE CDS FDR = 0.00027, LFQ si-eIF1 FDR = 0.00044, LFQ eIF1KO FDR = 0.00066) and in the TCA cycle (TE CDS FDR = 0.00051, LFQ eIF1KO FDR = 0.0021). In light of these findings, we decided to measure cellular ATP levels upon eIF1 knockdown (Figure 8A). We confirmed that reduced expression of glycolytic genes was accompanied by decreased cellular ATP levels, hinting to a more general impairment of energy metabolism induced by eIF1 deficiency. Further analysis of gene subsets affected by translationally active 5΄ leaders (significant Z-score TE uORF/CDS) against a background of all quantified genes was performed using GOrilla (Gene Ontology enrichment analysis and visualization tool) (60). This analysis revealed translational downregulation of mitochondrial outer membrane translocase complex components (including TOMM7, 20 and 70A; P = 0.000027). To test mitochondrial activity upon si-eIF1 treatment, we stained HCT116 cells with JC-10, a dye useful for determining mitochondrial membrane potential by flow cytometry. CCCP pre-treatment was performed as a positive control of absolute membrane depolarization. Overall, our results indicated, that si-eIF1 treated cells may suffer from decreased mitochondrial activity (Figure 8B).
Figure 7.
Gene annotation term enrichment analysis. Selected terms significantly associated with downregulated genes (upper panel) and upregulated genes (lower panel) upon eIF1 knockdown were shown. Bars represent log10 Benjamini–Hochberg false discovery rate (FDR) range detected across OMICS levels, using darker color for minimal FDR and lighter color for maximal FDR. Different OMICS levels were considered: Z-score CDS, Z-score TE CDS, Z-score mRNA, protein fold change or log2(LFQ si-eIF1/LFQ si-Ctrl) for HCT116 cells and log2(LFQ eIF1 KO/LFQ WT) for HAP1 cells.
Figure 8.
Energy status upon eIF1 deprivation. (A) Chemiluminescent readout of cellular ATP levels. Glucose deprivation was additionally performed as a positive control for ATP level reduction. (B) Measurement of mitochondrial membrane potential by FACS. **t test significance P < 0.01.
Similar GO-based analysis of up-regulated genes pointed to the increased expression of ribosomal proteins and thus increased ribosomal biogenesis, enhanced aminoacyl-tRNA synthesis and amino acid transport throughout all OMICS levels. More specifically, ribosome biogenesis (CDS FDR = 0.0065, TE CDS FDR = 0.0019), ribosomal protein (CDS FDR = 0.011, mRNA FDR = 0.00006, LFQ si-eIF1 FDR = 5.08e–11), aminoacyl tRNA biosynthetic process (mRNA FDR = 0.0083, CDS FDR = 0.0035, LFQ si-eIF1 FDR = 0.0025, LFQ eIF1KO FDR = 0.0019) and amino acid transport (mRNA FDR = 0.017, CDS FDR = 0.00077, LFQ eIF1KO FDR = 0.0073) were significantly upregulated terms (Figure 7, Supplementary Table S2). Ingenuity Pathway Analysis (IPA) of ribo-seq data (Z-score fold change CDS) further identified a causative relationship between the transcription factor ATF4 and the upregulation of processes related to amino acid metabolism (Figure 9A). In line, our data point to the increased expression of ATF4, which was linked to the translational downregulation of AUG uORFs in the 5΄ leader of ATF4 upon eIF1 knockdown (Z-score TE uORF/CDS ≤ –2.58).
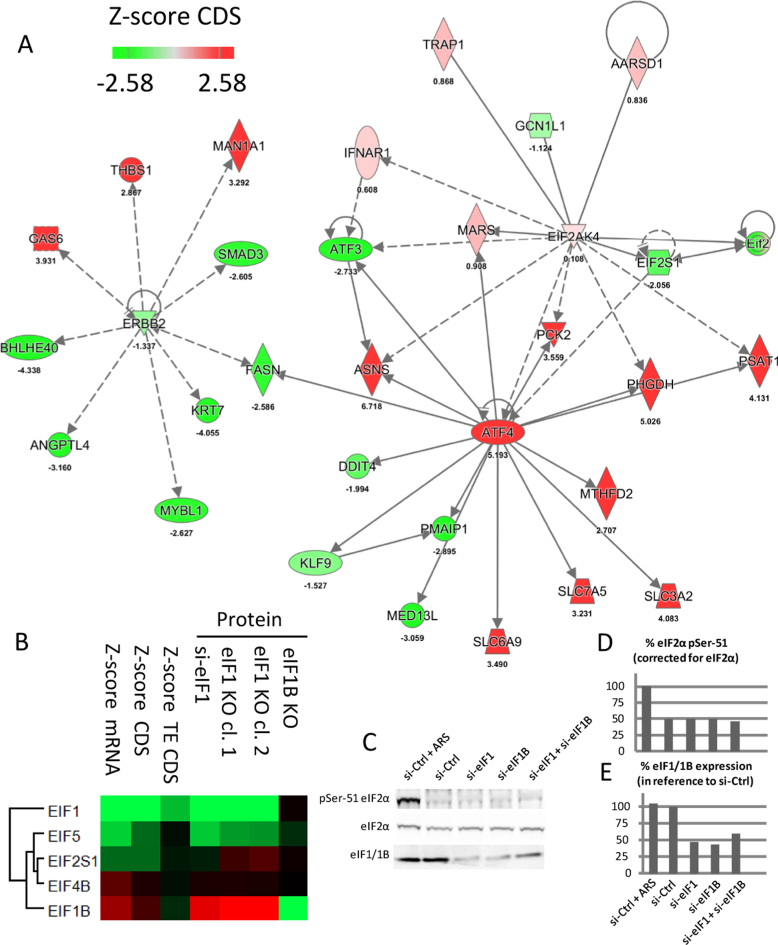
Figure 9.
OMICS analysis highlights the effect of eIF1 deficiency on the ATF4 interaction network and suggests a widespread regulation of the translational machinery. (A) Upregulation of ATF4 expression coincided with increased expression of its downstream targets involved in amino acid synthesis (ASNS Asparagine Synthetase (Glutamine-Hydrolyzing), PHGDH Phosphoglycerate Dehydrogenase involved in the early steps of L-serine synthesis and PSAT1 Phosphoserine Aminotransferase 1 that participates in glycine, serine and threonine metabolism) next to genes involved in amino acid transport (SLC3A2, SLC6A9 and SLC7A5 Solute Carrier Family members that transport L-type amino acids). Additionally, the expression pattern of ATF4 upstream regulators, such as EIF2S1, EIF2AK1 and GCN1L1, were visualized. IPA analysis indicated a common node (FASN Fatty Acid Synthase) between ATF4 and ERBB2 interaction network, which pointed to a possible impairment of adherent junction formation (due to downregulated KRT7 Keratin 7, SMAD Family Member 3 and ERBB2 Receptor Tyrosine Kinase 2). In contrast to the ATF4 network, differential regulation of proteins implicated in the formation of adherent junctions could not be confirmed at the steady-state protein level. (B) eIF proteins significantly regulated upon eIF1 knockdown were also found to be significantly regulated in eIF1KO cells. Differential expression values were subjected to hierarchical clustering using normalized protein expression fold changes next to displaying TE CDS, CDS and mRNA Z-score fold changes. (C) eIF1 knockdown does not affect eIF2α phosphorylation. Western blot analysis of phospho-eIF2α, total eIF2α and eIF1/1B were performed in HCT116 cells upon siRNA-mediated knockdown of eIF1 and/or knockdown of its paralog gene eIF1B. Control cells were transfected with a non-targeting siRNA (si-Ctrl) and treated with 500 μM arsenite (ARS) for 1 h to induce eIF2α phosphorylation (10). Quantification of phosho-eIF2α and eIF1/1B is presented in panel. (D and E), respectively. Of note, the primary antibody against eIF1 also recognizes the highly similar in sequence eIF1B. Discrimination of eIF1 from eIF1B on Western blot is additionally hindered due to their similar molecular weight (difference of 0.1 kDa).
Previous studies have shown that deviations in eIF1 expression levels occur in physiological conditions and that eIF1 expression is responsive to nutrient availability. Andreev et al. reported a two-fold upregulation of eIF1 in human PC12 cells during glucose and oxygen deprivation by means of ribo-seq (10). These results are corroborated by publicly available mRNA expression array data in HCT116 cells (61,62). To restore the stringency of start codon selection during glucose and oxygen deficiency, over two-fold upregulation of eIF1 was accompanied by decreased eIF5 levels, pushing the equilibrium of translation regulation the opposite direction as compared to what is observed upon eIF1 knockdown. Therefore, unsurprisingly, in support of the physiological relevance of eIF1 knockdown conditions used in our study, we found a high negative correlation between 26 genes significantly regulated during eIF1 knockdown and during nutritional stress reported by Andreev et al. (3) (Pearson correlation coefficient of –0.57 and Spearman correlation coefficient of –0.51), including 18 genes which displayed opposite regulation (10) (Supplementary Figure S7).
Equilibrium of eIFs
Interestingly, knockdown of eIF1 elicited a synergic response from other eIFs. Our proteomics data suggested a significant regulation of four other eukaryotic initiation factors (or subunits thereof) upon eIF1 knockdown, including EIF1B, EIF2S1 (encoding eIF2α), EIF5 and EIF4B (Figure 9B). Previous reports indicate a major role of eIF2α phosphorylation by upstream kinases in response to stress stimuli (8,15). eIF2α phosphorylation was shown to affect translation of many genes, including ATF4 as the flagship example (8,15). To test whether eIF2 contributes to the translational response observed in our study, we measured eIF2α phosphorylation levels upon eIF1 and/or eIF1B knockdown (Figure 9C and D). Overall no increase in the basal phosho-eIF2α levels could be observed upon eIF1 deprivation. To put our findings in a broader context, we also investigated the expression of EIF2S1 (encoding eIF2α subunit) and EIF2AK4 (encoding a kinase that phosphorylates eIF2α in response to amino-acid deprivation (15)). Although eIF1 knockdown coincided with a 10% decreased EIF2S1 protein expression, we have observed no significant expression change in case of EIF2AK4, while GCN1L1 encoding a positive activator of the EIF2AK4 protein kinase activity, was clearly downregulated (Figure 9A). In contrast however, and despite the overall good agreement between the knockdown and knockout experiments, both eIF1KO cell lines showed increased eIF2α expression (Figure 9B).
In both eIF1 knockdown and knockout conditions, we observed a significant downregulation of eIF5, while eIF4B and eIF1B were upregulated. eIF5 and eIF1 were previously reported to have opposing effects regarding the stringency of start codon selection (32). eIF5 expression is regulated by uORFs with poor context AUG TIS and therefore expected to decrease when translational initiation equilibrium is shifted towards more flexible start codon selection (32). In consequence, and in line with our results, the feedback loop discovered between eIF1 and eIF5 (32) leads to a decreased eIF5 expression upon eIF1 knockdown. eIF1B, on the other hand, is an eIF1 paralog gene (i.e. eIF1 and eIF1B share 92% sequence identity, Supplementary Figure S8) whose role in translation initiation remains to be determined. Viewing the fact that both eIF1 and eIF1B were here proven to be non-essential genes in HAP1 cells (both produced viable knockout cell lines), certain functional redundancy may be expected between these proteins. In consequence, eIF1B upregulation could thus possibly (in part) counteract eIF1 deficiency. To test if eIF1B becomes indispensable for cell growth upon eIF1 deprivation, we performed a double knockdown of eIF1 and eIF1B in both WT and eIF1BKO HAP1 cells. We followed the growth of cells by cell counting and total protein concentration measurements in the corresponding cell lysates at several time points post-transfection. However, no notable differences in cell proliferation could observed when knocking down eIF1 and eIF1B in WT or eIF1B KO HAP-1 cells (Supplementary Figure S9). Analogous experiments were performed in HCT116 cells, also showing no apparent effect of eIF1B and eIF1 double knockdown on cell proliferation (Supplementary Figure S10, Figure 9E).
DISCUSSION
By combining tailored proteomic strategies with next generation sequencing (11,12,63) we aimed at identifying the biological targets of translation control exerted by eIF1. Integrative OMICS studies face the problem of limited correlation observed between protein and mRNA levels. We improved on this aspect by relying on the ribosome-profiling readout for translation. We however detected 31 genes with significant changes at the protein level, independent of mRNA expression and translation efficiency. Further, in numerous cases, proteins levels only moderately corresponded to their synthesis rates (Figure 3C, clusters 2 and 6). There may be many reasons as to why such discrepancies are observed. First of all, protein synthesis is delayed compared to transcription, which reduces the overall correlation between mRNA and protein fluctuations (51), especially when sampling RNA and protein at the same time point after knockdown. Additionally, ribo-seq provides a snapshot of translational engagement of ribosomes, which may correspond to a transient state, while the magnitude of expression changes might be insufficient to impact on steady-state protein levels. Finally, compensatory effects might be at play.
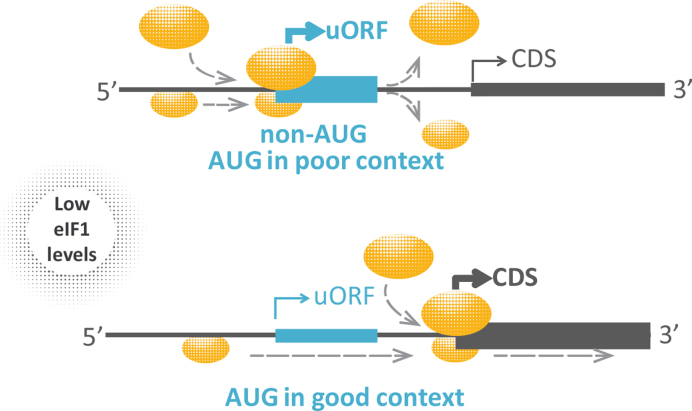
Using complementary high-throughput technologies, we here confirmed that eIF1 levels determine the stringency of start codon selection at the genome wide scale and thereby orchestrate the rates of leaky ribosomal scanning and uORF translation. More specifically, low eIF1 levels promote translation initiation at near-cognate codons and start sites embedded in a suboptimal nucleotide context (Figure 10). The initiation context was previously also found to determine aTIS versus downstream TIS (dTIS) selection (11). Although our data suggests, that the direction (and likely also the degree) of eIF1-induced regulation is dependent on the cellular availability of the translation factor and on the nature of upstream and downstream start codons, eIF1 modification status (33), not monitored in this study, is another potential factor involved.
Figure 10.
Translation initiation upon eIF1 deficiency. Our data suggest that eIF1 knockdown reduces the stringency of start codon selection and preference for an optimal nucleotide context, overall leading to translational upregulation of uORFs with suboptimal start (upper panel) and decreased translation initiation rates at upstream AUG codons embedded in more optimal nucleotide context (lower panel).
Although uORFs are omnipresent throughout protein-coding transcripts, uORF mediated regulation significantly affects only a subset of genes and tends to have an overall moderate effect on absolute expression levels (average fold change in CDS expression of about ±25% in our study), a range similar to miRNA effects (25). Therefore, we may consider that the predominant role of uORFs involves the fine-tuning of proteome homeostasis, buffering the effects of stress conditions for most genes while providing the capacity for stress response to particular effectors (9). Although the expression and functionality of uORF-derived peptides is debated with only a limited number of active peptides identified and characterized so far (64), proteogenomics strategies hold promise in expanding our knowledge in this field. We searched our shotgun proteomics data against a ‘custom database’ enriched for ribo-seq delineated reading frames, leading to the identification of peptides derived from one uORF in the CYP4F11 transcript and six u-oORFs (in MFGE8, POLR2M, SAMD1, PSMG4, SLC39A13 and RSU1 transcripts) (see Supplementary Figure S3, Supplementary Table S3 and Supporting File 2 for these and other examples of novel and non-synonymous proteoforms identified in our study). Additionally, our ribo-seq data has predicted the expression of 14 short ORF-encoded polypeptides previously identified by Slavoff et al. (65) and 60 alternative ORFs with peptide evidence reported by Vanderperre et al. (66). Of note however, 10 peptides in the Slavoff et al. dataset and 118 peptides from the Vanderperre et al. study that were attributed to alternative proteoforms, belonged to either Ensembl or SwissProt annotated proteins. These discrepancies were likely due to differences between Ensembl/SwissProt and NCBI (RefSeq) annotation (versions) used as the reference database by the other studies. Despite our efforts, the great majority of uORF-derived peptides remained undetected. Next to their lower MS detectability, their low identification rate may in part, also be attributed to the recently reported mechanism of co-translational degradation of rapidly translated polypeptides (67).
Reliable quantification of uORF expression by ribo-seq may be challenging due to their short length and the bias in ribosome signal at 5΄ leader introduced by the antibiotic treatment (27). Nevertheless, potential biases are similar across all samples analysed, and thus unlikely to affect our differential expression analysis, a finding corroborated by the fact that some of the regulatory uORFs identified in our study were previously reported in studies that specifically avoided antibiotic pretreatment (9,10) (see Supplementary Figure S3). Our data analysis pipeline also minimized the impact of ribosome accumulation at start and stop sites by adjusting the region used for measuring translation (see Supporting File 1: Supporting Methods). Analysis of overlapping ORFs may be increasingly difficult. For example, 68 of differentially regulated u-oORF/CDS pairs identified in our study were characterised by shorter than average overlaps (Supplementary Figure S4C), a likely consequence of including the region shared with the CDS for calculating u-oORF expression. Although our approach may underestimate the number of regulated u-oORFs, namely u-oORFs extensively overlapping with CDS, currently precise expression measurement of highly overlapping ORFs remains challenging.
Our findings suggest the role of eIF1 and its cellular levels as a mediator for translational regulation, but also underline the high interconnectivity of the translational machinery (1,10,51). The non-essential nature of eIF1 gene in HAP1 cells was not originally anticipated (68) and the nearly 3-fold upregulation of eIF1B in eIF1KO cells suggested a possible functional overlap between these paralog genes. On the other hand, no clear translational impairment, no apparent change in eIF1 expression, the overall mildly perturbed to unperturbed proteome expression profile of eIF1BKO cells observed, and the apparent lack of combined eIF1 and eIF1B knockdown on cell viability prohibit from drawing any conclusion regarding eIF1B activity. Overall, our results do not provide direct evidence for the indispensability of eIF1B for the growth of human cells during eIF1 deprivation. However, the functional overlap between eIF1 and eIF1B is not completely precluded, especially viewing the incomplete knockdown of both eIF1 and eIF1B (Figure 9E). Besides, the possibility remains that (next to eIF1B) other eIFs may substitute eIF1 activity. Recently, density regulated protein (DENR) bearing a SUI1 domain (SUI1 is a yeast analogue of eIF1) was shown to act as a non-canonical initiation factor indispensable for proliferation and tissue growth (69). In our ribo-seq dataset, DENR expression was unaffected by eIF1 knockdown. On the other hand, the Dikstein group has recently demonstrated that eIF1 may orchestrate translation via the TISU regulatory element (translation initiator of short 5΄UTR mRNAs) found in mitochondrial genes. While eIF1 and eIF1B deprivation was shown to severely impair TISU-driven translation, the canonical translation initiation was supported even upon combined eIF1/eIF1B knockdown in HEK293T cells. The authors suggested that eIF1 depletion may be partially compensated by eIF1A, supported by the redundant activity of these factors in translation assays using in-vitro reconstituted 48S ribosome complexes and TISU-containing mRNAs (70).
While independent of eIF2α phosphorylation, the effects of eIF1 deprivation impacted a widespread regulation of the eIF network, including eIF5, eIF1B and eIF4B. Previous studies demonstrated, that deviations in eIF1 levels occur in physiological conditions in response to nutrient availability, as a two-fold upregulation of eIF1 is observed in human cells during glucose and oxygen deprivation (10,61,62). Our study also points to a link between eIF1 levels and the regulation of genes involved in energy metabolism, as energy production and amino acid demand seemed to be significantly perturbed. In particular, lower cellular ATP levels and decreased mitochondrial respiration were detected, however the impact of this phenotype needs further examination. Additionally, we found a significant negative correlation between 26 genes regulated during eIF1 knockdown and upon nutritional stress reported by Andreev et al. (3), including 18 genes which displayed opposite regulation (Supplementary Figure S7; (10)), supporting the physiological relevance of our experimental conditions. Interestingly, changes in gene expression were frequently achieved by engaging u(-o)ORFs. u(-o)ORF repressiveness (perceived as the TE u(-o)ORF/TE CDS change upon eIF1 knockdown) was increased in the case of less favourable AUG context efficiencies, near cognate initiation codons and reduced distance between uORF and CDS. Translation efficiency of CDS also depended on the number of u(-o)ORFs and increased with more favorable aTIS context and weaker secondary structure.
AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (71) partner repository with the dataset identifier PXD004980. Ribo-seq and RNA-seq sequencing data has been deposited in NCBI's Gene Expression Omnibus (72) and is accessible through GEO Series accession number GSE87328. Besides, a BED file of peptides identified in the ‘custom database’ search of the HCT116 label-free shotgun proteomics data and mapped onto the human GRCh38 reference genome (Ensembl annotation bundle 82) was provided as Supporting File 2.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Prof. Kris Gevaert for conceptual assistance, manuscript revision and financial support of this research. The authors also thank José Van Der Heyden and Ali Adiby for experimental assistance.
Author contributions. D.F. performed experiments, analyzed data, drafted and revised the manuscript. S.V. and E.N. analyzed data and revised the manuscript. V.J. performed experiments. G.M. drafted and revised the manuscript. P.V.D conceived the study, analyzed data, drafted and revised the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research Foundation—Flanders (FWO-Vlaanderen) [G.0440.10 to K.G., G.0269.13N to P.V.D.]; Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen) [121171 to D.F.]. Funding for open access charge: Research Foundation—Flanders (FWO-Vlaanderen) [G.0440.10, G.0269.13N].
Conflict of interest statement. None declared.
REFERENCES
- 1. Wiita A.P., Ziv E., Wiita P.J., Urisman A., Julien O., Burlingame A.L., Weissman J.S., Wells J.A.. Global cellular response to chemotherapy-induced apoptosis. Elife. 2013; 2:e01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maier T., Schmidt A., Guell M., Kuhner S., Gavin A.C., Aebersold R., Serrano L.. Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol. Syst. Biol. 2011; 7:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kristensen A.R., Gsponer J., Foster L.J.. Protein synthesis rate is the predominant regulator of protein expression during differentiation. Mol. Syst. Biol. 2013; 9:689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Archer S.K., Shirokikh N.E., Beilharz T.H., Preiss T.. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature. 2016; 535:570–574. [DOI] [PubMed] [Google Scholar]
- 5. Ingolia N.T., Lareau L.F., Weissman J.S.. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011; 147:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith L.M., Kelleher N.L., Consortium for Top Down P.. Proteoform: a single term describing protein complexity. Nat. Methods. 2013; 10:186–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gawron D., Gevaert K., Van Damme P.. The proteome under translational control. Proteomics. 2014; 14:2647–2662. [DOI] [PubMed] [Google Scholar]
- 8. Sonenberg N., Hinnebusch A.G.. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009; 136:731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Andreev D.E., O’Connor P.B., Fahey C., Kenny E.M., Terenin I.M., Dmitriev S.E., Cormican P., Morris D.W., Shatsky I.N., Baranov P.V.. Translation of 5΄ leaders is pervasive in genes resistant to eIF2 repression. Elife. 2015; 4:e03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Andreev D.E., O’Connor P.B., Zhdanov A.V., Dmitriev R.I., Shatsky I.N., Papkovsky D.B., Baranov P.V.. Oxygen and glucose deprivation induces widespread alterations in mRNA translation within 20 minutes. Genome Biol. 2015; 16:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Van Damme P., Gawron D., Van Criekinge W., Menschaert G.. N-terminal proteomics and ribosome profiling provide a comprehensive view of the alternative translation initiation landscape in mice and men. Mol. Cell Proteomics. 2014; 13:1245–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koch A., Gawron D., Steyaert S., Ndah E., Crappe J., De Keulenaer S., De Meester E., Ma M., Shen B., Gevaert K. et al. A proteogenomics approach integrating proteomics and ribosome profiling increases the efficiency of protein identification and enables the discovery of alternative translation start sites. Proteomics. 2014; 14:2688–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Branca R.M., Orre L.M., Johansson H.J., Granholm V., Huss M., Perez-Bercoff A., Forshed J., Kall L., Lehtio J.. HiRIEF LC-MS enables deep proteome coverage and unbiased proteogenomics. Nat. Methods. 2014; 11:59–62. [DOI] [PubMed] [Google Scholar]
- 14. Kim M.S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S. et al. A draft map of the human proteome. Nature. 2014; 509:575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wek R.C., Jiang H.Y., Anthony T.G.. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006; 34:7–11. [DOI] [PubMed] [Google Scholar]
- 16. Calkhoven C.F., Muller C., Leutz A.. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 2000; 14:1920–1932. [PMC free article] [PubMed] [Google Scholar]
- 17. Calkhoven C.F., Muller C., Martin R., Krosl G., Pietsch H., Hoang T., Leutz A.. Translational control of SCL-isoform expression in hematopoietic lineage choice. Genes Dev. 2003; 17:959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tzani I., Ivanov I.P., Andreev D.E., Dmitriev R.I., Dean K.A., Baranov P.V., Atkins J.F., Loughran G.. Systematic analysis of the PTEN 5΄ leader identifies a major AUU initiated proteoform. Open Biol. 2016; 6:150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002; 299:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vattem K.M., Wek R.C.. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Calvo S.E., Pagliarini D.J., Mootha V.K.. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barbosa C., Peixeiro I., Romao L.. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 2013; 9:e1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ingolia N.T., Brar G.A., Stern-Ginossar N., Harris M.S., Talhouarne G.J., Jackson S.E., Wills M.R., Weissman J.S.. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014; 8:1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chew G.L., Pauli A., Schier A.F.. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat. Commun. 2016; 7:11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnstone T.G., Bazzini A.A., Giraldez A.J.. Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J. 2016; 35:706–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bazzini A.A., Johnstone T.G., Christiano R., Mackowiak S.D., Obermayer B., Fleming E.S., Vejnar C.E., Lee M.T., Rajewsky N., Walther T.C. et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014; 33:981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gerashchenko M.V., Gladyshev V.N.. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 2014; 42:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mamane Y., Petroulakis E., Martineau Y., Sato T.A., Larsson O., Rajasekhar V.K., Sonenberg N.. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS One. 2007; 2:e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Passmore L.A., Schmeing T.M., Maag D., Applefield D.J., Acker M.G., Algire M.A., Lorsch J.R., Ramakrishnan V.. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007; 26:41–50. [DOI] [PubMed] [Google Scholar]
- 30. Ivanov I.P., Loughran G., Sachs M.S., Atkins J.F.. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc. Natl. Acad. Sci. U.S.A. 2010; 107:18056–18060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheung Y.N., Maag D., Mitchell S.F., Fekete C.A., Algire M.A., Takacs J.E., Shirokikh N., Pestova T., Lorsch J.R., Hinnebusch A.G.. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 2007; 21:1217–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Loughran G., Sachs M.S., Atkins J.F., Ivanov I.P.. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res. 2012; 40:2898–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zach L., Braunstein I., Stanhill A.. Stress-induced start codon fidelity regulates arsenite-inducible regulatory particle-associated protein (AIRAP) translation. J. Biol. Chem. 2014; 289:20706–20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pestova T.V., Borukhov S.I., Hellen C.U.. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998; 394:854–859. [DOI] [PubMed] [Google Scholar]
- 35. Ju J., Lim S.K., Jiang H., Seo J.W., Shen B.. Iso-migrastatin congeners from Streptomyces platensis and generation of a glutarimide polyketide library featuring the dorrigocin, lactimidomycin, migrastatin, and NK30424 scaffolds. J. Am. Chem. Soc. 2005; 127:11930–11931. [DOI] [PubMed] [Google Scholar]
- 36. Schneider-Poetsch T., Ju J., Eyler D.E., Dang Y., Bhat S., Merrick W.C., Green R., Shen B., Liu J.O.. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 2010; 6:209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guo H., Ingolia N.T., Weissman J.S., Bartel D.P.. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010; 466:835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gawron D., Ndah E., Gevaert K., Van Damme P.. Positional proteomics reveals differences in N-terminal proteoform stability. Mol. Syst. Biol. 2016; 12:858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Crappe J., Ndah E., Koch A., Steyaert S., Gawron D., De Keulenaer S., De Meester E., De Meyer T., Van Criekinge W., Van Damme P. et al. PROTEOFORMER: deep proteome coverage through ribosome profiling and MS integration. Nucleic Acids Res. 2015; 43:e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cox J., Mann M.. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics. 2012; 13(Suppl. 16):S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M.. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011; 10:1794–1805. [DOI] [PubMed] [Google Scholar]
- 42. Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M.. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteomics. 2014; 13:2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J.. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016. [DOI] [PubMed] [Google Scholar]
- 44. Passerini V., Ozeri-Galai E., de Pagter M.S., Donnelly N., Schmalbrock S., Kloosterman W.P., Kerem B., Storchova Z.. The presence of extra chromosomes leads to genomic instability. Nat Commun. 2016; 7:10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee S., Liu B., Lee S., Huang S.X., Shen B., Qian S.B.. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci U S A. 2012; 109:E2424–E2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tress M.L., Abascal F., Valencia A.. Alternative Splicing May Not Be the Key to Proteome Complexity. Trends Biochem Sci. 2016; 42:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Calviello L., Mukherjee N., Wyler E., Zauber H., Hirsekorn A., Selbach M., Landthaler M., Obermayer B., Ohler U.. Detecting actively translated open reading frames in ribosome profiling data. Nat Methods. 2016; 13:165–170. [DOI] [PubMed] [Google Scholar]
- 48. Kozak M. An analysis of 5΄-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987; 15:8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M.. Global quantification of mammalian gene expression control. Nature. 2011; 473:337–342. [DOI] [PubMed] [Google Scholar]
- 50. Nagaraj N., Wisniewski J.R., Geiger T., Cox J., Kircher M., Kelso J., Paabo S., Mann M.. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011; 7:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Robles M.S., Cox J., Mann M.. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 2014; 10:e1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987; 7:3438–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Noderer W.L., Flockhart R.J., Bhaduri A., Diaz de Arce A.J., Zhang J., Khavari P.A., Wang C.L.. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol. Syst. Biol. 2014; 10:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kochetov A.V., Ahmad S., Ivanisenko V., Volkova O.A., Kolchanov N.A., Sarai A.. uORFs, reinitiation and alternative translation start sites in human mRNAs. FEBS Lett. 2008; 582:1293–1297. [DOI] [PubMed] [Google Scholar]
- 55. Ivanov I.P., Firth A.E., Michel A.M., Atkins J.F., Baranov P.V.. Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res. 2011; 39:4220–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Crooks G.E., Hon G., Chandonia J.M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:8301–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pestova T.V., Kolupaeva V.G.. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002; 16:2906–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z.. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009; 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C.L., Haase J., Janes J., Huss J.W. 3rd et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009; 10:R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Onnis B., Fer N., Rapisarda A., Perez V.S., Melillo G.. Autocrine production of IL-11 mediates tumorigenicity in hypoxic cancer cells. J. Clin. Invest. 2013; 123:1615–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Menschaert G., Van Criekinge W., Notelaers T., Koch A., Crappe J., Gevaert K., Van Damme P.. Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol. Cell. Proteomics. 2013; 12:1780–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Andrews S.J., Rothnagel J.A.. Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 2014; 15:193–204. [DOI] [PubMed] [Google Scholar]
- 65. Slavoff S.A., Mitchell A.J., Schwaid A.G., Cabili M.N., Ma J., Levin J.Z., Karger A.D., Budnik B.A., Rinn J.L., Saghatelian A.. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 2013; 9:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vanderperre B., Lucier J.F., Bissonnette C., Motard J., Tremblay G., Vanderperre S., Wisztorski M., Salzet M., Boisvert F.M., Roucou X.. Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PLoS One. 2013; 8:e70698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ha S.W., Ju D., Hao W., Xie Y.. Rapidly translated polypeptides are preferred substrates for cotranslational protein degradation. J. Biol. Chem. 2016; 291:9827–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Blomen V.A., Majek P., Jae L.T., Bigenzahn J.W., Nieuwenhuis J., Staring J., Sacco R., van Diemen F.R., Olk N., Stukalov A. et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015; 350:1092–1096. [DOI] [PubMed] [Google Scholar]
- 69. Schleich S., Strassburger K., Janiesch P.C., Koledachkina T., Miller K.K., Haneke K., Cheng Y.S., Kuchler K., Stoecklin G., Duncan K.E. et al. DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature. 2014; 512:208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sinvani H., Haimov O., Svitkin Y., Sonenberg N., Tamarkin-Ben-Harush A., Viollet B., Dikstein R.. Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab. 2015; 21:479–492. [DOI] [PubMed] [Google Scholar]
- 71. Gevaert K., Van Damme P., Martens L., Vandekerckhove J.. Diagonal reverse-phase chromatography applications in peptide-centric proteomics: ahead of catalogue-omics. Anal. Biochem. 2005; 345:18–29. [DOI] [PubMed] [Google Scholar]
- 72. Edgar R., Domrachev M., Lash A.E.. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.