Abstract
5΄-Vinylphosphonate modification of siRNAs protects them from phosphatases, and improves silencing activity. Here, we show that 5΄-vinylphosphonate confers novel properties to siRNAs. Specifically, 5΄-vinylphosphonate (i) increases siRNA accumulation in tissues, (ii) extends duration of silencing in multiple organs and (iii) protects siRNAs from 5΄-to-3΄ exonucleases. Delivery of conjugated siRNAs requires extensive chemical modifications to achieve stability in vivo. Because chemically modified siRNAs are poor substrates for phosphorylation by kinases, and 5΄-phosphate is required for loading into RNA-induced silencing complex, the synthetic addition of a 5΄-phosphate on a fully modified siRNA guide strand is expected to be beneficial. Here, we show that synthetic phosphorylation of fully modified cholesterol-conjugated siRNAs increases their potency and efficacy in vitro, but when delivered systemically to mice, the 5΄-phosphate is removed within 2 hours. The 5΄-phosphate mimic 5΄-(E)-vinylphosphonate stabilizes the 5΄ end of the guide strand by protecting it from phosphatases and 5΄-to-3΄ exonucleases. The improved stability increases guide strand accumulation and retention in tissues, which significantly enhances the efficacy of cholesterol-conjugated siRNAs and the duration of silencing in vivo. Moreover, we show that 5΄-(E)-vinylphosphonate stabilizes 5΄ phosphate, thereby enabling systemic delivery to and silencing in kidney and heart.
INTRODUCTION
Small interfering RNAs (siRNAs) guide the sequence-specific cleavage of targeted mRNAs (1,2). The ability to design and chemically synthesize an siRNA against virtually any target gene offers a powerful therapeutic strategy to treat genetic diseases, particularly those for which small molecule drugs do not exist (such as Huntington΄ s disease and other neurodegenerative diseases). The sequence of an siRNA determines its target, but the chemical architecture determines its pharmacokinetic behavior (3). Thus, siRNAs can readily be tailored to fit the needs of personalized medicine.
The clinical utility of siRNA therapeutics has been limited by in vivo stability and safe, efficient delivery to tissues. Both challenges are being met by advances in oligonucleotide chemistry (4–7). Hydrophobic conjugates—e.g. cholesterol—drive efficient cellular uptake of siRNA via a general mechanism (4,8), which may enable targeting of a wide range of tissues. Extensive chemical modification of conjugated siRNAs improves stability and activity in vivo (4,9,10). siRNA compounds currently in clinical studies are modified using a combination of 2΄-fluoro (2΄-F) and 2΄-O-methyl (2΄-O-Me) modifications (5,9,11).
The guide strand of an siRNA duplex must bear a 5΄-phosphate to bind the effector protein of the RNA-induced silencing complex Argonaute 2 (AGO2) (12–15). The in vivo phosphorylation state of a synthetic siRNA depends on the balance of kinase and phosphatase activity. A dephosphorylated siRNA must be phosphorylated for effectiveness in vivo; however, fully chemically modified siRNAs are poor substrates for intracellular kinases (16). Therefore, to preserve proper 5΄ phosphorylation, phosphonates can be used as metabolically stable phosphate analogs. The stability resides in the carbon-phosphorus bond of phosphonates that resists phosphatases, which hydrolyze oxygen-phosphorus bonds (17). Among phosphonates tested, 5΄-(E)-vinylphosphonate appears to be the most effective phosphate analog in siRNAs (18–21). Indeed, both single stranded siRNAs and GalNAc-conjugated double stranded siRNAs benefit from 5΄-(E)-vinylphosphonate modification (22–25).
Here we evaluate how chemical phosphorylation of hydrophobically modified siRNAs (hsiRNAs) with either phosphate, or the metabolically stable phosphate analog 5΄-(E)-vinylphosphonate, impacts efficacy and duration of effect in vitro and in vivo. We show that 5΄-phosphate and 5΄-(E)-vinylphosphonate equally enhance hsiRNA activity in vitro. When administered in vivo, 5΄-phosphate hsiRNAs are de-phosphorylated within hours, but metabolic stabilization with 5΄-(E)-vinylphosphonate significantly increases retention of hsiRNAs in tissues, silencing activity, and duration of effect. The 5΄-(E)-vinylphosphonate, cholesterol-conjugated hsiRNAs remain active in liver and kidneys for at least 6 weeks after single administration. 5΄-(E)-vinylphosphonate hsiRNAs silences target genes in the heart, a tissue previously not accessible by conjugated siRNAs. Finally, we show that 5΄-(E)-vinylphosphonate not only resists phosphatases in vivo, but also resists 5΄-phosphate-dependent exonucleolytic destruction by XRN1, contributing to overall stabilization of the guide strand in tissues.
MATERIALS AND METHODS
mRNA quantification from cells and tissue punches
HeLa cells (ATCC, #CCL-2) were plated in DMEM (Cellgro, #10-013CV) supplemented with 6% fetal bovine serum (FBS; Gibco, #26140) at 10 000 cells per well in 96-well tissue culture plates. hsiRNA was diluted in OptiMEM (Gibco, #31985-088) and added to cells, resulting in 3% FBS. Cells were incubated for 72 h at 37°C, 5% CO2. Cells were lysed and mRNA quantification was performed using the QuantiGene 2.0 assay kit (Affymetrix, #QS0011) as described previously (26). For in vivo experiments, mice were euthanized and organs placed in RNAlater (Sigma, #R0901) at 4°C overnight. Then tissue punches (∼10 mg) were taken using 1.5 mm disposable biopsy punch with plunger (Integra, Miltex, # 33-31A-P/25). Tissue punches were lysed and mRNA quantification was performed using the QuantiGene 2.0 assay kit (Affymetrix, #QS0011) as described previously (26). Catalog numbers for probes used in QuantiGene 2.0 assay kit are as follows: human HTT (Affymetrix, #SA-50339), mouse Htt (Affymetrix, #SB-14150), human PPIB (Affymetrix, #SA-10003), mouse Ppib (Affymetrix, #SB-10002), human HPRT (Affymetrix, #SA-10030), mouse Hprt (Affymetrix, #SB-15463). Data sets were normalized to housekeeping gene HPRT.
In both in vitro and in vivo experiments, hsiRNAPPIB was used as non-targeting control (NTC) for HTT silencing, and hsiRNAHTT was used as non-targeting control (NTC) for PPIB silencing.
PNA (peptide nucleic acid) based assay for quantitation of hsiRNA and detection of hsiRNA metabolites in mouse tissues
Tissues punches (10 mg) were lysed in 100 μl MasterPure™ Tissue Lysis Solution (EpiCentre®) in the presence of proteinase K (2 mg/ml; Invitrogen, #25530-049) in TissueLyser II (Qiagen). SDS was precipitated from the lysate using KCl (3M) and pelleted at 5000 × g for 15 min. hsiRNA in cleared supernatant was annealed to a Cy3-labeled PNA that was fully complementary to the guide strand (PNABio, Thousand Oaks, CA, USA) by heating to 95°C for 15 min, incubating at 50°C for 15 min, and cooling to room temperature. Tissue lysates containing PNA-guide strand hybrids were injected into HPLC DNAPac® PA100 anion-exchange column (Thermo Fisher Scientific Inc.), Cy3 fluorescence was monitored, and peaks were integrated. The mobile phase for HPLC was Buffer A (50% water, 50% acetonitrile, 25 mM Tris–HCl, pH 8.5, 1 mM EDTA) and Buffer B (800 mM NaClO4 in buffer A). For hsiRNA guide strand quantitation, a steep gradient of Buffer B (10–100% in 2.5 min) was used, and for hsiRNA guide strand metabolite detection a shallow gradient of Buffer B (10–100% in 18 min) was applied. For calibration curve, known amounts of hsiRNA duplex was spiked into the tissue lysis solution derived from untreated mice before annealing to PNA. Fluorescent peaks (excitation 550 nm, emission 570 nm) corresponding to hsiRNA-guide strand–PNA hybrid were recorded, integrated and calibration curve generated by correlating the area under the curve (AUC) of the hsiRNA-PNA fluorescent peak with the spiked amounts of hsiRNA duplex.
Animal experiments
Animal experiments were performed in accordance with guidelines of University of Massachusetts Medical School Institutional Animal Care and Use Committee (IACUC, protocol number A-2411). Mice were 6- to 10-week old at the time of experiments. All animals were kept on a 12-h light/dark cycle in a pathogen-free facility, with food and water provided ad libitum.
For systemic administration of hsiRNA, FVBNj female mice were injected with either phosphate buffered saline (PBS) or with different amounts of hsiRNA resuspended in PBS, either through the tail vein or subcutaneously at the nape of the neck.
After a time of incubation—indicated on individual figures—mice were deeply anesthetized with 0.1% Avertin, and after cervical dislocation, tissues were harvested and stored in RNAlater (Sigma, #R0901) for later use. We used a single injection per mouse in each experiment.
Statistical analysis
Data were analyzed using GraphPad Prism 7 software. IC50 curves were fitted using log(inhibitor) versus response—variable slope (four parameters). For in vivo systemic silencing, the significance was calculated using One-way ANOVA with Bonferroni΄ s multiple comparisons. To compare hsiRNA guide strand concentrations measured by PNA assay, the data were analyzed by One-Way ANOVA with Tukey΄ s multiple comparisons. For in vivo duration of effect, the data were analyzed by two-way ANOVA with Holm–Sidak correction. Differences were considered significant at P values <0.05 compared to the PBS injected group.
In vitro XRN1 resistance assay
hsiRNA guide strands (30 pmol) were incubated in water or with 1 μl Terminator™ (EpiCentre) exonuclease overnight at 37°C in buffer A (EpiCentre, provided with Terminator™ enzyme). Then Novex® high-density TBE sample buffer (5×) (Thermo Fisher Scientific) was added to samples and loaded to polyacrylamide gel. Denaturing polyacrylamide gels (24%) were made in house using a mixture of 4 ml 10× TBE (Tris/borate/EDTA buffer), 17 g urea, 14 ml acrylamide:bis-acrylamide (19:1) 40% solution (Bio-Rad), 400 μl of 10% APS (ammonium persulfate) and 30 μl of TEMED (tetramethylethylanediamine). Urea–PAGE was performed in 1× TBE at 500 V at room temperature (SE600 system, Hoefer) for ∼6 h. Gels were stained with SYBR® Gold Nucelic Acid Gel Stain (Thermo Fisher Scientific) and imaged with Typhoon FLA 9000 (GE Healthcare).
RESULTS
5΄ Chemical phosphorylation enhances hsiRNA efficacy in vitro
Hydrophobically modified hsiRNAs (Figure 1A) are asymmetric siRNAs with alternating 2΄-F and 2΄-O–Me modification of each ribose to resist endonucleases and to avoid innate immune activation. The 2΄-F and 2΄-O–Me modifications offset each other in the short (15 base-pair) double-stranded region. Phosphorothioate linkages at the ends of both strands and throughout the single-stranded 3΄ tail of the guide strand enhance cellular uptake of hsiRNAs. A cholesterol group linked to the 3΄ end of the passenger strand drives unassisted cellular uptake. hsiRNAs rapidly (within minutes) and efficiently enter cells through an EEA1-related endocytosis pathway (27), and they show in vivo efficacy after local administration (4,28,29).
Figure 1.
5΄ phosphorylation of hsiRNAs increases activity in vitro. (A) Cartoon of hsiRNA chemical scaffold. (B) Graphs showing the levels of PPIB (left) or HTT mRNA (right) in HeLa cells treated with 5΄-hydroxyl (5΄-OH) or 5΄-phosphate (5΄-P) hsiRNAPPIB (left) or hsiRNAHTT (right) or non-targeting control (NTC) hsiRNAs. mRNA levels measured by QuantiGene® 2.0 assay are normalized to the level of a control HPRT mRNA and shown as percent of the untreated (UNT) control. The IC50 for the experimental hsiRNAs are indicated in the legend. N = 3 for each datapoint.
hsiRNAs targeting PPIB (peptidylprolyl isomerare B or cyclophilin B) and HTT (huntingtin) were synthesized with 5΄-hydroxyl and 5΄-phosphate on the guide strand (see Supplementary Table S1 for sequences and chemical modification patterns of hsiRNAs used in this study). Chemical phosphorylation at the 5΄ end significantly increased the level of target mRNA silencing and hsiRNA potency (2.7-fold for hiRNAPPIB, p < 0.001 and 1.6-fold hiRNAHTT, p = 0.01) compared to 5΄-hydroxyl hsiRNA. Thus, chemical phosphorylation significantly contributes to the overall potency of hsiRNAs in vitro in passive uptake, with the level of impact showing sequence dependence.
hsiRNAs are quickly dephosphorylated in vivo after systemic administration
Efficient delivery of conjugated hsiRNA requires chemical modifications that are resistant against metabolic cleavage. To evaluate metabolic stability of chemically introduced 5΄-phosphate in vivo, we administered 10 mg/kg of 5΄-phosphate hsiRNA systemically into mice by tail vein injection. We analyzed the livers of injected mice after 2, 24 or 120 hours for the presence of hsiRNA guide strand metabolites. For comparison, we also analyzed a series of predicted guide strand metabolites (references), which we added into liver lysates from PBS treated animals (Supplementary Figure S1A and Supplementary Figure S2). Tissues were lysed and hsiRNA guide strand metabolites were detected using a peptide nucleic acid (PNA)-based hybridization assay (30) and anion exchange chromatography. Correlation of elution times between the references and metabolites extracted from mouse livers identified major hsiRNA degradation products.
We failed to detect intact hsiRNA guide strands in livers harvested 2 hours after injection (Supplementary Figure S1B), indicating that systemically administered hsiRNAs, in spite of being fully modified by 2΄-F and 2΄-O–Me, are rapidly metabolized in vivo. At each time point, we observed two major chromatography peaks whose elution times correlated with that of a 5΄-hydroxyl full-length (20-nucleotide) guide strand and a 5΄-hydroxyl 19-nucleotide metabolite with one nucleotide removed from the 3΄ end (Supplementary Figure S1B). Mass spectrometry analysis confirmed the identity of both metabolites (Supplementary Figure S1C), and resolved additional products, including a 5΄-phosphate metabolite trimmed at the 3΄ end by 1 nucleotide (product 2, Supplementary Figure S1A) and further 5΄-hydroxyl products trimmed at the 3΄ end by two or three nucleotides (products 4 and 5 in Supplementary Figure S1A). At 2 hours after injection, most of the hsiRNA guide strand was dephosphorylated at the 5΄ end, and by 24 hours the levels of full-length 5΄-phosphate guide strand could not be distinguished from noise. Thus, the primary degradation events in vivo appear to be eliminating of the 5΄-phosphate and trimming the 3΄ end. Although RISC should be able to load a phosphorylated guide strand trimmed by one or two 3΄ nucleotides (31), dephosphorylated guide strands are expected to be significantly less active, thereby limiting in vivo efficacy.
5΄-(E)-Vinylphosphonate modification could be added to all sequences of hsiRNA and is fully active in vitro
We sought to compare the stability and activity of hsiRNAs with 5΄-phosphate or a metabolically stabilized 5΄-(E)–vinylphosphonate. The unsaturated C–C bond of 5΄-(E)–vinylphosphonate restricts the torsion angle to 180°, and the resulting trans- or E-configuration mimics the optimal electronic and spatial positioning of 5΄-phosphate (20) (Figure 2A). We synthesized the phosphoramidite of 5΄-(E)-vinylphosphonate 2΄-O–Me-uridine and incorporated it into the guide strand as a last coupling during the 3΄-5΄ oligonucleotide chemical synthesis. Since 5΄ terminal base is not involved in RISC - target mRNA interaction (25,32), the 5΄-(E)-vinylphosphonate 2΄-O–Me-uridine phosphoramidite can be used to incorporate 5΄-(E)-vinylphosphonate in any siRNA independently of target gene and sequence (Figure 2 and Supplementary Figure S3). When tested in vitro by passive uptake, 5΄-(E)-vinylphosphonate hsiRNAs were just as effective as 5΄-phosphate hsiRNAs, confirming that the 5΄-(E)-vinylphosphonate modification is well tolerated by the RNA-induced silencing complex (RISC) assembly (Figure 2B).
Figure 2.
Stabilization of the 5΄ end by 5΄-(E)-vinylphosphonate supports RISC loading and activity. (A) Chemical structures of 5΄phosphate (5΄-P) and 5΄-(E)-vinylphosphonate (5΄-VP). (B) Except that HeLa cells treated with 5΄-P or 5΄ -VP modified hsiRNAs, as indicated in the legend.
5΄-(E)-Vinylphosphonate hsiRNA outperforms 5΄-phosphate hsiRNA in kidney and heart, and is comparable in liver and spleen
Current clinical stage siRNAs accumulate in liver (7,10). Therefore, we first compared the efficacy of 5΄-hydroxyl, 5΄-phosphate and 5΄-(E)-vinylphosphonate hsiRNAs in the liver after intravenous or subcutaneous administration. We used previously identified hsiRNA sequences that efficiently silence Ppib or Htt mRNAs (Supplementary Table S1; (4,28)). All 5΄ variants of hsiRNAHtt reduced Htt mRNA levels in liver by ∼60% when administered subcutaneously and by ∼40% when administered intravenously (Figure 3). In contrast, 5΄-(E)-vinylphosphonate modified hsiRNAPpib silenced Ppib mRNA substantially better (66% when administered subcutaneously and 57% when administered intravenously) than 5΄-phosphate or 5΄-hydroxyl hsiRNAs (30–50% when administered subcutaneously and 25–30% when administered intravenously)(Figure 3).
Figure 3.
5΄ -(E)-vinylphosphonate modification enhances hsiRNA efficacy in liver, kidneys and heart column scatter plots showing Ppib or Htt mRNA levels in the livers, kidneys and hearts of mice (n = 5 per group) treated with 5΄-hydroxyl (5΄-OH), 5΄-phosphate (5΄-P), or and 5΄-(E)-vinylphosphonate (5΄-VP) hsiRNAs by intravenous (IV) of subcutaneous (SC) injection. mRNA levels measured by QuantiGene® 2.0 assay, were normalized to Hprt mRNA and expressed as percent of mRNA levels in PBS-treated animals. NTC, non-targeting control. T, targeting hsiRNA. Significance calculated by ANOVA with Bonferroni΄ s correction: ns, non-significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 and ****P ≤ 0.0001.
The cholesterol moiety conjugated to hsiRNAs is highly hydrophobic and drives non-specific uptake of hsiRNA by many cell and tissue types such as muscle (8) and eye (4). We therefore asked whether subcutaneous delivery of hsiRNA could promote silencing in other organs. We specifically looked at kidney as this is prime location for drug clearance, spleen as a major member of the reticuloendothelial system, and heart as a clinically interesting target organ. Since hsiRNA accumulation was expected to be substantially lower in these organs (i.e. kidneys, heart and spleen) than in liver, we used a higher dose (20 mg/kg) to ensure sufficient hsiRNA accumulation in secondary tissues to support silencing. Subcutaneous administration of 5΄phosphate hsiRNAPPIB silenced Ppib mRNA by 28% in spleen (Supplementary Figure S4) but did not lead to silencing in kidney and heart. 5΄-phosphate hsiRNAHTT did not result in silencing Htt mRNAs in kidney, spleen or heart (Figure 3). The 5΄-hydroxyl hsiRNAs supported 50% silencing of Ppib and 20% silencing of Htt in kidney, 15% Ppib silencing but no Htt silenicing in spleen (Supplementary Figure S4) and no Ppib silencing but 35% Htt silencing in heart (Figure 3). The metabolically stabilized 5΄-(E)-vinylphosphonate hsiRNAs showed the highest activity, showing 64% Ppib mRNA silencing and 40% Htt mRNA silencing in kidney (Figure 3), 32% Ppib silencing and 25% Htt silencing (P = 0.06) in spleen (Supplementary Figure S4), as well as 46% Ppib silencing and 51% Htt silencing in heart (Figure 3).
5΄-(E)-Vinylphosphonate improves hsiRNA accumulation in multiple tissues
Using the PNA-based hybridization assay to quantify hsiRNA levels in tissue lysates, we found that the metabolically stable 5΄-(E)-vinylphosphonate hsiRNAs accumulated to significantly higher concentrations (up to 22-fold) than 5΄-hydroxyl hsiRNAs in liver, kidney, heart, and spleen (Figure 4A and B). When administered intravenously, the positive impact of 5΄-(E)-vinylphosphonate modification on hsiRNA concentration was the highest in heart (Figure 4B). In all tissues examined, 5΄-(E)-vinylphosphonate hsiRNAs accumulated to higher levels than 5΄-phosphate hsiRNAs, regardless of the administration route or hsiRNA sequence (Figure 4A and B). Hence, the metabolic stabilization of the 5΄-phosphate via 5΄-(E)-vinylphosphonate has resulted in overall increase of guide strand tissue accumulation, which could explain improved in vivo activity of 5΄-(E)-vinylphosphonate hsiRNAs.
Figure 4.
Metabolic stabilization of the 5΄ phosphate increases retention in both primary and secondary tissues. (A) Bar graphs showing the concentrations of 5΄-hydroxyl (5΄-OH), 5΄-phosphate (5΄-P), or and 5΄-(E)-vinylphosphonate (5΄-VP) hsiRNA guide strands and their metabolites in liver (primary tissue), kidneys, heart and spleen (secondary tissues), one week after intravenous (IV) or subcutaneous (SC) injection. Concentrations measured by PNA hybridization assay. Bar graphs show the mean ± SD, n = 5 mice per group. (B) Summary of fold change in concentrations of 5΄-P and 5΄-VP hsiRNA guide strands compared to 5΄-OH hsiRNA guide strand. Fold changes are color-coded: high fold changes in red and low fold changes in white. Fold changed were calculated by dividing average hsiRNA concentrations in respective organs from n = 5 mice per group.
5΄-(E)-vinylphosphonate modification of hsiRNA confers resistance to phosphatases and to 5΄-to-3΄ exoribonuclease XRN1
The improved retention and silencing activity of 5΄-(E)-vinylphosphonate hsiRNAs in vivo could reflect protection from natural phosphatases and nucleases.
Phosphonate bonds have been shown to resist snake venom phosphatase (17,20) and 5΄-(E)-vinylphosphonate siRNAs have performed better in vivo than 5΄-hydroxyl siRNAs (Figure 3, (23,24)) consistent with phosphatase resistance. However, no direct evidence exists to date that 5΄-(E)-vinylphosphonate resist phosphatases in vivo in a mouse. We therefore used a PNA-based hybridization assay (30) to resolve phosphorylated and dephosphorylated hsiRNA metabolites in liver lysates from mice injected with 20 mg/kg 5΄-hydroxyl, 5΄-phosphate or 5΄-(E)-vinylphosphonate hsiRNAs. As a control, we analyzed untreated liver lysates, into which we added intact hsiRNAs with 5΄-hydroxyl, 5΄-phosphate, or 5΄-(E)-vinylphosphonate end. The control chromatography shows 5΄-phosphate and 5΄-(E)-vinylphosphonate hsiRNAs elute with overlapping profiles, and 5΄-hydroxyl hsiRNAs elute earlier, reflecting the difference of one charge (the phosphate) absent in the 5΄-hydroxyl hsiRNA (Figure 5A, left panels). In livers harvested from mice 1 week after injection, however, we found that chromatography profiles of 5΄-phosphate hsiRNA converged with the profile of 5΄-hydroxyl hsiRNA, whereas 5΄-(E)-vinylphosphonate hsiRNA guide strand eluted later (Figure 5A, middle and left graphs), regardless of administration route or sequence. These findings are consistent with rapid dephosphorylation (and 3΄ trimming) of 5΄-phosphate hsiRNA and protection of 5΄-(E)-vinylphosphonate hsiRNAs from dephosphorylation.
Figure 5.
5΄ -(E)-vinylphosphonate infers resistance against phosphatases in vivo and 5΄-3΄ exonuclease in vitro. (A) HPLC traces of Cy3-PNA/hsiRNA hybrids in liver lysates from mice harvested 1 week after intravenous (IV) or subcutaneous (SC) injection with 5΄-hydroxyl (5΄-OH), 5΄-phosphate (5΄-P), or and 5΄-(E)-vinylphosphonate (5΄-VP) hsiRNA. The ‘spike-in’ (left) panels show control traces of Cy3-PNA/hsiRNA hybrids after full-length guide strands were spiked into liver lysates from untreated mice. 5΄-P spike-in guide strands elute more slowly than 5΄-OH guide strands, corresponding to the difference of one charge (phosphate). Metabolite profile in liver lysates clearly show dephosphorylation of 5΄-P hsiRNA but partially intact 5΄ end of 5΄-VP hsiRNA. (B) Urea-PAGE of 5΄-hydroxyl (5΄-OH), 5΄-phosphate (5΄-P), or and 5΄-(E)-vinylphosphonate (5΄-VP) hsiRNAs resolved on an 7 M urea/24% polyacrylamide gel after 12 h incubation in the absence (–) or presence (+) of Terminator™ enzyme. 5΄-P and 5΄-VP (no PS) compound had the same nucleotide modification pattern (2΄-F, 2΄-O-Me) as 5΄-P, 5΄-OH and 5΄ -VP, but did not contain phosphorothioate (PS) internucleotide linkages. 5΄-VP hsiRNA is protecting against degradation by Terminator™ enzyme, whereas 5΄-P hsiRNA is degraded, and phosphorothioate internucleotide linkage interferes with enzyme processivity.
Nucleic acids with a 5΄ phosphate are sensitive to the 5΄-to-3΄ exoribonuclease XRN1 (33). Furthermore, XRN1 and XRN2 play a role in miRNA stability (34–36), especially if miRNA is taken up from the extracellular space (37). XRN1 also regulates accumulation of viral dsRNA (38). Taken together with the observation that 5΄-(E)-vinylphosphonate hsiRNA showed higher concentrations in several tissues than 5΄-phosphate hsiRNA, we hypothesized that 5΄-(E)-vinylphosphonate hsiRNA may have improved stability due to XRN1 resistance. To test whether 5΄-(E)-vinylphosphonate modification protects against 5΄-to-3΄ exoribonuclease-mediated degradation, we treated 5΄-phosphate, 5΄-hydroxyl, and 5΄-(E)-vinylphosphonate hsiRNA guide strands with Terminator™ 5΄-phosphate-dependent exoribonuclease (i.e. recombinant XRN1) overnight and then resolved potential metabolites by denaturing polyacrylamide gel electrophoresis. Whereas we detected a 5΄-phosphate hsiRNA guide strand metabolite shortened by ∼1 nt, the 5΄-hydroxyl and 5΄-(E)-vinylphosphonate hsiRNA guide strands—regardless of sequence—remained intact (Figure 5B). This finding indicates that 5΄-(E)-vinylphosphonate protects against turnover by 5΄-phosphate-specific exoribonucleases. We hypothesized that the 19mer metabolite of 5΄-phosphate hsiRNA could be a result of interference between phosphorothioate internucleotide linkage with XRN1 processivity. Therefore, we synthesized 5΄-phosphate and 5΄-(E)-vinylphosphonate hsiRNAs fully modified with 2΄-O-Me and 2΄-F but containing no phosphorothioates. The guide strand of 5΄phosphate hsiRNA lacking phosphorothioates was fully degraded by Terminator™, whereas 5΄-(E)-vinylphosphonate hsiRNA guide strand containing no phosphorothioates was protected (Figure 5B). These data suggest that 5΄-(E)-vinylphosphonate modification is sufficient to protect hsiRNA guide strand from degradation by 5΄-to-3΄ exoribonuclease. Overall, resistance to phosphatases and to the exoribonuclease XRN1 underlie the improved in vivo stability of 5΄-(E)-vinylphosphonate -modified hsiRNAs, leading to improved silencing performance.
5΄-(E)-vinylphosphonate modification improves hsiRNA duration of effect in vivo
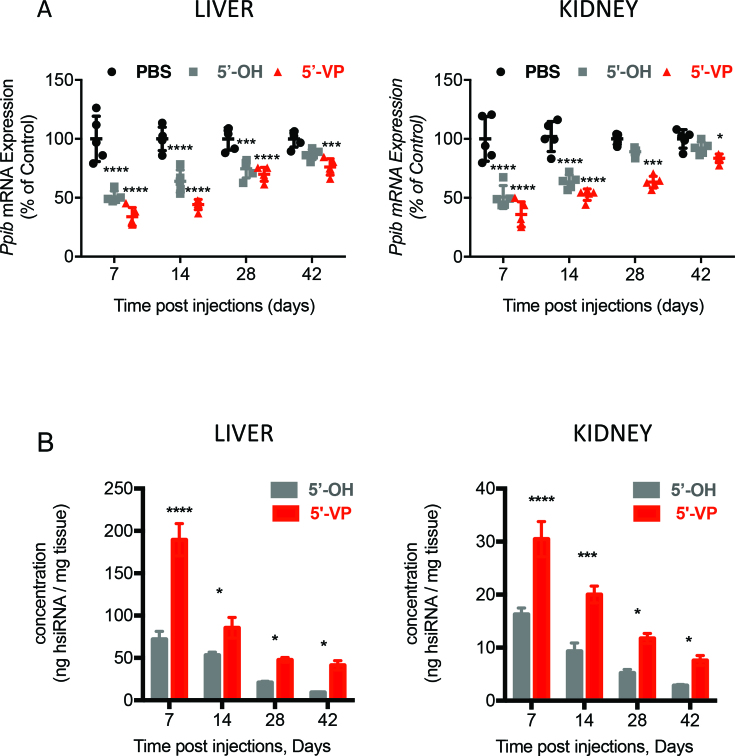
The improved in vivo stability and silencing performance of 5΄-(E)-vinylphosphonate hsiRNA could translate into improved duration of effect. We tested this hypothesis by subcutaneously injecting a single 20 mg/kg dose of 5΄-hydroxyl or 5΄-(E)-vinylphosphonate hsiRNAPpib and monitoring the levels of Ppib mRNA and hsiRNA in mouse kidney and liver at different time points. We found that the silencing of Ppib mRNA and hsiRNAPpib concentration in kidney and liver gradually decreased with time, but the level of Ppib mRNA silencing and retention of 5΄-(E)-vinylphosphonate hsiRNA exceeded that of 5΄-hydroxyl hsiRNA at each time point (Figure 6). Metabolically stable phosphate containing 5΄-(E)-vinylphosphonate hsiRNA maintained significant silencing in liver and kidney, from ∼70% Ppib mRNA silencing in both tissues after 1 week to ∼24% (liver) and ∼17% (kidney) silencing after 6 weeks. The 5΄-hydroxyl hsiRNA showed ∼50% target mRNA silencing after 1 week in both liver and kidney, but lost significant silencing activity within 4 weeks in liver and within 2 weeks in kidney (Figure 6A). The 5΄-(E)-vinylphosphonate hsiRNA concentration remained two to four times higher than the concentration of 5΄-hydroxyl hsiRNA in liver and kidney at each time point (Figure 6B). Hence, stabilization of 5΄ end by 5΄-(E)-vinylphosphonate supports increased tissue content, increased silencing activity and increased duration of effect when compared to 5΄hydroxyl hsiRNA.
Figure 6.
Stabilization of 5΄ phosphate increases duration of effect and corresponding tissue accumulation of hsiRNA in vivo. (A) Column scatter plots showing Ppib mRNA levels in the livers and kidneys of mice (n = 5 per group) at the indicated times after subcutaneous injection of PBS or 5΄-OH or 5΄-VP hsiRNAs. mRNA levels measured by QuantiGene® assay were normalized to Hprt mRNA and expressed as a percent of mRNA levels in PBS-treated animals. Significance calculated by ANOVA with Bonferroni΄ s correction, significance was calculated in comparison to PBS-treated animals: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 and ****P ≤ 0.0001. Mean ± SD. (B) Bar graphs showing the concentration of 5΄-OH and 5΄-VP hsiRNA guide strands in liver and kidney, as measured by the PNA-based hybridization assay at the indicated time points after subcutaneous injection. Mean ± SD, N = 50.
DISCUSSION
Here, we have shown that 5΄-(E)-vinylphosphonate modification confers clinically useful properties to siRNA drugs. These properties include increase in siRNA tissue concentration and duration of silencing, which in turn may allow lowering of dose and frequency of administration. We have identified two underlying mechanisms: resistance to phosphatases and resistance to 5΄-to-3΄ exonucleases. We and others have shown that 5΄-(E)-vinylphosphonate is recognized as a phosphate mimic by AGO2 (a nuclease in the RNA-induced silencing complex, RISC) and allows or even facilitates loading of siRNA guide strand into RISC (23,25). However, our data suggests that 5΄-(E)-vinylphosphonate is not recognized as a phosphate mimic by the main cytoplasmic 5΄-to-3΄ exonuclease, XRN1. Hence, 5΄-(E)-vinylphosphonate modification improves both the pharmacodynamic (better loading to RISC) and pharmacokinetic (slower degradation) behavior of an siRNA through altering protein binding.
The impact of 5΄-(E)-vinylphosphonate on siRNA silencing activity showed sequence-dependence in liver but not in kidney and heart. Different hsiRNA-to-target mRNA ratios could explain this observation. In mouse liver, Ppib mRNA is expressed at a fifty times higher level than Htt mRNA (FPKM ∼184 versus FPKM ∼4; Proteinatlas.org), whereas hsiRNAPPIB and hsiRNAHTT concentrations were very similar (∼200 ng/mg). Therefore, the injected dose of 5΄-phosphate or 5΄-hydroxyl hsiRNA may be above the level needed to fully silence Htt in liver, but below the level needed to fully silence Ppib. Increased concentration of 5΄-(E)-vinylphosphonate hsiRNA would therefore improve silencing of Ppib but not that of Htt. hsiRNA concentrations are approximately six times lower in kidney and twenty times lower in heart than in liver, while mRNA expression levels are within the same range. We propose that 5΄-(E)-vinylphosphonate only improves silencing activity when hsiRNA-to-target mRNA ratios are below the level of saturation.
We observed that modification of the 5΄ end of hsiRNAs influenced silencing activity differently in vitro and in vivo. First, 5΄-(E)-vinylphosphonate hsiRNA was equally active in vitro compared to 5΄-phosphate hsiRNA, but significantly more active in vivo. This is explained by rapid dephosphorylation of 5΄-phosphate hsiRNA in vivo (Supplementary Figure S1 and (39)) In parallel, 5΄-hydroxyl hsiRNA was less active in vitro but more active in vivo compared to 5΄-phosphate hsiRNA. Indeed, unmodified 5΄-hydroxyl siRNA has been found to be more efficacious in vivo than unmodified 5΄-phosphate siRNA (40). Furthermore, all siRNAs showing efficacy to date in clinical trials have 5΄-hydroxyl ends (11,41,42). The mechanism why 5΄-hydroxyl siRNA performs superior to 5΄-phosphate siRNA in vivo is unclear. We propose that the 5΄ end of the guide strand may influence protein binding profile of the siRNAs in subcutaneous extracellular space or in serum or in lymph. siRNA-bound proteins or peptides can alter the trafficking (43) and phosphorylation (39) of siRNAs. The different impact of 5΄end of the guide strand on silencing activity in in vitro and in vivo experimental settings may be explained by exposure to a different protein environment.
Concentration of 5΄-(E)-vinylphosphonate hsiRNA was 4–22-fold higher than concentration of 5΄-hydroxyl hsiRNA, while silencing of 5΄-(E)-vinylphosphonate hsiRNA was only 0.9–3.3-fold higher than silencing of 5΄-hydroxyl hsiRNA. This phenomenon has been observed before (23) and indicates a non-linear relationship between tissue concentration and silencing activity. Resistance to exonucleases shown in this paper is only one contributor to the concentration—silencing activity relationship. Other factors may be proteins or peptides differentially binding to the different 5΄ends of hsiRNAs and influence phosphorylation (39) or endosomal release (44).
We found better silencing activity following subcutaneous administration compared to intravenous administration, as has been observed in the case of GalNac-conjugated siRNAs as well (7). Subcutaneous administration results in slower release and longer residence time of the hsiRNA compared to intravenous administration. We speculate that certain serum proteins carry hsiRNAs to the organs and the hsiRNA binding capacity of the serum is saturated upon intravenous administration. Binding to the proper carrier protein may enable a productive cellular entry pathway.
In this study, we used hsiRNA modified with a combination of 2΄-F, 2΄-O-Me and phosphorothioates. These siRNA modifications can support a duration of silencing up to 6 months in vivo (11). The mechanism may be explained by enhanced nuclease stability and formation of an intracellular depot, which is slowly releasing siRNAs for continuous reloading of AGO2 (45). However, recent findings suggest that reduction of 2΄-F modification of an siRNA could improve safety profiles (11,46,47). Thus, carefully fine-tuning the amount of 2΄-F modifications in 5΄-(E)-vinylphosphonate hsiRNA might be crucial to ensure clinical safety and success.
5΄-(E)-Vinylphosphonate hsiRNA leads to long-lasting Huntingtin mRNA silencing in liver, heart and kidneys. Lowering Huntingtin mRNA could be beneficial in treating peripheral symptoms and improving quality of life of patients with Huntington's disease (48–50).
DEFINITIONS
Conjugated siRNA: sense strand of siRNA is covalently conjugated to a small molecule (i.e. cholesterol), Fully modified siRNA: all nucleotides are modified at the 2΄ position with either 2΄-F or 2΄-O-methyl, Metabolically stabilized phosphate: 5΄-(E)-vinylphosphonate.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Darryl Conte for help in editing manuscript. We thank Mike Moazami for contribution in mouse work and Matthew R. Hassler for maintaining infrastructure for oligonucleotide synthesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CHDI Foundation [A-6119, JSC A6367]; National Institute of Health [RO1GM10880302, RO1NS03819415, S10 OD020012]; Milton-Safenowitz Post-Doctoral Fellowship from the Amytrophic Lateral Sclerosis Association (to B.M.D.C.G.). The open access publication charge for this paper has been waived by Oxford University Press—NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. A.K. owns stock at RXi Pharmaceuticals and Advirna LLC, which holds a patent and license on asymmetric, hydrophobically modified siRNAs. Other authors do not have any competing financial interest to disclose.
REFERENCES
- 1. Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C.. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998; 391:806–811. [DOI] [PubMed] [Google Scholar]
- 2. Zamore P.D. RNA interference: listening to the sound of silence. Nat. Struct. Biol. 2001; 8:746–750. [DOI] [PubMed] [Google Scholar]
- 3. Khvorova A., Watts J.K.. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017; 35:238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Byrne M., Tzekov R., Wang Y., Rodgers A., Cardia J., Ford G., Holton K., Pandarinathan L., Lapierre J., Stanney W. et al. Novel hydrophobically modified asymmetric RNAi compounds (sd-rxRNA) demonstrate robust efficacy in the eye. J. Ocul. Pharmacol. Ther. 2013; 29:855–864. [DOI] [PubMed] [Google Scholar]
- 5. Allerson C.R., Sioufi N., Jarres R., Prakash T.P., Naik N., Berdeja A., Wanders L., Griffey R.H., Swayze E.E., Bhat B.. Fully 2΄-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005; 48:901–904. [DOI] [PubMed] [Google Scholar]
- 6. Deleavey G.F., Watts J.K., Alain T., Robert F., Kalota A., Aishwarya V., Pelletier J., Gewirtz A.M., Sonenberg N., Damha M.J.. Synergistic effects between analogs of DNA and RNA improve the potency of siRNA-mediated gene silencing. Nucleic Acids Res. 2010; 38:4547–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nair J.K., Willoughby J.L., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel’in A.V., Milstein S. et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014; 136:16958–16961. [DOI] [PubMed] [Google Scholar]
- 8. Khan T., Weber H., DiMuzio J., Matter A., Dogdas B., Shah T., Thankappan A., Disa J., Jadhav V., Lubbers L. et al. Silencing Myostatin Using Cholesterol-conjugated siRNAs Induces Muscle Growth. Mol. Ther. Nucleic Acids. 2016; 5:e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Matsuda S., Keiser K., Nair J.K., Charisse K., Manoharan R.M., Kretschmer P., Peng C.G., A V.K.i., Kandasamy P., Willoughby J.L. et al. siRNA conjugates carrying sequentially assembled trivalent N-acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chem. Biol. 2015; 10:1181–1187. [DOI] [PubMed] [Google Scholar]
- 10. Sehgal A., Vaishnaw A., Fitzgerald K.. Liver as a target for oligonucleotide therapeutics. J. Hepatol. 2013; 59:1354–1359. [DOI] [PubMed] [Google Scholar]
- 11. Fitzgerald K., White S., Borodovsky A., Bettencourt B.R., Strahs A., Clausen V., Wijngaard P., Horton J.D., Taubel J., Brooks A. et al. A highly durable RNAi therapeutic inhibitor of PCSK9. N. Engl. J. Med. 2016; 376:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martinez J., Patkaniowska A., Urlaub H., Luhrmann R., Tuschl T.. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002; 110:563–574. [DOI] [PubMed] [Google Scholar]
- 13. Ma J.B., Yuan Y.R., Meister G., Pei Y., Tuschl T., Patel D.J.. Structural basis for 5΄-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005; 434:666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frank F., Sonenberg N., Nagar B.. Structural basis for 5[prime]-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010; 465:818–822. [DOI] [PubMed] [Google Scholar]
- 15. Weitzer S., Martinez J.. The human RNA kinase hClp1 is active on 3΄ transfer RNA exons and short interfering RNAs. Nature. 2007; 447:222–226. [DOI] [PubMed] [Google Scholar]
- 16. Kenski D.M., Cooper A.J., Li J.J., Willingham A.T., Haringsma H.J., Young T.A., Kuklin N.A., Jones J.J., Cancilla M.T., McMasters D.R. et al. Analysis of acyclic nucleoside modifications in siRNAs finds sensitivity at position 1 that is restored by 5΄-terminal phosphorylation both in vitro and in vivo. Nucleic Acids Res. 2010; 38:660–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Engel R. Phosphonates as analogues of natural phosphates. Chem. Rev. 1977; 77:349–367. [Google Scholar]
- 18. Lima W.F., Prakash T.P., Murray H.M., Kinberger G.A., Li W., Chappell A.E., Li C.S., Murray S.F., Gaus H., Seth P.P. et al. Single-stranded siRNAs activate RNAi in animals. Cell. 2012; 150:883–894. [DOI] [PubMed] [Google Scholar]
- 19. Yu D., Pendergraff H., Liu J., Kordasiewicz H.B., Cleveland D.W., Swayze E.E., Lima W.F., Crooke S.T., Prakash T.P., Corey D.R.. Single-Stranded RNAs Use RNAi to Potently and Allele-Selectively Inhibit Mutant Huntingtin Expression. Cell. 2012; 150:895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hampton A., Kappler F., Perini F.. Evidence for the conformation about the C(5΄)-O(5΄) bond of AMP complexed to AMP kinase: Substrate properties of a vinyl phosphonate analog of AMP. Bioorg. Chem. 1976; 5:31–35. [Google Scholar]
- 21. Kappler F., Hai T.T., Hampton A.. Use of a vinyl phosphonate analog of ATP as a rotationally constrained probe of the C5΄·O5΄ torsion angle in ATP complexed to methionine adenosyl transferase. Bioorg. Chem. 1985; 13:289–295. [Google Scholar]
- 22. Prakash T.P., Lima W.F., Murray H.M., Elbashir S., Cantley W., Foster D., Jayaraman M., Chappell A.E., Manoharan M., Swayze E.E. et al. Lipid nanoparticles improve activity of single-stranded siRNA and gapmer antisense oligonucleotides in animals. ACS Chem. Biol. 2013; 8:1402–1406. [DOI] [PubMed] [Google Scholar]
- 23. Parmar R., Willoughby J.L., Liu J., Foster D.J., Brigham B., Theile C.S., Charisse K., Akinc A., Guidry E., Pei Y. et al. 5΄-(E)-Vinylphosphonate: a stable phosphate mimic can improve the RNAi activity of siRNA-GalNAc conjugates. Chembiochem. 2016; 17:985–989. [DOI] [PubMed] [Google Scholar]
- 24. Prakash T.P., Kinberger G.A., Murray H.M., Chappell A., Riney S., Graham M.J., Lima W.F., Swayze E.E., Seth P.P.. Synergistic effect of phosphorothioate, 5΄-vinylphosphonate and GalNAc modifications for enhancing activity of synthetic siRNA. Bioor. Med. Chem. Lett. 2016; 26:2817–2820. [DOI] [PubMed] [Google Scholar]
- 25. Elkayam E., Parmar R., Brown C.R., Willoughby J.L., Theile C.S., Manoharan M., Joshua-Tor L.. siRNA carrying an (E)-vinylphosphonate moiety at the 5΄ end of the guide strand augments gene silencing by enhanced binding to human Argonaute-2. Nucleic Acids Res. 2016; 45:3528–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Coles A.H., Osborn M.F., Alterman J.F., Turanov A.A., Godinho B.M., Kennington L., Chase K., Aronin N., Khvorova A.. A high-throughput method for direct detection of therapeutic oligonucleotide-induced gene silencing in vivo. Nucleic Acid Ther. 2016; 26:86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ly S., Navaroli D.M., Didiot M.C., Cardia J., Pandarinathan L., Alterman J.F., Fogarty K., Standley C., Lifshitz L.M., Bellve K.D. et al. Visualization of self-delivering hydrophobically modified siRNA cellular internalization. Nucleic Acids Res. 2016; 45:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alterman J.F., Hall L.M., Coles A.H., Hassler M.R., Didiot M.C., Chase K., Abraham J., Sottosanti E., Johnson E., Sapp E. et al. Hydrophobically modified siRNAs silence Huntingtin mRNA in primary neurons and mouse brain. Mol. Ther. Nucleic Acids. 2015; 4:e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nikan M., Osborn M.F., Coles A.H., Godinho B.M., Hall L.M., Haraszti R.A., Hassler M.R., Echeverria D., Aronin N., Khvorova A.. Docosahexaenoic acid conjugation enhances distribution and safety of siRNA upon local administration in mouse brain. Mol. Ther. Nucleic Acids. 2016; 5:e344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roehl I., Schuster M., Seiffert S.. 2011; Vol. US20110201006 A1. [Google Scholar]
- 31. Birmingham A., Anderson E., Sullivan K., Reynolds A., Boese Q., Leake D., Karpilow J., Khvorova A.. A protocol for designing siRNAs with high functionality and specificity. Nat. Protoc. 2007; 2:2068–2078. [DOI] [PubMed] [Google Scholar]
- 32. Birmingham A., Anderson E.M., Reynolds A., Ilsley-Tyree D., Leake D., Fedorov Y., Baskerville S., Maksimova E., Robinson K., Karpilow J. et al. 3΄ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006; 3:199–204. [DOI] [PubMed] [Google Scholar]
- 33. Ruegger S., Grosshans H.. MicroRNA turnover: when, how, and why. Trends Biochem. Sci. 2012; 37:436–446. [DOI] [PubMed] [Google Scholar]
- 34. Chatterjee S., Fasler M., Bussing I., Grosshans H.. Target-mediated protection of endogenous microRNAs in C. elegans. Dev. Cell. 2011; 20:388–396. [DOI] [PubMed] [Google Scholar]
- 35. Bail S., Swerdel M., Liu H., Jiao X., Goff L.A., Hart R.P., Kiledjian M.. Differential regulation of microRNA stability. RNA. 2010; 16:1032–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chatterjee S., Grosshans H.. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009; 461:546–549. [DOI] [PubMed] [Google Scholar]
- 37. Zangari J., Ilie M., Rouaud F., Signetti L., Ohanna M., Didier R., Romeo B., Goldoni D., Nottet N., Staedel C. et al. Rapid decay of engulfed extracellular miRNA by XRN1 exonuclease promotes transient epithelial-mesenchymal transition. Nucleic Acids Res. 2016; 45:4131–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Burgess H.M., Mohr I.. Cellular 5΄-3΄ mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host Microbe. 2015; 17:332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Trubetskoy V.S., Griffin J.B., Nicholas A.L., Nord E.M., Xu Z., Peterson R.M., Wooddell C.I., Rozema D.B., Wakefield D.H., Lewis D.L. et al. Phosphorylation-specific status of RNAi triggers in pharmacokinetic and biodistribution analyses. Nucleic Acids Res. 2017; 45:1469–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kenski D.M., Willingham A.T., Haringsma H.J., Li J.J., Flanagan W.M.. In vivo activity and duration of short interfering RNAs containing a synthetic 5΄-phosphate. Nucleic Acid Ther. 2012; 22:90–95. [DOI] [PubMed] [Google Scholar]
- 41. Coelho T., Adams D., Silva A., Lozeron P., Hawkins P.N., Mant T., Perez J., Chiesa J., Warrington S., Tranter E. et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 2013; 369:819–829. [DOI] [PubMed] [Google Scholar]
- 42. Sehgal A., Barros S., Ivanciu L., Cooley B., Qin J., Racie T., Hettinger J., Carioto M., Jiang Y., Brodsky J. et al. An RNAi therapeutic targeting antithrombin to rebalance the coagulation system and promote hemostasis in hemophilia. Nat. Med. 2015; 21:492–497. [DOI] [PubMed] [Google Scholar]
- 43. Wolfrum C., Shi S., Jayaprakash K.N., Jayaraman M., Wang G., Pandey R.K., Rajeev K.G., Nakayama T., Charrise K., Ndungo E.M. et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 2007; 25:1149–1157. [DOI] [PubMed] [Google Scholar]
- 44. Lonn P., Kacsinta A.D., Cui X.S., Hamil A.S., Kaulich M., Gogoi K., Dowdy S.F.. Enhancing endosomal escape for intracellular delivery of macromolecular biologic therapeutics. Sci. Rep. 2016; 6:32301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Khvorova A. Oligonucleotide therapeutics—a new class of cholesterol-lowering drugs. N. Engl. J. Med. 2017; 376:4–7. [DOI] [PubMed] [Google Scholar]
- 46. Shen W., Liang X.H., Sun H., Crooke S.T.. 2΄-Fluoro-modified phosphorothioate oligonucleotide can cause rapid degradation of P54nrb and PSF. Nucleic Acids Res. 2015; 43:4569–4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Garber K. Alnylam terminates revusiran program, stock plunges. Nat. Biotech. 2016; 34:1213–1214. [DOI] [PubMed] [Google Scholar]
- 48. Carroll J.B., Bates G.P., Steffan J., Saft C., Tabrizi S.J.. Treating the whole body in Huntington's disease. Lancet Neurol. 2015; 14:1135–1142. [DOI] [PubMed] [Google Scholar]
- 49. Albin R.L. Out of one mutation, many Huntington's disease effects. Lancet Neurol. 2015; 14:1071–1072. [DOI] [PubMed] [Google Scholar]
- 50. Martin B., Golden E., Keselman A., Stone M., Mattson M.P., Egan J.M., Maudsley S.. Therapeutic perspectives for the treatment of Huntington's disease: treating the whole body. Histol. Histopathol. 2008; 23:237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.