Abstract
AID/APOBEC family enzymes are best known for deaminating cytosine bases to uracil in single-stranded DNA, with characteristic sequence preferences that can produce mutational signatures in targets such as retroviral and cancer cell genomes. These deaminases have also been proposed to function in DNA demethylation via deamination of either 5-methylcytosine (mC) or TET-oxidized mC bases (ox-mCs), which include 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine. One specific family member, APOBEC3A (A3A), has been shown to readily deaminate mC, raising the prospect of broader activity on ox-mCs. To investigate this claim, we developed a novel assay that allows for parallel profiling of activity on all modified cytosines. Our steady-state kinetic analysis reveals that A3A discriminates against all ox-mCs by >3700-fold, arguing that ox-mC deamination does not contribute substantially to demethylation. A3A is, by contrast, highly proficient at C/mC deamination. Under conditions of excess enzyme, C/mC bases can be deaminated to completion in long DNA segments, regardless of sequence context. Interestingly, under limiting A3A, the sequence preferences observed with targeting unmodified cytosine are further exaggerated when deaminating mC. Our study informs how methylation, oxidation, and deamination can interplay in the genome and suggests A3A's potential utility as a biotechnological tool to discriminate between cytosine modification states.
INTRODUCTION
Members of the AID/APOBEC family of cytosine deaminases are nucleic acid editing enzymes that function in both adaptive and innate immune contexts (1). Activation-induced deaminase (AID), a founding member of the family, targets host genome immunoglobulin loci to initiate antibody somatic hypermutation and class switching (2). The APOBEC3 subfamily targets foreign genomes, including retroviruses and retrotransposons, rendering them unable to replicate or targeting them for degradation (3). Each deaminase family member has distinctive sequence preferences and therefore leaves a different mutational signature (4). Nonetheless, the canonical reaction for AID and the APOBEC3 enzymes in these settings is the same: deamination of the 4-position of the cytosine base in single-stranded DNA (ssDNA), forming uracil.
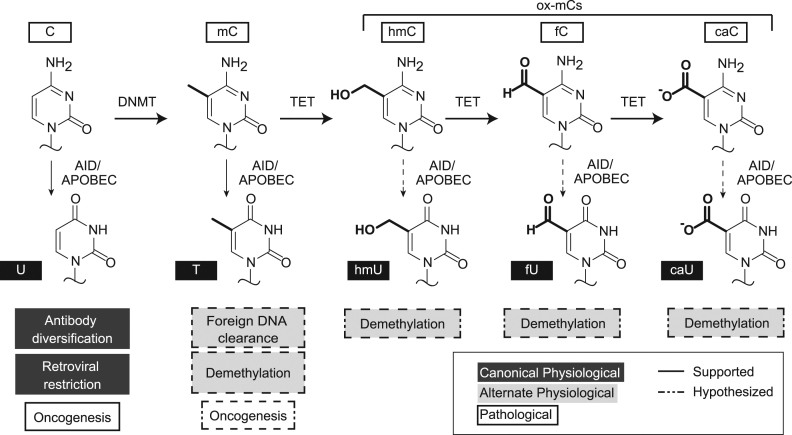
Aside from their established roles in immunity, several other functions have been proposed for these DNA deaminases, mainly involving their activities on noncanonical substrates (Figure 1). Several family members have been shown to deaminate 5-methylcytosine (mC), a common gene silencing mark, although activity on mC is 5- to 15-fold less than on unmodified cytosine (5–8). Human APOBEC3A (A3A) has garnered particular attention as it can more efficiently deaminate mC, with rates only slightly reduced relative to unmodified cytosine in both biochemical and cellular assays (9–11). This enhanced mC deamination activity by A3A has been proposed to function in restriction of foreign methylated DNA (12); however, whether this efficient activity on mC extends to cytosines with even bulkier modifications has yet to be explored.
Figure 1.
Physiological and pathological roles of AID/APOBEC enzymes. In the genome, cytosine can be found in various forms: unmodified (C), methylated (mC), or oxidized mC bases (hmC, fC, and caC). The canonical function of AID/APOBECs in both innate and adaptive immunity is deamination of unmodified cytosine to uracil (dark gray). Mutational signatures in cancer have implicated AID/APOBEC activity on the human genome, on unmodified cytosine or possibly on mC (white). Alternative physiological roles for APOBECs have also been proposed (light gray), including in active DNA demethylation. These proposed pathways involve deamination of modified cytosine bases to their corresponding uracil analogs, followed by base excision repair to revert the original base to an unmodified cytosine.
This question is particularly relevant as AID/APOBEC deaminases have been proposed to play a role in active DNA demethylation (13,14). Currently, the most widely accepted view of DNA demethylation first involves oxidation of mC by ten-eleven translocation (TET) family enzymes, producing 5-hydroxymethylcytosine (hmC), 5-formylcytosine (fC), and 5-carboxylcytosine (caC) via stepwise oxidation (15,16). The latter two oxidized mC (ox-mC) bases can be removed by thymine DNA glycosylase (TDG), a base excision repair (BER) enzyme, and resultant repair provides a feasible complete cycle for DNA demethylation (15–18). However, since TDG and other DNA glycosylases have also been shown to excise modified uracils, an alternative, accessory model for demethylation exists involving AID/APOBEC deamination. In this model, the modified cytosine is deaminated to generate a genomic mismatch, which can be repaired to unmodified cytosine by BER (19–21). Pathways invoking mC deamination (5,22–24), hmC deamination (22,25,26), or caC deamination (27) have all been proposed as possible contributors to DNA demethylation (Figure 1). Among ox-mCs, only deamination of hmC has been directly examined with select AID/APOBEC family members. Prior biochemical work showed that human AID, mouse APOBEC1, and mouse APOBEC3 all discriminate against this bulky modification (6,8): deamination of hmC was at least 100-fold slower than unmodified C but could not be further quantified due to the detection limit. Importantly, no study has yet tested AID/APOBEC activity on fC and caC. 5-hydroxymethyluracil (hmU) and 5-formyluracil (fU) have been detected in genomes (28–31); however, whether these bases are derived from enzymatic deamination, oxidative damage or other processes such as TET-mediated oxidation of thymidine rather than mC remains unclear (30,31). Therefore, though often cited as plausible, demethylation involving oxidation and enzymatic deamination in concert has yet to be validated biochemically.
Recent work has also called attention to the pathological roles of AID/APOBEC enzymes, as many cancer genomes were found to harbor clusters of strand-coordinated mutations that were predominantly C-to-T transitions in a 5΄-TC sequence context (32–35). This phenomenon, known as kataegis, is thought to arise from enzymatic deamination in areas of exposed single-stranded DNA (36,37). The 5΄-TC sequence preference implicated A3A and its closely-related family member APOBEC3B (A3B) in these clustered mutations (33,38–40). Although the 5΄-TC sequence preference of A3A and A3B is well established, the impact of methylation status on these deamination preferences has not been previously investigated. This question is important because the interplay between sequence context dependence and C/mC deamination activity could aid in deciphering these deaminases’ mutational footprint on cancer genomes.
In this study, we devised and applied a novel restriction enzyme-coupled assay that allows for parallel determination of the steady-state kinetic parameters for A3A on all epigenetic cytosine modifications. Our results reveal that, despite its efficient activity on mC, A3A potently discriminates against hmC at rates that can now be quantified as 5600-fold slower than for unmodified cytosine. We newly determine that fC is a similarly poor substrate (with 3700-fold discrimination) and that there is no detectable activity of A3A on caC. This level of enzymatic selectivity suggests little cross-talk between A3A and TET-generated oxidized mC bases (ox-mCs) in the genome. We further show that A3A is remarkably proficient under conditions of enzyme excess, deaminating all C and mC bases to completion in a long ssDNA substrate including non-preferred sequence contexts. By contrast, under limiting enzyme conditions, we find that the well-characterized sequence preferences with unmodified C deamination are exaggerated with mC. Taken together, our study provides a rigorous, quantitative framework for understanding the interplay between cytosine methylation, oxidation, and deamination.
MATERIALS AND METHODS
Synthesis of DNA substrates
Oligonucleotides with modified bases (including appropriate modified uracil controls, aside from fU and 5-carboxyluracil (caU)) were synthesized in-house using an ABI 384 DNA/RNA synthesizer using standard phosphoramidite chemistry and supplier deprotection protocols (Glen Research). Where necessary, polyacrylamide gel electrophoresis purifications were performed on the oligonucleotide substrates; otherwise, DMT-on purifications (GlenPak, Glen Research) were performed and masses were validated by MALDI-TOF mass spectrometry (Supplementary Table S1). As phosphoramidites for fU and caU are not commercially available, the fU- and caU-containing control oligonucleotides were instead synthesized by primer extension (Supplementary Figure S1) using 5-formyl-dUTP or 5-carboxyl-dUTP (Trilink Biotechnologies). Complementary strands for the activity assay were purchased from Integrated DNA Technologies (IDT). Aside from fU and caU controls, oligonucleotides were either synthesized containing a 3΄ 6-fluorescein (FAM) group or were post-synthetically labeled with 3΄ fluorescein-12-dideoxyuridine (ddU-FAM) using terminal transferase (TdT) from New England Biolabs (NEB) and ddUTP-FAM (Enzo Life Sciences). Oligonucleotides used in sequence context experiments were purchased from IDT and subsequently 3΄-ddU-FAM-labeled using TdT.
Expression and purification of recombinant A3A
Human A3A was amplified from the cDNA plasmid repository (FLH257586.01L, DNASU) and cloned into a pET41 expression vector with a maltose-binding protein (MBP) N-terminal tag and a C-terminal 8X histidine tag, both separated from A3A by TEV protease cleavage sites. The plasmid was transformed into a BL21(DE3) bacterial expression line containing a plasmid encoding trigger factor to aid solubility (41). Expression in liquid culture was induced with 1 mM IPTG at OD 0.6. Cells were grown overnight at 16°C, pelleted, and lysed by sonication, and MBP-A3A-His was purified using His-Pur resin (ThermoFisher). Both the N- and C-terminal tags were removed by treatment with TEV protease, and the resulting protein was purified over a HiTrap heparin column (GE Healthcare) using an AKTA FPLC. Purified A3A was dialyzed into storage buffer containing 50 mM Tris-Cl (pH 7.5), 50 mM NaCl, 10% glycerol, 0.5 mM dithiothreitol (DTT) and 0.01% Tween-20, and stored at −80°C.
Qualitative deaminase activity using restriction enzyme-based method
A total of 500 nM oligonucleotide substrate was treated with 10-fold dilutions of A3A (from 1 nM to 1 μM) and allowed to react for 30 min in 20 mM Tris-Cl (pH 7.0) and 0.1% Tween-20 (‘reaction buffer’) at 37°C. Deamination was terminated by incubation at 95°C for 20 min. DNA was ethanol precipitated, resuspended in water, and annealed to a 1.5-fold excess of complementary strand (S35-C-Comp, Supplementary Table S1). SwaI (NEB) was added with appropriate buffer and incubated overnight at room temperature. The next day, the samples were heat denatured, run on a preheated 20% acrylamide/Tris-Borate-EDTA(TBE)/urea gel at 50°C, and imaged using FAM filters on a Typhoon imager (GE Healthcare).
Steady-state kinetic studies of A3A
For improved sensitivity, kinetic assays were performed with substrate DNA (10 pmol) that was 5΄-end radiolabeled using T4 polynucleotide kinase (NEB) and 32P-γ-ATP, and purified using Illustra Microspin G-50 columns. Unlabeled DNA at a concentration of 5 μM was added to ∼100 nM radiolabeled substrate. For fC and caC substrate, 3΄-6-FAM-labeled substrates (like in qualitative assays above) were used instead of radiolabeled substrates. SwaI assays were performed as described above, except that substrate concentrations were varied from 80 nM to 1.25 μM (in 2-fold dilutions). A3A concentrations and time intervals for each substrate were chosen to yield ∼20% deamination with 80 nM substrate at 37°C in reaction buffer (Supplementary Figure S2). The protocol above was followed, except that after ethanol precipitation, reactions were resuspended in different volumes of water to dilute all samples to 80 nM before annealing complement. In the case of the substrates with poor reactivity, we chose time intervals that would allow us to approximate initial reaction velocities at high substrate concentrations (2 μM), and then titrated A3A up to 1.4 μM. Specific conditions for each substrate are provided in Supplementary Figure S2. In the case of 5-iodocytosine (iC), we observed a small portion of unmodified C (validated by MALDI-TOF) in the sample that arose via deiodonation. As such, we performed digest controls with uracil DNA glycosylase (UDG, from NEB) on each of the reactions, as UDG does not excise 5-iodouracil, and subtracted the signal resulting from C deamination from each reaction. Also, the fC substrate was found to undergo some degradation overnight in the SwaI assay, so SwaI reactions for kinetic determinations were performed for 3 h at 25°C, which still permitted the fU control to efficiently cleave. For radiolabeled substrates, the gels were exposed to a phosphor screen for 0.5–4 h after electrophoresis. The screens were scanned on a Typhoon imager. For FAM-labeled substrates, the gels were imaged as noted in qualitative assays above. Percent deamination was calculated by quantifying the intensity of the substrate and product gel bands using BioRad Quantity One software and then taking the ratio of intensities: (product)/(product+substrate), with background correction as needed to account for trace substrate degradation with fC. Michaelis-Menten plots and A3A titration plots were generated using Prism graphing software. kcat and KM values for C, mC, flC, and brC were calculated through nonlinear regression fitting to the Michaelis-Menten equation: rate = (kcat) ([S])/(KM + [S]). kcat values for iC, hmC, fC, and caC were determined by linear regression fit on Prism (Supplementary Figure S3). Reactions were performed in triplicate, and errors are reported as standard deviations.
Local sequence context preference profile
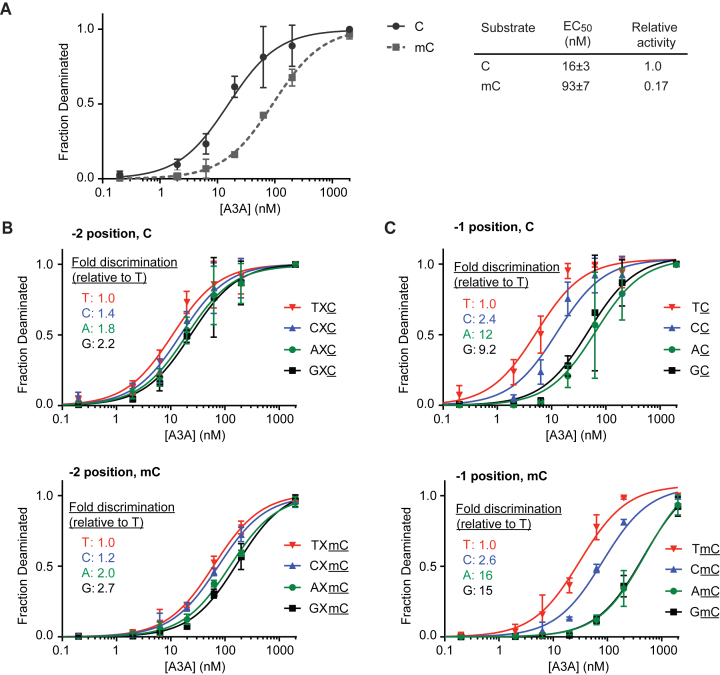
A 636-bp gene block (IDT) designed to minimize secondary structure was used to generate single-stranded 636-mer DNAs for the sequence profile experiments using a modified linear-after-the-exponential (LATE)-PCR protocol (49) with either unmodified dNTPs or 5-methyl-dCTP (Trilink) in place of dCTP, and subsequent λ-exonuclease treatment to degrade the 5΄-phosphorylated complementary strand (Supplementary Figure S4). Single-stranded 636-mer DNA substrates (1 ng), in which all cytosines were either C or mC, were incubated with various concentrations of A3A (ranging from 200 pM to 2 μM) in reaction buffer for 30 min at 37°C and then heat quenched at 95°C for 10 min. A total of 1 μl of the reaction was used to template polymerase chain reaction (PCR) amplification of the DNA using Taq polymerase and internal primers F2 and R2 (Supplementary Figure S4). Amplified products were run on a 1% agarose gel, extracted, and purified (Gel Extraction Kit, Zymo). Products were then TA cloned using the TOPO TA cloning kit (Invitrogen), transformed into Turbo High-Efficiency Cells (NEB), and plated on LB kanamycin plates containing 80 μg/ml X-gal for blue-white screening. After overnight growth, representative white colonies were miniprepped and sequenced (Genewiz). Deamination events were evidenced by C→T mutations, and local sequence context was recorded for each deamination event. For the analyses shown in Figure 3B and Supplementary Figure S5, data were binned based on the identity of either the −2 or the −1 base only, without regard for the identity of any other positions. For the analysis in Supplementary Figure S6, where two positions were combined in analysis, only the cytosine bases in a TTC/mC, TGC/mC, GAC/mC sequence context were analyzed. Independent reactions under each condition (each substrate and A3A concentration) were performed in triplicate, with each replicate incorporating data from 2–4 separate clones per experiment, with errors calculated as standard deviation between experiments. The fraction deamination was plotted against A3A concentration, and the effective enzyme concentration to achieve 50% deamination (EC50) was calculated using hyperbolic fit parameters in Prism graphing software. Under each condition, the fold discrimination was calculated by taking the ratio of EC50 values for a given base relative to the most preferred base at that position. For example, at the −1 position, the fold discrimination is given by the EC50 for A, G or C at the −1 position divided by that of T.
Figure 3.
Deamination of long, single-stranded substrates. (A) A total of 1 ng of single-stranded 636-mer DNA substrates containing either unmodified bases or mC in place of all cytosines were treated with various concentrations of A3A and then were clonally sequenced. Plotted are the fractions of deaminated bases as a function of A3A concentration. Each data point represents the average from at least three independent experiments, with each experiment containing 2–4 clones sequenced under that condition. The standard deviation is calculated between the independent experiments. Both C- and mC-containing substrates can be fully deaminated at the highest concentrations of A3A. The EC50 is the enzyme concentration to achieve 50% deamination of all C or mC bases and was calculated by fitting each curve to a hyperbolic function. The effect of local sequence context was examined by separately analyzing deamination efficiency for (B) the −2 position relative to the target base or (C) the −1 position relative to the target base. Each position is considered independently of the other position, and X denotes any base. The fold discrimination is the ratio of EC50 for a given base, relative to that of the preferred T at the same position.
Sequence context kinetics
For steady-state kinetic experiments on short C substrates in different sequence contexts, reactions were prepared similarly to conditions detailed above (see ‘Steady-state kinetic studies of A3A’), except that ∼100 nM radiolabeled substrates were added to 20 μM unlabeled DNA and substrate concentrations ranged from 80 nM to 5 μM in assays. Conditions were chosen that would approximate initial reaction velocities for each substrate. For the UDG-coupled assay, after incubation with A3A for a given time at 37°C, reactions were quenched at 95°C for 10 min and then 5 units of UDG were added and incubated at 37°C for 2 h under recommended buffer conditions. Samples were boiled in formamide loading dye containing 300 mM NaOH and 5 mM ethylenediaminetetraacetic acid (EDTA) and loaded on a 20% acrylamide/TBE/urea gel for analysis. For A3A titration experiments on C and mC substrates, a TDG-coupled assay was performed. Substrate was fixed at 9 μM (>5-fold above the highest calculated KM) and A3A was titrated in serial dilutions. Reactions proceeded at 37°C for 45 min before inactivation. Oligonucleotides were ethanol precipitated and annealed to their respective complement strands (at 1.5× concentrations). TDG, purified as previously described (42), was then added in 8× excess of DNA substrate along with the following 10× buffer: 20 mM Tris pH 8.0, 1 mM DTT, 5 mM EDTA, 0.1 mg/ml bovine serum albumin. Reactions were incubated at 16°C for 12 h and samples were analyzed as described in the SwaI assay. All reactions were performed in triplicate on different days, and errors were calculated as standard deviations.
RESULTS
Restriction enzyme-based assay for assessing deaminase activity
The catalytic activities of the AID/APOBEC family members have typically been measured in vitro by exploiting various DNA glycosylases that differentiate between deaminated cytosine products (uracil analogs) and cytosine substrates. The major limitation of this technique lies in the glycosylases themselves: many recognize only a subset of deamination products and some, such as TDG, can also excise modified cytosine bases in addition to their deaminated products. The requirement for multiple different coupling enzymes for probing more than one modified cytosine functionally makes comparisons between substrates more difficult. Although some glycosylases such as the thermostable mismatch glycosylase MtMig (8) have been adapted to examine deamination of several modified cytosine bases, a suitably selective glycosylase, capable of discriminating deamination of C, mC, hmC, fC, and caC, has not previously been described.
To overcome these limitations, we developed a uniform method to analyze A3A-catalyzed deamination of all modified cytosine (xC) bases that maintains the selectivity previously afforded by glycosylases, ensuring efficient detection of uracil analog products (xU) while minimizing background activity on their cytosine substrates. To this end, we synthesized a series of oligonucleotides, 35 bases in length, containing either the TTxCA substrate sequence or TTxUA as the corresponding product. We focused on this sequence context, as TTCA is a preferred substrate for A3A (40). We surveyed a series of restriction enzymes that displayed altered cleavage patterns in response to a TTxCA to TTxUA change to find one that retained this capability regardless of the modification. We found the desired properties in SwaI, which normally cuts at ATTTAAAT recognition sequences (Figure 2A). Interestingly, when the complementary strand contains the matched complementary SwaI restriction site intact, both the xC-containing and xU-containing duplexes are cleaved to some extent. However, when the complementary oligonucleotide contains a mismatched G on the opposite strand, sensitive discrimination between all uracil and cytosine analogs is achieved (Supplementary Figure S1). This phenomenon of mismatch recognition of some restriction enzymes has been observed previously (43). Interestingly, consistent with the tolerance for various xU modifications we observe, a recently solved structure of SwaI in complex with DNA (44) reveals a lack of steric interactions with the 5-methyl group of T at the center of the cleavage recognition site (see additional discussion provided in Supplementary Figure S1).
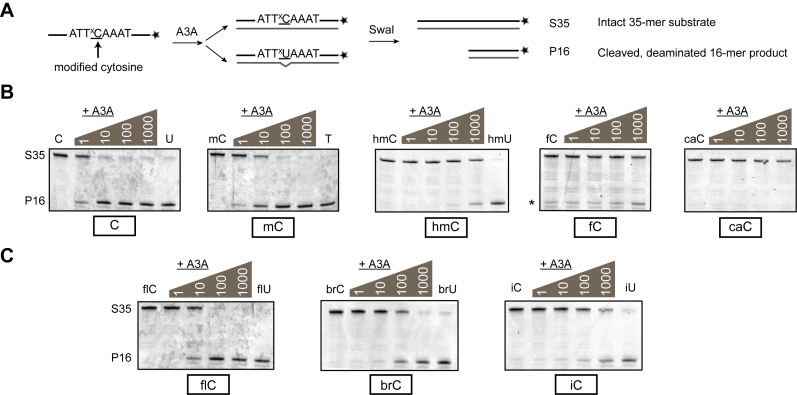
Figure 2.
Qualitative activity of A3A using a restriction-enzyme based assay. (A) 3΄-FAM-labeled single-stranded oligonucleotides containing one substrate cytosine in an ATTxCAAAT sequence context are treated with A3A. A complementary strand is annealed, with G paired across from the substrate xC (mismatched if deaminated to xU). SwaI recognizes and specifically cleaves only the 5΄-ATTxUAAAT in this context, allowing the labeled, unreacted 35-mer substrate (S35) to be separated from cleaved, deaminated 16-mer product (P16) on a denaturing polyacrylamide gel imaged with FAM filters. (B) A3A titrations on substrates containing C, mC or ox-mCs. In each gel, from left to right are incubations of each substrate (500 nM) with 0, 1, 10, 100 and 1000 nM A3A (37°C for 30 min). When noted, the rightmost lane is a product control without A3A. Product controls for the fC/caC reactions are shown in Supplementary Figure S1. Degradation of the fC substrate with prolonged incubation produces a band (*) at the same size as P16 product, requiring background correction in quantification of deamination. Deamination of fC is above background at 1000 nM A3A. (C) A3A reactions with substrates containing with 5-halogenated cytosines are shown in an analogous format.
Qualitative activity of APOBEC3A on modified cytosines
Using our SwaI-based assay, we first qualitatively examined the activity of A3A on the five physiological cytosine modification states in DNA (C, mC, hmC, fC, and caC). A total of 500 nM of each labeled ssDNA substrate was treated with serial 10-fold dilutions of A3A. After the reaction, strands were annealed to the mismatched complement. The duplexes were then digested with SwaI, and the cleaved xU-containing products were separated from uncleaved xC-containing substrates on denaturing gels (Figure 2B). As expected, the unmodified C substrate showed partial deamination at even the lowest A3A concentration chosen (1 nM) and was fully deaminated with 10 nM A3A under these conditions. In line with previous reports showing high mC activity (9–11), the mC substrate also showed signs of deamination at low A3A concentration, though complete deamination was not achieved until treatment with 100 nM A3A. hmC- and fC-containing oligos were resistant to deamination at low concentrations of A3A, but discernable deamination was detected at the highest concentrations of A3A tested (1 μM). In contrast, no deamination of caC-containing substrate was detected, even at the highest A3A concentrations evaluated.
In general, the pattern of activity by A3A follows a steric trend of discrimination previously observed with human AID, mouse APOBEC1, and mouse APOBEC3, in which larger 5-substituents were disfavored (6). In order to probe this steric trend further, we synthesized analogous substrates containing 5-halogenated cytosines of various sizes and electrochemical properties. When these substrates were tested with the SwaI-based assay, we find that the patterns observed with natural cytosine substrates are maintained at a qualitative level (Figure 2C). Iodo-C (iC) shows a similar level of deamination to hmC, which correlates with its size, while bromo-C (brC) shows an expected intermediate level of deamination between mC and hmC/fC. In line with its small size, 5-fluorocytosine (flC) is deaminated proficiently, to a roughly similar extent as mC.
Steady-state kinetics of APOBEC3A on modified cytosines
Given the overall high activity of A3A on C and several 5-modified cytosines, we posited that our SwaI-based assay should permit steady-state kinetic analyses for rigorous quantitative comparisons between substrates. Also, since our qualitative assays are the first to note low-level A3A activity on hmC and fC, a sensitive approach could newly allow for accurate quantification of the extent of discrimination against ox-mCs by A3A. To this end, reactions were set up to achieve steady-state conditions and meet necessary assumptions to fit the data with Michaelis-Menten parameters (Supplementary Figure S3). Rates of A3A-mediated deamination were obtained for a range of concentrations of the smaller modified substrates (C, mC, flC and brC), and fitted to obtain kcat and KM values (Table 1). Our quantitative analysis demonstrates high A3A activity on unmodified cytosine (kcat/KM = 79 μM−1min−1), and decreasing kcat/KM values with increasing size of substituent. We noted that the differences in activity were predominantly due to a catalytic effect: the kcat values changed significantly across the substrate series while the KM values remained relatively similar (310–550 nM). Intriguingly, a subtle but quantifiable exception to the observed steric trend manifested in the calculated kcat/KM values of flC and mC. Though our qualitative assay showed little difference in activity between these two substrates, we calculated a lower kcat/KM for the smaller flC (16 μM−1min−1) than for the larger mC (25 μM−1min−1), supporting the previous observations that A3A has a particular predilection for mC deamination.
Table 1. Kinetic constants for A3A acting on modified cytosine substrates.
| Substrate | Substituent | Size* (Å3) | k cat † (min−1) | K M (μM) | k cat/KM (μM−1min−1) | Relative activity†† (norm to –H sub) |
|---|---|---|---|---|---|---|
| C | -H | 10.5 | 41 ± 7 | 0.52 ± 0.19 | 79 | 1.0 |
| flC | -F | 16.8 | 4.9 ± 0.3 | 0.31 ± 0.05 | 16 | 0.12 ± 0.02 |
| mC | -CH3 | 30.5 | 10 ± 1 | 0.41 ± 0.13 | 25 | 0.25 ± 0.05 |
| brC | -Br | 33.0 | 0.33 ± 0.03 | 0.55 ± 0.11 | 0.060 | 8.0 (±1.5) × 10−3 |
| fC | -CHO | 36.3 | 0.011 ± 0.001 | n.c. | 2.7 (±0.5) × 10−4 | |
| iC | -I | 37.5 | 0.015 ± 0.001 | n.c. | 3.7 (±0.7) × 10−4 | |
| hmC | -CH2OH | 40.8 | 0.0073 ± 0.0004 | n.c. | 1.8 (±0.3) × 10−4 | |
| caC | -COO- | 43.1 | <0.002 | n.c. | < 4.9 (±0.8) × 10−5 |
Substrates differ only in the identity of the substituent at the 5-position of the target cytosine and are listed in order of increasing size.
* The size of each substituent (in Å3) was calculated using SPARTAN molecular modeling software.
† kcat and KM values were determined for the smallest four substrates (C, flC, mC, and brC) by performing steady-state kinetic measurements. kcat values were approximated for the four largest substrates (fC, iC, hmC, and caC) by determining the observed rate at saturating substrate concentrations. As no activity was detected on caC, the value reported corresponds to the calculated detection limit (determined in Supplementary Figure S2C). n.c., not calculated. Errors are reported as standard deviations from three independent kinetic replicates.
†† Relative activities are calculated as the ratio of kcat values, with associated error calculated from propagation of error in kcat values.
Due to the lower activity on the larger, oxidized substrates, steady-state assumptions could not be readily achieved at low substrate concentrations. However, extrapolating from our observation that the KM values are not substantially altered across the different substrates tested, we fixed substrate concentrations at a saturating level (2 μM) and titrated enzyme to calculate the presumptive kcat values (Table 1). Most of the larger and/or oxidized modifications show very little, though detectable, activity and there is no detectable activity on caC. The kinetic analysis suggests that, relative to deamination of unmodified C, deamination of hmC is ∼5600-fold less proficient and fC deamination is ∼3700-fold less proficient. Even with large excess of A3A and long incubation time, caC showed no detectable deamination, which suggests a >20 000-fold discrimination against caC based on our calculated limits of detection for caU (Supplementary Figure S2C). Thus, despite its notable activity on mC, our kinetic analysis provides the first evidence that A3A potently discriminates against all three TET-oxidized products of mC.
Activity of A3A on Cs and mCs across all sequence contexts
While discrimination against ox-mCs holds significant implications for models of DNA demethylation involving coupled oxidation and deamination, the proficiency of mC deamination raises the question of how such activity might manifest on genomic substrates. The target substrates of DNA deaminase activity are conjectured to be long ssDNA stretches that arise from processes such as DNA replication, DNA repair, or telomere crisis (45–48), and sequence preferences have been one of the most important features defining AID/APOBEC activity at these sites. We therefore next asked if proficiency with mC deamination would extend to more complex substrates and if local sequence context would differentially influence A3A's ability to target C versus mC for deamination. To this end, we generated long 636-mer single-stranded DNA substrates (Supplementary Figure S4) containing exclusively either C or mC using a modified version of linear-after-the-exponential PCR (49). These extended ssDNA substrates (1 ng) were treated with increasing concentrations of A3A, and individual strands were then PCR amplified, cloned, and sequenced.
This assay approach readily demonstrates the efficiency of A3A in both C and mC deamination. In case of the C-containing substrate, we were surprised to find that at the highest concentration of A3A evaluated, every single C could be deaminated to completion in all clones (Figure 3A; all 183 C bases in each of 9 clones), a phenomenon that has yet to be described in the literature with any AID/APOBEC deaminase. We observed similar levels of deamination in the mC-containing substrate at the highest A3A concentrations, although complete deamination was not as consistent across all clones. Complete deamination was observed in 6 of 18 clones and the other clones were deaminated to near-completion (83.1–99.5%).
Examining deamination at lower concentrations of A3A permitted a more nuanced analysis of the proficiency of C versus mC deamination in various sequence contexts. For AID/APOBEC family enzymes, the upstream bases are known to be the strongest determinants of deamination efficiency, with a dominant role for the −1 base and a secondary role for the −2 base; the immediate downstream +1 position is also known to contribute to specificity to a lesser extent (40,50). We therefore categorized each target cytosine based upon the identity of the −2, −1, and +1 bases and then plotted the fraction of deaminated cytosines in each context (Figure 3B and C; Supplementary Figure S5). For a quantitative comparison of A3A activity, we defined the EC50 as the effective concentration of A3A required to achieve deamination of 50% of C or mC bases in that context; we normalized each EC50 relative to that for the preferred base at each respective site. As anticipated, the ability of A3A to deaminate cytosines depends most heavily on the −1 base. In the case of −1 preferences with C substrates, we observe a 2.4-fold discrimination of CC compared to TC and an even more pronounced 9.2- or 12-fold discrimination against GC and AC, respectively. In the case of the mC substrates, the relative EC50 values shift to 2.6-fold, 15-fold and 16-fold for CC, GC and AC, respectively. Thus, the influence of the −1 substituent is mildly exaggerated in the case of mC relative to C, with a wider span of EC50 values. The data for the −2 base, which has a less significant influence on activity, shows a subtle but similar trend, with a wider span of EC50 values for mC than for C (Figure 3B). The +1 base also shows only a minor influence on deamination efficiency, with similar patterns of EC50 values for C and mC (Supplementary Figure S5). Given the dominant role for upstream bases, we also examined the effect of combining the −2 and −1 positions in analysis rather than treating the two positions independently (Supplementary Figure S6). In the preferred sequence context (5΄-TTC/mC) the EC50 ratio reflects a 5.9-fold preference for C versus mC deamination, while this ratio increases to 10-fold for deamination in the least preferred GAC/mC context.
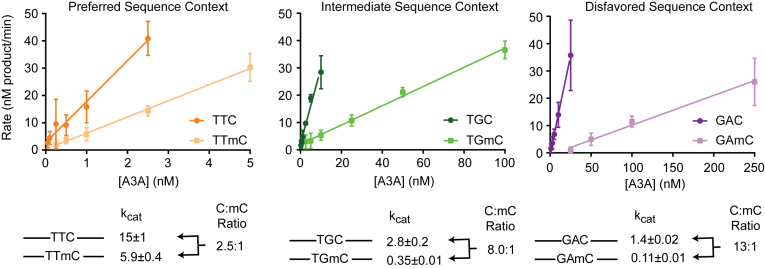
The observed shift in sequence preferences with C versus mC substrates prompted us to ask how sequence context and methylation status combine to determine deamination efficiency in shorter oligonucleotide substrates. To this end, we synthesized a matched set of six oligonucleotides that differed in the local sequence context—preferred (TTC/mC), intermediate (TGC/mC) or disfavored (GAC/mC)—along with the associated product controls. With the cytosine-containing substrates, we again verified that the major differences in deamination efficiency were largely attributable to kcat (Supplementary Figure S7), and turned to a TDG-coupled assay under saturating substrate concentrations to compare the relative reactivity of C- and mC-containing substrates (Figure 4). Similar to the results seen with the SwaI-based assay, we observed a 2.5-fold drop in kcat with the TTmC-containing substrate relative to TTC-containing substrate. In the intermediate and disfavored sequence contexts, we noted 8.0-fold discrimination between TGC and TGmC and 13-fold discrimination between GAC and GAmC. The skew in sequence preferences with the long 636-mer (5.9-fold with TTC/mC versus 10-fold with GAC/mC) is even more apparent with oligonucleotide substrates (2.5-fold in TTC/mC versus 13-fold in GAC/mC). This difference could be attributable to the nature of A3A interactions with a long substrate containing multiple cytosine bases that could alter rate-determining steps in deamination. Nonetheless, as we consistently observed a larger C/mC discrimination in non-preferred substrates, we conclude that the sequence context and the modification state of the cytosine base together influence the overall activity of A3A on target bases.
Figure 4.
Enzyme titrations on C and mC substrates in various sequence contexts. A total of 9 μM oligonucleotide substrate in a preferred (TTC/mC), intermediate (TGC/mC), or disfavored (GAC/mC) sequence context was incubated with various amounts of A3A and product formation quantified using a TDG-coupled assay. Experiments were performed in triplicate, with error bars indicating standard deviation between experiments, and values were fit with linear regression. Observed rates were calculated as the slope, reflecting the presumptive kcat values.
DISCUSSION
Recent studies have implicated enzymatic deamination of cytosine and epigenetically-modified cytosine bases in diverse processes including DNA demethylation and cancer mutagenesis. We focused on characterizing the activities of A3A specifically given prior evidence of enhanced activity on mC. To this end, we devised a deamination assay that permitted parallel, quantitative comparisons of A3A activity on the cytosine variants in DNA. Using this approach, we found, in line with prior studies, that mC is efficiently deaminated by A3A, with only a ∼4-fold decrease in kcat relative to unmodified cytosine in the preferred sequence context. While prior studies on AID/APOBEC family members using less sensitive or semi-quantitative assays suggested that hmC cannot be deaminated by DNA deaminases (6,8,10), our assays indicate that deamination can occur, albeit at rates ∼5600-fold reduced relative to unmodified cytosine. Our approach also allowed us to evaluate deamination of fC and caC for the first time with any DNA deaminase. We report strong discrimination against fC, and no detectable activity on caC.
We can gain mechanistic insight into our findings by considering the recent crystal structure of A3A solved in complex with ssDNA (51). The structure revealed that the cytosine base is rotated away from stacking with neighboring bases and inserted into a deep active site pocket where the C5-C6 side of the base abuts a hydrophobic phenolic ring of tyrosine (Y130). This Y130 residue likely serves as the ‘steric gate’ in the active site, providing a mechanism for the observed discrimination against bulkier 5-position substituents. Although the trend of steric discrimination generally holds, A3A deaminates mC more efficiently than the smaller flC. This minor deviation could be explained by enhanced hydrophobic interactions between the methyl substituent and Y130 that are lost in the case of the electronegative flC. Such active site steric constraints on deamination of 5-modified cytosines are not universal in the larger deaminase superfamily beyond AID/APOBECs. Unlike A3A, cytidine deaminase (CDA), a related enzyme that deaminates free cytidine nucleosides rather than cytosine bases in DNA, can efficiently deaminate the nucleosides of hmC and fC (52). Given that AID/APOBECs bind and deaminate cytosines as part of longer DNA substrates, it is likely that the flexibility constraints on the cytosine base prevent access to the alternative orientations that permit CDA to deaminate ox-mC nucleobases (51).
While prior cell-based studies have suggested that deamination of ox-mCs could contribute to DNA demethylation (22,25,27), our work indicates that these pathways are biochemically disfavored. As A3A has the highest activity on mC among the AID/APOBEC family members, the enzyme's potent discrimination against ox-mCs is particularly compelling. Given these results, we therefore suggest that it is unlikely that DNA deaminases play a major role in demethylation via an oxidation-deamination mechanism (15). This conclusion does not eliminate the possibility that other factors, such as protein interaction partners or post-translational modifications, could change the local concentration of the enzyme near a given substrate or otherwise impact activity; however, such factors have yet to be demonstrated. Also, it remains plausible that AID/APOBECs could play a role in active DNA demethylation via activity on mC bases, as suggested in certain cell and tissue types (5,22–24). Such demethylation could involve direct deamination of mC by DNA deaminases, or, alternatively, patch repair of bases neighboring the site of either C or mC deamination (53).
To characterize C and mC deamination activity further, we expanded from simple oligonucleotide substrates with a single target base to longer, more complex ssDNA substrates. Importantly, in these assays on lengthy single-stranded C- and mC-containing substrates, we observed that A3A can fully deaminate substrates containing Cs or mCs across all sequence contexts in vitro. To our knowledge this is the first demonstration of such proficiency with any AID/APOBEC deaminase, raising the prospect of its utility as a biotechnological tool to distinguish between epigenetic cytosine modification states. Shortly after the initial discovery of AID, the concept of using DNA deaminases to discriminate between C and mC bases as part of epigenetic sequencing approaches was suggested (5); however, the poor overall activity of the enzymes and lack of sufficient discrimination between C and mC across all sequence contexts prevented their application as bisulfite-like mC sequencing tools (54). Revisiting those suggestions in light of our new results, efficient deamination of C/mC by A3A and potent discrimination against ox-mCs could now be exploited to localize modified bases at single-base resolution in genomic DNA.
Our study on both long and short DNA substrates in various sequence contexts demonstrates that sequence preferences of C deamination are exaggerated with mC deamination. In oligonucleotide substrates, while the C:mC deamination ratio was ∼3:1 when in the preferred sequence context, it increased to ∼13:1 when in a highly-disfavored context. This phenomenon is potentially important to account for when interpreting the AID/APOBEC mutational signatures in cancer sequencing projects. First, while most sequencing studies attribute deamination in CpG contexts predominantly to spontaneous deamination of mC, enzymatic deamination resulting from aberrant expression of A3A could also play a role in deamination of mC, especially when mC is in A3A's preferred sequence contexts (i.e. YmCpGs). As most methylation exists in a CpG context in the genome, it was particularly notable that deamination of mCpG was only ∼1.4-fold less efficient relative to the most preferred mCpA and mCpC contexts. Second, separately evaluating the mutational signature in CpG and non-CpG contexts might provide an added means to distinguish the impact of A3A from that of A3B (33,55,56), especially since A3B has been shown to have minimal activity on mC (57,58).
In mammalian genomes, cytosine bases are the focus of potentially intersecting modifications, with various permutations of cytosine methylation, deamination, and oxidation. Overall, our results reveal the intrinsic preferences of a DNA deaminase, helping to narrow the scope of potentially relevant in vivo modifications. Unmodified C can be methylated or deaminated; mC can be deaminated or oxidized; but the ox-mC bases—hmC, fC, and caC—are not subject to significant deamination. While the local sequence context is one feature which further impacts the nature of modifications on a target base, ongoing studies to understand the interplay of these various DNA-modifying processes in immunity, epigenetics, and cancer biology remain an important priority.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful for critical feedback from Matt Weitzman and Monica Liu.
Footnotes
Present address: Christopher S. Nabel, Department of Medicine, Brigham and Women's Hospital, Boston, MA 02115, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01-GM118501 to R.M.K]; Rita Allen Foundation Scholar Award (to R.M.K.); National Science Foundation Fellowship (to E.K.S., J.E.D.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Siriwardena S.U., Chen K., Bhagwat A.S.. Functions and malfunctions of mammalian DNA-cytosine deaminases. Chem. Rev. 2016; 116:12688–12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T.. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000; 102:553–563. [DOI] [PubMed] [Google Scholar]
- 3. Harris R.S., Bishop K.N., Sheehy A.M., Craig H.M., Petersen-Mahrt S.K., Watt I.N., Neuberger M.S., Malim M.H.. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003; 113:803–809. [DOI] [PubMed] [Google Scholar]
- 4. Conticello S.G., Langlois M.A., Yang Z., Neuberger M.S.. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv. Immunol. 2007; 94:37–73. [DOI] [PubMed] [Google Scholar]
- 5. Morgan H.D., Dean W., Coker H.A., Reik W., Petersen-Mahrt S.K.. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: Implications for epigenetic reprogramming. J. Biol. Chem. 2004; 279:52353–52360. [DOI] [PubMed] [Google Scholar]
- 6. Nabel C.S., Jia H., Ye Y., Shen L., Goldschmidt H.L., Stivers J.T., Zhang Y., Kohli R.M.. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat. Chem. Biol. 2012; 8:751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abdouni H., King J.J., Suliman M., Quinlan M., Fifield H., Larijani M.. Zebrafish AID is capable of deaminating methylated deoxycytidines. Nucleic Acids Res. 2013; 41:5457–5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rangam G., Schmitz K.M., Cobb A.J., Petersen-Mahrt S.K.. AID enzymatic activity is inversely proportional to the size of cytosine C5 orbital cloud. PLoS One. 2012; 7:e43279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wijesinghe P., Bhagwat A.S.. Efficient deamination of 5-methylcytosines in DNA by human APOBEC3A, but not by AID or APOBEC3G. Nucleic Acids Res. 2012; 40:9206–9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suspene R., Aynaud M.M., Vartanian J.P., Wain-Hobson S.. Efficient deamination of 5-methylcytidine and 5-substituted cytidine residues in DNA by human APOBEC3A cytidine deaminase. PLoS One. 2013; 8:e63461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carpenter M.A., Li M., Rathore A., Lackey L., Law E.K., Land A.M., Leonard B., Shandilya S.M., Bohn M.F., Schiffer C.A. et al. . Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J. Biol. Chem. 2012; 287:34801–34808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stenglein M.D., Burns M.B., Li M., Lengyel J., Harris R.S.. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat. Struct. Mol. Biol. 2010; 17:222–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mayer W., Niveleau A., Walter J., Fundele R., Haaf T.. Demethylation of the zygotic paternal genome. Nature. 2000; 403:501–502. [DOI] [PubMed] [Google Scholar]
- 14. Oswald J., Engemann S., Lane N., Mayer W., Olek A., Fundele R., Dean W., Reik W., Walter J.. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 2000; 10:475–478. [DOI] [PubMed] [Google Scholar]
- 15. Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y.. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011; 333:1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L. et al. . Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011; 333:1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kohli R.M., Zhang Y.. Tet, TDG and the dynamics of DNA demethylation. Nature. 2013; 502:472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maiti A., Drohat A.C.. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 2011; 286:35334–35338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nabel C.S., Manning S.A., Kohli R.M.. The curious chemical biology of cytosine: Deamination, methylation, and oxidation as modulators of genomic potential. ACS Chem. Biol. 2012; 7:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bhutani N., Burns D.M., Blau H.M.. DNA demethylation dynamics. Cell. 2011; 146:866–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Drohat A.C., Coey C.T.. Role of base excision “repair" enzymes in erasing epigenetic marks from DNA. Chem. Rev. 2016; 116:12711–12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cortellino S., Xu J., Sannai M., Moore R., Caretti E., Cigliano A., Le Coz M., Devarajan K., Wessels A., Soprano D. et al. . Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011; 146:67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bhutani N., Brady J.J., Damian M., Sacco A., Corbel S.Y., Blau H.M.. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010; 463:1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rai K., Huggins I.J., James S.R., Karpf A.R., Jones D.A., Cairns B.R.. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008; 135:1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo J.U., Su Y., Zhong C., Ming G.L., Song H.. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011; 145:423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hashimoto H., Hong S., Bhagwat A.S., Zhang X., Cheng X.. Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: Its structural basis and implications for active DNA demethylation. Nucleic Acids Res. 2012; 40:10203–10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xue J.H., Xu G.F., Gu T.P., Chen G.D., Han B.B., Xu Z.M., Bjoras M., Krokan H.E., Xu G.L., Du Y.R.. Uracil-DNA glycosylase UNG promotes tet-mediated DNA demethylation. J. Biol. Chem. 2016; 291:731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Globisch D., Munzel M., Muller M., Michalakis S., Wagner M., Koch S., Bruckl T., Biel M., Carell T.. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010; 5:e15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gackowski D., Zarakowska E., Starczak M., Modrzejewska M., Olinski R.. Tissue-specific differences in DNA modifications (5-hydroxymethylcytosine, 5-formylcytosine, 5-carboxylcytosine and 5-hydroxymethyluracil) and their interrelationships. PLoS One. 2015; 10:e0144859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Samson-Thibault F., Madugundu G.S., Gao S., Cadet J., Wagner J.R.. Profiling cytosine oxidation in DNA by LC-MS/MS. Chem. Res. Toxicol. 2012; 25:1902–1911. [DOI] [PubMed] [Google Scholar]
- 31. Pfaffeneder T., Spada F., Wagner M., Brandmayr C., Laube S.K., Eisen D., Truss M., Steinbacher J., Hackner B., Kotljarova O. et al. . Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat. Chem. Biol. 2014; 10:574–581. [DOI] [PubMed] [Google Scholar]
- 32. Roberts S.A., Lawrence M.S., Klimczak L.J., Grimm S.A., Fargo D., Stojanov P., Kiezun A., Kryukov G.V., Carter S.L., Saksena G. et al. . An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013; 45:970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burns M.B., Lackey L., Carpenter M.A., Rathore A., Land A.M., Leonard B., Refsland E.W., Kotandeniya D., Tretyakova N., Nikas J.B. et al. . APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013; 494:366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nik-Zainal S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L.A. et al. . Mutational processes molding the genomes of 21 breast cancers. Cell. 2012; 149:979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Borresen-Dale A.L. et al. . Signatures of mutational processes in human cancer. Nature. 2013; 500:415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taylor B.J., Nik-Zainal S., Wu Y.L., Stebbings L.A., Raine K., Campbell P.J., Rada C., Stratton M.R., Neuberger M.S.. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife. 2013; 2:e00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lada A.G., Dhar A., Boissy R.J., Hirano M., Rubel A.A., Rogozin I.B., Pavlov Y.I.. AID/APOBEC cytosine deaminase induces genome-wide kataegis. Biol. Direct. 2012; 7:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vartanian J.P., Guetard D., Henry M., Wain-Hobson S.. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science. 2008; 320:230–233. [DOI] [PubMed] [Google Scholar]
- 39. Nik-Zainal S., Wedge D.C., Alexandrov L.B., Petljak M., Butler A.P., Bolli N., Davies H.R., Knappskog S., Martin S., Papaemmanuil E. et al. . Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat. Genet. 2014; 46:487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Logue E.C., Bloch N., Dhuey E., Zhang R., Cao P., Herate C., Chauveau L., Hubbard S.R., Landau N.R.. A DNA sequence recognition loop on APOBEC3A controls substrate specificity. PLoS One. 2014; 9:e97062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kohli R.M., Abrams S.R., Gajula K.S., Maul R.W., Gearhart P.J., Stivers J.T.. A portable hotspot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J. Biol. Chem. 2009; 284:22898–22904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu M.Y., Torabifard H., Crawford D.J., DeNizio J.E., Cao X.J., Garcia B.A., Cisneros G.A., Kohli R.M.. Mutations along a TET2 active site scaffold stall oxidation at 5-hydroxymethylcytosine. Nat. Chem. Biol. 2017; 13:181–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Langhans M.T., Palladino M.J.. Cleavage of mispaired heteroduplex DNA substrates by numerous restriction enzymes. Curr. Issues Mol. Biol. 2009; 11:1–12. [PMC free article] [PubMed] [Google Scholar]
- 44. Shen B.W., Heiter D.F., Lunnen K.D., Wilson G.G., Stoddard B.L.. DNA recognition by the SwaI restriction endonuclease involves unusual distortion of an 8 base pair A:T-rich target. Nucleic Acids Res. 2017; 45:1516–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roberts S.A., Sterling J., Thompson C., Harris S., Mav D., Shah R., Klimczak L.J., Kryukov G.V., Malc E., Mieczkowski P.A. et al. . Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol. Cell. 2012; 46:424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kazanov M.D., Roberts S.A., Polak P., Stamatoyannopoulos J., Klimczak L.J., Gordenin D.A., Sunyaev S.R.. APOBEC-induced cancer mutations are uniquely enriched in early-replicating, gene-dense, and active chromatin regions. Cell Rep. 2015; 13:1103–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Green A.M., Landry S., Budagyan K., Avgousti D.C., Shalhout S., Bhagwat A.S., Weitzman M.D.. APOBEC3A damages the cellular genome during DNA replication. Cell Cycle. 2016; 15:998–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Maciejowski J., Li Y., Bosco N., Campbell P.J., de Lange T.. Chromothripsis and kataegis induced by telomere crisis. Cell. 2015; 163:1641–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sanchez J.A., Pierce K.E., Rice J.E., Wangh L.J.. Linear-after-the-exponential (LATE)-PCR: an advanced method of asymmetric PCR and its uses in quantitative real-time analysis. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:1933–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Langlois M.A., Beale R.C., Conticello S.G., Neuberger M.S.. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 2005; 33:1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shi K., Carpenter M.A., Banerjee S., Shaban N.M., Kurahashi K., Salamango D.J., McCann J.L., Starrett G.J., Duffy J.V., Demir O. et al. . Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat. Struct. Mol. Biol. 2017; 24:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zauri M., Berridge G., Thezenas M.L., Pugh K.M., Goldin R., Kessler B.M., Kriaucionis S.. CDA directs metabolism of epigenetic nucleosides revealing a therapeutic window in cancer. Nature. 2015; 524:114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Franchini D.M., Chan C.F., Morgan H., Incorvaia E., Rangam G., Dean W., Santos F., Reik W., Petersen-Mahrt S.K.. Processive DNA demethylation via DNA deaminase-induced lesion resolution. PLoS One. 2014; 9:e97754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bransteitter R., Pham P., Scharff M.D., Goodman M.F.. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:4102–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chan K., Roberts S.A., Klimczak L.J., Sterling J.F., Saini N., Malc E.P., Kim J., Kwiatkowski D.J., Fargo D.C., Mieczkowski P.A. et al. . An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat. Genet. 2015; 47:1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Starrett G.J., Luengas E.M., McCann J.L., Ebrahimi D., Temiz N.A., Love R.P., Feng Y., Adolph M.B., Chelico L., Law E.K. et al. . The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nat. Commun. 2016; 7:12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fu Y., Ito F., Zhang G., Fernandez B., Yang H., Chen X.S.. DNA cytosine and methylcytosine deamination by APOBEC3B: enhancing methylcytosine deamination by engineering APOBEC3B. Biochem. J. 2015; 471:25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Siriwardena S.U., Guruge T.A., Bhagwat A.S.. Characterization of the catalytic domain of human APOBEC3B and the critical structural role for a conserved methionine. J. Mol. Biol. 2015; 427:3042–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.