Abstract
Identifying extrapulmonary legionellosis is difficult due to the lack of clinical suspicion and limitations of conventional microbiologic methods. We present a case series of hematopoietic cell transplant recipients with extrapulmonary legionellosis diagnosed via molecular diagnostics: 16S ribosomal ribonucleic acid gene Sanger sequencing and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.
Keywords: 16S ribosomal ribonucleic acid gene sequencing, allogeneic hematopoietic cell transplant, extrapulmonary legionellosis, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, molecular diagnostics
Diagnosing extrapulmonary legionellosis is challenging due to the lack of clinical suspicion and limitations of diagnostic tools for nonrespiratory specimens. We present a single-center case series of allogeneic hematopoietic cell transplant (HCT) recipients diagnosed with extrapulmonary legionellosis via 16S ribosomal ribonucleic acid (rRNA) gene Sanger sequencing (case 1 and 2) [1] or matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (case 3) [2].
CASE 1
A 3.5-year-old boy with familial hemophagocytic lymphohistiocytosis underwent haploidentical, T-cell depleted (TCD) peripheral blood stem cell transplant (PBSCT) from his mother. His posttransplant course was notable for graft-versus-host disease (GVHD) of the skin and gastrointestinal (GI) tract, human herpesvirus 6 (HHV6), and adenovirus viremia.
On day +795, he was hospitalized for a nodular skin rash of his lower extremities for 3 weeks and new fever. His CD4 count was 0 cells/µL. A skin shave biopsy showed spongiotic psoriasiform dermatitis without granulomas and negative Gram, Periodic Acid-Schiff-Diastase (PAS-D), and acid-fast bacilli (AFB) stains. Routine bacterial, fungal, and mycobacterial cultures of skin tissue were also negative. The fever resolved after 9 days of empiric intravenous (IV) cefepime, followed by an additional week of amoxicillin-clavulanate upon discharge.
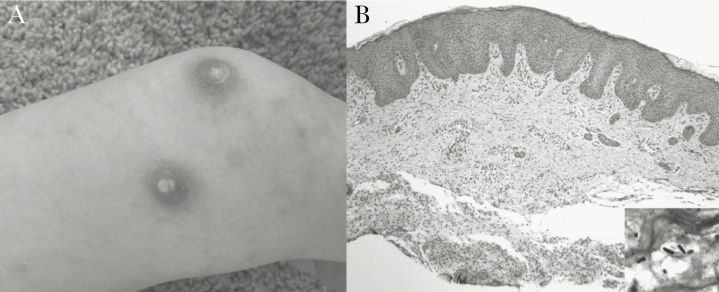
Twenty-three days later, he developed new fever, nonbloody diarrhea, emesis, dry cough, and worsening nodular skin lesions (Figure 1A). He was restarted on empiric IV cefepime and vancomycin.
Figure 1.
(A) Case 1, right lower extremity with nodulo-pustular rash. (B) Case 1, hematoxylin and eosin stained skin biopsy (×100) shows nonspecific dermal mononuclear inflammation and edema, within which bacilli were identified with a modified Steiner stain (inset, ×1000 oil immersion).
A repeat skin biopsy showed similar findings with negative routine stains and cultures. Tissue from repeat skin biopsy was sent for 16S rRNA gene Sanger sequencing [3], which identified Legionella anisa 7 days later. Upon molecular diagnosis, the skin tissue was re-examined using modified Steiner stain that showed scattered Gram-negative bacilli compatible with Legionella (Figure 1B). None of the antimicrobials used previously had activity against Legionella, so he was started on levofloxacin, and his GI symptoms and skin rash subsequently resolved.
CASE 2
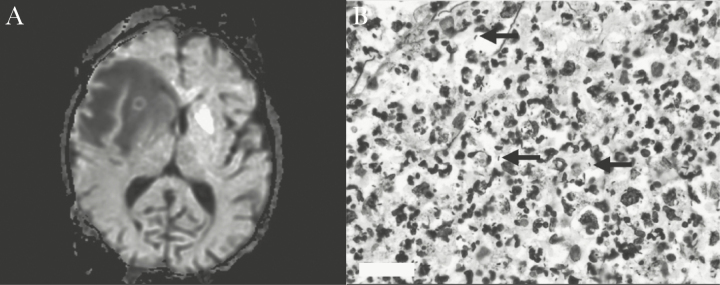
A 54-year-old female with relapsed multiple myeloma underwent allogeneic TCD PBSCT from a matched, unrelated donor (MUD) that was complicated by GI GVHD and HHV6 viremia. On day +120, she was admitted for 48 hours of nonbloody diarrhea, headaches, and lethargy. Her CD4 count was 0 cells/µL. Brain magnetic resonance imaging revealed an irregular necrotic right frontal lobe mass with extensive edema (Figure 2A). Chest computed tomography (CT) also showed a new right lower lobe (RLL) lung lesion. She was started on IV vancomycin, cefepime, metronidazole, liposomal amphotericin B, and trimethoprim/sulfamethoxazole (TMP/SMX) for empiric coverage of bacterial or fungal brain abscess and toxoplasmosis. Initial workup, including blood cultures, urinary Legionella antigen, serum fungal markers, and cultures from bronchoalveolar lavage fluid, was negative. A brain biopsy showed acute necrotizing polymorphonuclear infiltration without granulomas or malignancy. Immunohistochemical staining for cytomegalovirus, adenovirus, and toxoplasma was negative. Gram, Grocott-Gomori’s methenamine silver (GMS), PAS-D, and AFB stains of brain tissue were negative, as well as brain tissue cultures. Twelve days later, 16S rRNA gene Sanger sequencing was performed on shavings from the formalin-fixed paraffin embedded (FFPE) brain tissue [3] and revealed Legionella jamestowniensis. After molecular diagnosis, the brain tissue was re-examined using modified Steiner stain and showed 35 Steiner-positive bacilli per ×100 field compatible with Legionella (Figure 2B). She was continued on TMP/SMX to treat legionellosis, her neurologic symptoms eventually resolved, and serial brain and chest imaging after 15 weeks of treatment showed successful improvement.
Figure 2.
(A) Case 2, right frontal lobe mass with extensive edema and leftward midline shift. (B) Case 2, modified Steiner preparation (×100) demonstrating rod forms (black arrows).
CASE 3
A 3-year-old girl with acute lymphocytic leukemia in second remission received an allogeneic TCD PBSCT from a MUD, but she experienced graft rejection within 27 days posttransplant. A second TCD PBSCT was performed 2 months later with a posttransplant course notable for cytomegalovirus, Epstein Barr virus, and adenovirus viremia. All subsequently resolved, and the patient was doing well.
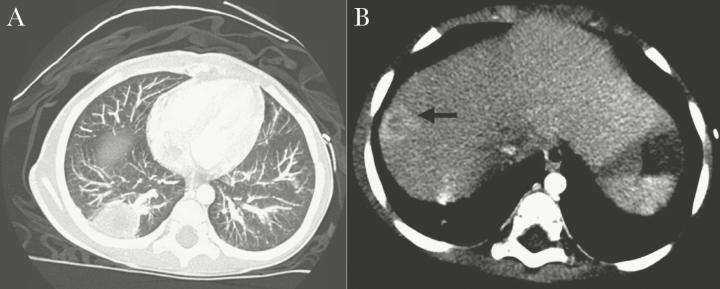
On day +63 after the second transplant, she was admitted for fever, emesis, nonbloody diarrhea, and right upper abdominal pain. Her CD4 count was 19 cells/µL. The CT imaging showed a lesion in the RLL of the lung and right hepatic lobe, both with central necrosis (Figure 3A and B). She was empirically started on IV vancomycin, piperacillin/tazobactam, and liposomal amphotericin B. The patient underwent RLL lung wedge resection and liver biopsy. Pathology showed benign lung and liver parenchyma with necrotizing abscess formation; negative Gram, GMS, PAS-D, and AFB stains; and negative immunohistochemical staining for adenovirus and cytomegalovirus. Gram stain of colonies from both lung and liver specimens showed Gram-negative rods identified as Legionella pneumophila via MALDI-TOF MS (VITEK MS InVitro Diagnostic [IVD], version 2.0; bioMérieux Inc., Durham, NC) within the first 48 hours of culture on buffered charcoal yeast extract (BCYE) agar. She was started on a 21-day course of levofloxacin, and CT imaging performed 44 days later showed radiographic resolution of both lung and liver lesions. The New York City Department of Health confirmed L pneumophila serotype 1 via polymerase chain reaction from the respiratory isolate recovered by culture.
Figure 3.
(A) Case 3, right lower lobe lung mass with central necrosis. (B) Case 3, right centrally necrotic hepatic lobe lesion within segment 8 (black arrow).
DISCUSSION
We report 3 cases of extrapulmonary legionellosis caused by different Legionella species in allogeneic HCT recipients from a single institution in New York City and without an identified spatial cluster of legionellosis. Two cases were caused by nonpneumophila species: L anisa and L jamestowniensis, the latter of which had not been previously reported to cause human disease until January 2017 [4]. The third case was caused by L pneumophila serotype 1. It is notable that all 3 patients had low CD4 counts and impaired cell-mediated immunity, which is a well known risk factor for legionellosis [5]. Fortunately, all responded well to targeted treatment without fatal outcomes.
Diagnosis of extrapulmonary legionellosis is challenging for several reasons. For one, clinicians may not be aware that Legionella can infect other tissues besides lungs. The ability of Legionella to disseminate was recognized in 1979 after the organism was recovered from blood in a patient with pneumonia [6]. Since then, rare cases of extrapulmonary legionellosis with or without pneumonia have been described. The most common extrapulmonary site is the heart, including reports of myositis, pericarditis, and prosthetic valve endocarditis [7]. There have also been cases of confined involvement of the brain, skin, genitourinary, and musculoskeletal systems [8]. Our patients had skin (case 1), brain (case 2), and lung and liver (case 3) abscesses. Legionella had not been considered in the differential diagnosis, although we are well versed in considering this entity in atypical pulmonary presentations in immunocompromised patients [9]. There were no features suggestive of legionellosis on routine tissue stains, and requisite Steiner staining was not performed until molecular diagnosis.
The difficulty in recovering this fastidious organism from culture is another diagnostic challenge. The standard media for Legionella isolation is BCYE agar, which is routinely used only for respiratory specimens [10]. It is plausible that many extrapulmonary cases are underdiagnosed if there is no clinical suspicion for Legionella. In addition, its size and intracellular nature make visualization difficult with Gram and GMS stains, hence the need to use specialized stains (ie, Steiner stain). For our cases in which the diagnosis was made by 16S rRNA gene sequencing, the organisms were seen on Steiner stain upon re-examination by pathology, which opens the debate whether earlier use of Steiner stains should be performed in immunocompromised patients. Furthermore, the urine Legionella antigen detects only L pneumophila serotype 1, so a negative result does not rule out infections by other L pneumophila serotypes or nonpneumophila species.
CONCLUSIONS
Our case series highlights the promising role of molecular techniques to aid in making the diagnosis. For instance, MALDI-TOF MS rapidly and accurately identifies most bacterial and yeast species after recovery from culture with decreased time-to-identification in comparison to conventional microbiologic methods [11]. A recent study found that MALDI-TOF MS was able to provide reliable identification of relevant Legionella species [12]. Legionella pneumophila is included in the US Food and Drug Administration-approved IVD version 2.0 database, and no deoxyribonucleic acid extraction is necessary. However, its utility is limited if no organism is recovered in culture or if its spectrum is not included in the reference databases. Alternatively, 16S rRNA gene sequencing is particularly useful to identify organisms with unusual phenotypes or directly from clinical specimens with slow-growing or uncultivable pathogens [1]. In cases where there are no microbiologic specimens left to culture, 16S rRNA gene sequencing can be performed on FFPE or fresh tissue if available [3]. Hurdles include cost, lack of process automation, and variability among methods of extraction. Nonetheless, one study demonstrated accurate identification of L pneumophila and close to 90% of non-pneumophila species using 16S rRNA gene sequencing [13]. Legionella is ubiquitous in the environment and may be present in reagents and sterile water. In our cases, negative controls were included for each sequencing run to rule out contamination. For nonsterile specimens such as skin, the Sanger technique is useful if the organisms exist dominantly in high frequencies.
In summary, extrapulmonary legionellosis is likely underdiagnosed due to lack of clinician awareness and limitations of conventional microbiologic tests. A high index of clinical suspicion in immunocompromised patients is needed. Adoption of rapid molecular diagnostics such as MALDI-TOF for organisms grown in the laboratory and 16S rRNA Sanger sequencing from tissue specimens increases diagnostic yield, potentially leading to timelier, appropriate treatment.
Acknowledgments
Financial support. This research was funded in part through the National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30 CA008748.
Potential conflicts of interest. All authors: No reported conflicts of interest.All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Clarridge JE., 3rd Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev 2004; 17:840–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. He Y, Chang TC, Li H et al. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry and database for identification of Legionella species. Can J Microbiol 2011; 57:533–8. [DOI] [PubMed] [Google Scholar]
- 3. Moncada PA, Budvytiene I, Ho DY et al. Utility of DNA sequencing for direct identification of invasive fungi from fresh and formalin-fixed specimens. Am J Clin Pathol 2013; 140:203–8. [DOI] [PubMed] [Google Scholar]
- 4. Edelstein PH. Legionella jamestowniensis fatal pneumonia in an immunosuppressed man. J Infect Chemother 2017; 23:59–61. [DOI] [PubMed] [Google Scholar]
- 5. Chow JW, Yu VL. Legionella: a major opportunistic pathogen in transplant recipients. Semin Respir Infect 1998; 13:132–9. [PubMed] [Google Scholar]
- 6. Edelstein PH, Meyer RD, Finegold SM. Isolation of Legionella pneumophila from blood. Lancet 1979; 1:750–1. [DOI] [PubMed] [Google Scholar]
- 7. Lowry PW, Tompkins LS. Nosocomial legionellosis: a review of pulmonary and extrapulmonary syndromes. Am J Infect Control 1993; 21:21–7. [DOI] [PubMed] [Google Scholar]
- 8. Stout JE, Yu VL. Legionellosis. N Engl J Med 1997; 337:682–7. [DOI] [PubMed] [Google Scholar]
- 9. del Castillo M, Lucca A, Plodkowski A et al. Atypical presentation of Legionella pneumonia among patients with underlying cancer: a fifteen-year review. J Infect 2016; 72:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baron EJ, Miller JM, Weinstein MP et al. Executive summary: a guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2013 recommendations by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM)(a). Clin Infect Dis 2013; 57:485–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang AM, Newton D, Kunapuli A et al. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis 2013; 57:1237–45. [DOI] [PubMed] [Google Scholar]
- 12. Gaia V, Casati S, Tonolla M. Rapid identification of Legionella spp. by MALDI-TOF MS based protein mass fingerprinting. Syst Appl Microbiol 2011; 34:40–4. [DOI] [PubMed] [Google Scholar]
- 13. Wilson DA, Reischl U, Hall GS, Procop GW. Use of partial 16S rRNA gene sequencing for identification of Legionella pneumophila and non-pneumophila Legionella spp. J Clin Microbiol 2007; 45:257–8. [DOI] [PMC free article] [PubMed] [Google Scholar]