Abstract
The coordinated temporal and spatial activation of gene expression is essential for proper stem cell differentiation. The Chromodomain Helicase DNA-binding protein 1 (CHD1) is a chromatin remodeler closely associated with transcription and nucleosome turnover downstream of the transcriptional start site (TSS). In this study, we show that CHD1 is required for the induction of osteoblast-specific gene expression, extracellular-matrix mineralization and ectopic bone formation in vivo. Genome-wide occupancy analyses revealed increased CHD1 occupancy around the TSS of differentiation-activated genes. Furthermore, we observed that CHD1-dependent genes are mainly induced during osteoblast differentiation and are characterized by higher levels of CHD1 occupancy around the TSS. Interestingly, CHD1 depletion resulted in increased pausing of RNA Polymerase II (RNAPII) and decreased H2A.Z occupancy close to the TSS, but not at enhancer regions. These findings reveal a novel role for CHD1 during osteoblast differentiation and provide further insights into the intricacies of epigenetic regulatory mechanisms controlling cell fate determination.
INTRODUCTION
Bone marrow-derived skeletal (also known as stromal or mesenchymal) stem cells (MSC) are self-renewable and multipotent adult stem cells residing perivascular in the bone marrow niche and can differentiate into adipocytes and osteoblasts (1–4). The differentiation of MSC is directed by lineage-specific transcription factors and is accompanied by changes within the chromatin around gene promoters and regulatory regions (5–7). These chromatin changes accompany gene expression regulation and include post-translational histone modifications, chromatin remodeling and exchange of histone variants (8). While the importance of chromatin remodeling for gene induction and maintenance of cell identity is well established, mechanistic and regulatory insights remain elusive (9). Concordantly, many proteins which slide, disrupt or replace nucleosomes are pivotal for various reprogramming and differentiation events (10,11). Moreover, during differentiation, the proximal region surrounding the transcriptional start site (TSS) undergoes changes in histone modifications and incorporation of the histone variants H2A.Z and H3.3 (12–15). In particular, the first nucleosome represents a barrier to the early elongating RNA Polymerase II (RNAPII) and can be modulated by the above mentioned chromatin changes, where its regulation is particularly important in controlling gene activity (16–19).
CHD1 is a highly conserved ATP-dependent chromatin remodeling protein which is associated with active genes (10,20) and interacts with transcription-related complexes such as Polymerase Associated Factor (PAF), Facilitates Chromatin Transcription (FACT), Mediator and the U2 snRNP splicing complex (21–23). Its binding to chromatin is enhanced by H3K4me2/3 (22), a histone modification located predominantly around the TSS, where CHD1 promotes nucleosome turnover and decreases the nucleosomal barrier to RNAPII (16,24). Concordantly, it was shown that CHD1 enhances gene activation after stimulation in normal and tumor cells (20,25) and elevates global transcriptional output in specific cell types (26). Furthermore, CHD1 is not only required for the maintenance of pluripotency in mouse embryonic stem cells (mESC) but also for efficient reprogramming as well as differentiation of hematopoietic stem cell progenitors (10,27). Although these studies clearly establish the importance of CHD1 for differentiation, its regulatory role in controlling gene expression changes during differentiation and the underlying molecular mechanisms remain unclear. Given the increasing interest in MSC for regenerative medicine purposes (28) as well as a previous study reporting that CHD1 is required for maintaining proper levels of histone H2B monoubiquitination (29), which we previously demonstrated to be crucial for MSC differentiation (30), we hypothesized that CHD1 may also have a role in controlling osteoblast differentiation.
Here, we show that CHD1 is required for transcriptional changes during osteoblast differentiation of MSC and for in vivo ectopic bone formation. Genome-wide binding studies revealed a significant increase in CHD1 occupancy near the TSS of differentiation-activated genes in osteoblasts as well as adipocytes. Interestingly, we observed that CHD1-dependent genes highly overlapped with differentiation-induced genes, which displayed a particular enrichment of CHD1 around their TSS. Finally, CHD1 depletion resulted in defective RNAPII elongation at these genes and a global increase in RNAPII pausing at the TSS accompanied by decreased H2A.Z occupancy near the TSS.
MATERIALS AND METHODS
Cell culture
Human fetal osteoblast (hFOB 1.19) and human skeletal (mesenchymal) stem cells (hMSC-Tert) were maintained as previously described (31,32). In brief, MSC were cultured in alpha-Minimum essential medium Eagle modification (α-MEM, Invitrogen) and hFOB 1.19 in phenol red-free high-glucose Dulbecco's modified Eagle's medium (DMEM/F12, Invitrogen), both supplemented with 10% fetal bovine serum (Thermo Fisher Scientific) and 1% penicillin/streptomycin (Sigma-Aldrich). For differentiation confluent cells were treated with a differentiation cocktail as described previously (30) every alternative day for 5 days in total. hFOB 1.19 cells were shifted to 39°C for 2 h before differentiation.
Generation of stable shRNA expressing cell lines
Individual pGIPZ plasmids encoding microRNA-adapted short hairpin RNAs (shRNA) which target the mRNA of CHD1 (shRNA-CHD1 #1—V2LHS_312675; shRNA-CHD1 #2—V2LHS_112971) or non-targeting (shRNA-nt—RHS4346) were purchased from OpenBiosystems. Lentiviral particles were produced by transfection of HEK-293T cells via linear polyethylenimine (PEI) with pGIPZ, psPAX2 packaging plasmid and pMD2.G envelope plasmid (Addgene plasmids #12260, #12259). The viral-containing supernatant was harvested 2 days after transfection and applied to MSC medium together with 8μg/ml polybrene. Two days after transduction cells were selected with 1 μg/ml puromycin and maintained in this medium for at least 10 days before performing further experiments.
Ectopic bone formation
Injection of cells, tissue preparation and quantification were performed as described previously (33). In brief, MSC cell lines expressing non-targeting (MSC-shRNA #nt) or CHD1 targeting shRNAs (MSC-shRNA CHD1 #1 and #2) as well as untransformed MSC were seeded onto a hydroxyapatite/tricalcium phosphate (HA/TCP, Zimmer Inc.) matrix and subcutaneously implanted into female 8-week old NOD.CB17-Prkdc<scid>/J (‘NOD-scid’) mice. The mice were sacrificed 8 weeks after implantation and the processed tissue sections were stained by H&E to measure the formed bone area (BA) relative to the total tissue area (TA). For statistical analysis, a single-factor ANOVA test was performed between the control group (MSC untransformed, n = 3; MSC-shRNA-NT, n = 2) and the CHD1-depleted group (MSC-shRNA CHD1 #1, n = 4; and MSC-shRNA CHD1 #2, n = 4) to analyze statistical differences among the groups. The groups were further analyzed by Tukey multiple comparison tests with a significance threshold set to 0.05.
siRNA transfection
siRNA was transfected using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. Control siRNA (Luciferase GL2 duplex; target sequence: CGUACGCGGAAUACUUCGA), single CHD1 targeting siRNA (siCHD1 #1—CAUCAAGCCUCAUCUAAUA; siCHD1 #2—GAAACAAGCUCUAGAUCAU) and a pool of four CHD1 targeting siRNAs (siCHD1) were purchased from Dharmacon, ThermoScientific. Cells were transfected 36 h before differentiation and again 48 h after differentiation.
Western Blot, qRT-PCR, alkaline phosphatase and Alizarin Red S staining
Isolation of RNA and proteins as well as western blot analysis, reverse transcription and qRT-PCR were performed as previously described (30,34). Antibodies and qRT-PCR primers used in this study are listed in Supplementary Table S5. For alkaline phosphatase (AP) staining, the Leukocyte Alkaline Phosphatase Kit (Sigma-Aldrich) was used according to the manufacturer's protocol. Alizarin Red S staining was performed as previously described (35). For AP and Alizarin Red S quantification the whole wells were scanned and the stained area was quantified with the Threshold Color Plugin on ImageJ software relative to the total measured area. Statistical analysis of qRT-PCR and stainings were performed with two-tailed two-sample Student's t-test.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described previously (30,34). In brief, cells were fixed in 1% or 1.42% formaldehyde (for CHD1 ChIP) for 10 min. Nuclear pellets were isolated in nuclear preparation buffer (150 mM NaCl, 50 mM Tris–HCl (pH 7.5), 5 mM EDTA, NP-40 (0.5%, v/v), Triton X-100 (1.0%, v/v)), pellets were washed in the same buffer and then sonicated in lysis buffer (150 mM NaCl, NP-40 (1%, v/v), sodium deoxycholate (0.5%, w/v), SDS (0.1%, w/v), 50 mM Tris–HCl (pH 8), 20 mM EDTA, 20 mM sodium fluoride and protease inhibitor cocktail) for 30 cycles (30 s on/off) using a Bioruptor Pico (Diagenode). The sheared chromatin was first precleared by adding lysis buffer supplemented with 50% Sepharose 4B beads (GE Healthcare) and then incubated separately with the corresponding antibodies for 16 h (Supplementary Table S6). Immunocomplexes were precipitated by Protein-A or -G sepharose (GE Healthcare) for 2 h and washed thereafter with lysis buffer, wash buffer (100 mM Tris (pH 8.5), 500 mM LiCl, NP-40 (1%, v/v), sodium deoxycholate (1%, w/v), 20 mM EDTA, 20 mM NaF) and finally twice with TE buffer. Samples were treated with RNase A (Qiagen) and incubated with 20 mM EDTA, 100 mM Tris–HCl (pH 8), SDS (2%, w/v) and 20 μg Proteinase K for 16 h at 65°C. The DNA was isolated via phenol–chloroform extraction and used for qRT-PCR or ChIP-seq analysis.
Formaldehyde-assisted isolation of regulatory elements
The Formaldehyde-Assisted Isolation of Regulatory Elements (FAIRE) protocol was performed as previously described with modifications in the isolation step of cell nuclei (36). Briefly, cells were crosslinked for 10 min with 1% formaldehyde at room temperature and cell nuclei were isolated as for ChIP. Samples were then sheared for 10 cycles (30 s on/off) in Bioruptor Pico Microtubes (Diagenode).
DNA and RNA library preparation
DNA libraries for sequencing were prepared using the MicroPlex Library Preparation Kit v2 (Diagenode) according to the manufacturer's protocol. Prior to library preparation the DNA was sheared for another 30 cycles (30 s on/off) to ensure optimal fragment size for sequencing. For CHD1 ChIP the NEBNext Ultra DNA Library Prep Kit (NEB) was used. Before RNA-seq library preparation RNA integrity was confirmed using a Bioanalyzer 2100 (Agilent) and mRNA was purified from 1 μg total RNA via the NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB) before proceeding with the NEBNext Ultra RNA Library Prep Kit for Illumina (NEB). Before sequencing DNA size was measured and found to be ca. 300 bp using the Bioanalyzer 2100 (Agilent). Single-end 50 bp sequencing was performed on an Illumina HiSeq2500.
RNA-seq bioinformatic analysis
RNA-seq data processing was performed as described previously (34). Reads were mapped to the human reference transcriptome (UCSC hg19) using Bowtie2 (version 2.1.0) (37). Read counts for samples and genes were aggregated using a custom Ruby script and differential gene expression was analyzed by DEseq (version 1.14.0) (38) in R 3.1.0. Significantly differentially regulated genes were selected by a Benjamini–Hochberg adjusted P-value below 0.05 and log2-fold change values above 0.5 or below –0.5 for up- and downregulated genes, respectively. Heatmaps were created with heatmap.2 from gplots R package (39). Grouping of gene expression into quintiles was done on all expressed genes having a basemean value >10. Experiments were performed in biological duplicates. The similarities between replicates were assessed via principle component analysis (PCA). The PCA plot was calculated and plotted based on the software package DESeq (38).
DAVID analysis
Gene ontology (GO) analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) tool (version 6.7) with default conditions (40) using gene identifiers (IDs) and Homo sapiens set as background. The top two enriched annotation clusters and selected terms were listed as indicated.
ChIP-seq bioinformatic analysis
Reads were mapped to the human reference genome (UCSC GRCh37/hg19) using Bowtie (version 1.1.1) with parameters set to -m 1 and -k 1 (41) and converted to BAM format via samtools (version 1.2) (42). BAM files were further processed using the BamCoverage tool within the deeptools2 software (43) and normalized to reads per genomic content (RPGC). All experiments were performed in biological triplicates except H3.3 ChIP-seq, which was performed in duplicates for siCHD1 and a single replicate for control siRNA transfected hFOB 1.19. For peak calling Model-based Analysis of ChIP-Seq (MACS2) (version 2.1.0) was used with input samples set as background and default parameters (q-value < 0.05) (44). Genomic localization of CHD1 peaks (MSC-Ob, n = 8853; K562, n = 59924) relative to annotation was analyzed using the cis-regulatory Element Annotation System (CEAS) software (version 1.0.2) with default parameters (45). Single gene profiles were visualized in the Integrated Genome Viewer (IGV; version 2.3.14) (46). Heatmaps, aggregate and correlation plots were created with the deeptools2 software. For correlation plots the regions from the TSS to 1 kb downstream were used. For illustrating average profiles over all genes the computeMatrix scale region mode was used by averaging genes to 60 kb with 10 kb up- and downstream of the TSS and TES, respectively. Gene coordinates as well as CpG-island information were downloaded from the UCSC Table Browser (UCSC genes GRCh37/hg19) (47). Genes with multiple TSS were selected for the TSS with highest CHD1 occupancy. For this purpose, occupancy values were calculated within a region from TSS to +1 kb using the computeMatrix tool. Differential binding analysis of ChIP-Seq peak data was performed for all CHD1 called peak regions identified with MACS2 in undifferentiated and osteoblast-differentiated conditions using DiffBind (version 1.10.2) (48). Significantly enriched regions in the osteoblast-differentiated condition (P-value < 0.05 and False Discovery Rate < 0.1) were used as an input for the Genomic Regions Enrichment Annotation Tool (GREAT) (49) with the setting ‘single nearest gene’.
Bivalent promoter regions were defined by overlapping H3K4me3- and H3K27me3-enriched regions in undifferentiated MSC identified by MACS2 with default parameters. Further, these regions were overlapped with TSS expanded in each direction for 1 kb. hFOB 1.19-specific enhancer regions were defined as recently described (35). Briefly, segments enriched for H3K4me1, H3K27ac and BRD4, but depleted for H3K27me3 or H3K4me3, were specified as enhancer sites (n = 5409 regions). Averaged ChIP-seq signals at these regions were calculated by scale-region mode over computeMatrix. Analysis of additional transcription factor binding was further performed using the ReMap tool (50).
RNAPII pausing ratio
The RNAPII pausing ratio was calculated as described previously (51). Briefly, the TSS region was defined as a window ranging from 300 bp upstream of the TSS to 30% of gene body length added downstream to the TSS. The gene body (GB) was set as 30% of gene body length downstream of TSS to 3 kb downstream of TES. Genes smaller than 600 bp were excluded from analysis to reduce bias between the signal of TSS and GB (52). The RNAPII occupancy was measured as the median value over the GB and as the mean value at TSS using the computeMatrix tool in deeptools2 via scaleRegion mode. Genes were considered for calculation of the RNAPII pausing ratio if the RNAPII signal was over 1.4 RPGC at the TSS and a median value at the gene body above 0.1 RPGC. In total, the pausing ratio was calculated for 11,502 genes. For statistical analysis pausing ratios were compared by Wilcoxon-rank sum test.
Publically available datasets
SRA data of DNaseI hypersensitivity (GEO: GSM1946442, GSM1946441) (7), H3K27ac in differentiated, RNA Polymerase II in undifferentiated and differentiated hFOB 1.19 (35) were processed as described above and used as indicated.
RESULTS
CHD1 is required for transcriptional programming during osteoblast differentiation
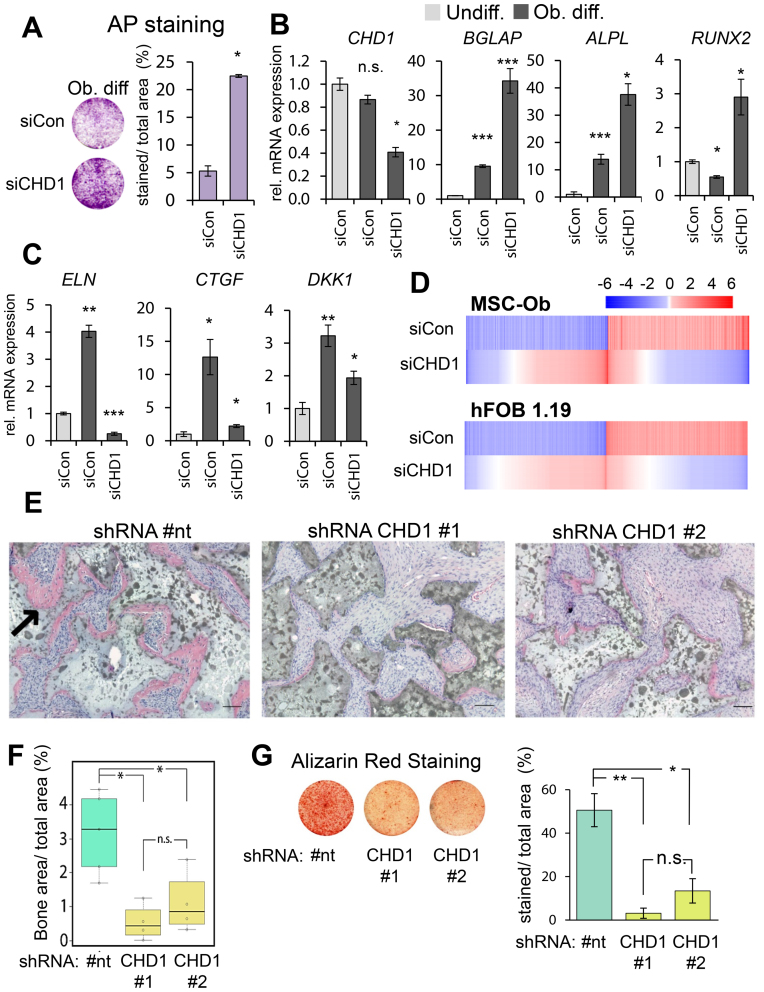
To elucidate the role of CHD1 in lineage specification we utilized the well-established MSC model of differentiation into the osteoblast lineage. Interestingly, depletion of CHD1 (Supplementary Figure S1A) during osteoblast differentiation of MSC led to increased alkaline phosphatase (AP) activity (Figure 1A) and mRNA levels (ALPL; Figure 1B). Furthermore, the expression of two other early osteoblast-specific genes BGLAP and RUNX2 were similarly increased upon CHD1 depletion. In contrast, other genes activated during osteoblastdifferentiation such as DKK1, ELN and CTGF displayed decreased expression levels following CHD1 depletion (Figure 1C). This initially suggested a gene-specific role of CHD1 in regulating transcription during osteoblast differentiation. Therefore, in order to further determine the extent of the effects of CHD1 depletion on genes regulated during osteoblast differentiation we performed transcriptome-wide mRNA sequencing analyses in MSC and human fetal osteoblasts (hFOB 1.19) (Figure 1D, Supplementary Figure S1C). In total, 1416 genes in MSC and 4749 genes in hFOB 1.19 were induced during osteoblast differentiation while 1292 and 4844 genes were repressed in MSC and hFOB 1.19, respectively. Notably, CHD1 depletion largely attenuated gene expression changes induced by differentiation (Figure 1C), as 550 differentiation-activated genes in MSC and 419 in hFOB 1.19 were less activated upon CHD1 depletion while 452 and 341 genes were more weakly repressed in MSC and hFOB 1.19, respectively (Supplementary Figure S1B). To further investigate the transcriptional changes of CHD1-regulated genes during differentiation we analyzed their gene expression and correlation between undifferentiated and osteoblast-differentiated states in control and CHD1-depleted conditions. Consistent with decreased mRNA levels upon CHD1-depletion, a higher correlation was observed between the gene expression patterns of undifferentiated cells to the CHD1-depleted osteoblast-differentiated condition compared to control osteoblast-differentiated condition (Supplementary Figure S1D). This supports the idea that CHD1 is required for transcriptional changes during osteogenesis and that loss of CHD1 maintains cells in a more undifferentiated state even in the presence of differentiation-inducing signals. Additionally, principle component analysis (PCA) revealed considerable differences between the various conditions (differentiation and with CHD1 depletion) at the transcriptome level, as well as good concordance between single replicates (Supplementary Figure S1D). Next, we performed gene ontology (GO) analyses of the CHD1-regulated genes in MSC and hFOB 1.19. This revealed an enrichment of genes associated with skeletal system development, developmental processes, response to stimuli and the extracellular matrix, which is strongly remodeled during osteogenesis and required for osteoblast differentiation (53) (Supplementary Table S1).
Figure 1.
CHD1 depletion impairs osteoblast differentiation in MSC and hFOB 1.19. (A) Osteoblast-differentiated mesenchymal stem cells (Ob. diff) transfected with control (siCon) or CHD1 (siCHD1) siRNAs were stained for alkaline phosphatase (AP) activity. Mean values ± SD (n = 2), with P-value shown. qRT-PCR gene expression analysis of CHD1 and the osteoblast-specific genes ALPL, BGLAP, RUNX2 (B) and DKK1, ELN, CTGF (C) in undifferentiated (Undiff.) and osteoblast-differentiated (Ob. diff) mesenchymal stem cells transfected with control (siCon) or CHD1 targeting siRNA (siCHD1). Values were normalized to 18S rRNA and quantified relative to control undifferentiated samples. Mean ± SD, (n = 3) (*P-value < 0.05; **P-value < 0.01; ***P-value < 0.001). (D) Heatmaps depict log2-fold changes of gene expression in MSC and human fetal osteoblasts (hFOB 1.19) during osteoblast differentiation. Relative changes upon siCHD1 treatment (Diff. +siCHD1) compared to differentiation-associated changes in gene expression are shown in the lower panel of each heatmap. Genes were sorted by log2-fold changes with siCHD1 treatment and differentiation-regulated genes were selected by adjusted P-value < 0.05 and ±0.5 log2-fold changes. (E) Representative H&E stained tissue sections of ectopic bone formation (pink area; see black arrow) in SCID mice implanted with MSC stably expressing a non-targeting (#nt) or CHD1-targeting shRNAs (#1, #2). Mice were sacrificed 8 weeks after implantation. Scale bars represent 100 μM. (F) Boxplots show the quantification of ectopic bone formation of control and CHD1-shRNA expressing MSC with each point representing one biological replicate. The ratio of bone area (BA) over total area (TA) was compared with Tukey multiple comparison test with a threshold at 0.05 (*). (G) Representative images of Alizarin Red S staining of MSC differentiated for 14 days in control (#nt) and CHD1-depleted (CHD1 #1, #2) are shown with the quantification of the relative percentage of stained area over total area (n = 3; * P-value < 0.05).
In order to examine the effect of CHD1 depletion on bone formation in vivo, two stable CHD1-shRNA and one non-targeting shRNA-expressing MSC cell lines were established and tested for their ectopic bone formation capacity in immune-deficient mice (Supplementary Figure S2A). Interestingly, both CHD1-depleted MSC cell lines formed significantly less bone tissue compared to control cells and showed less extracellular matrix (Figure 1E and F). As we observed defects in expression of extracellular matrix genes and decreased bone formation we investigated if mineralization, a later step in osteoblast differentiation, and dependent on the extracellular matrix, was also impaired. Indeed, as assessed by Alizarin Red S staining, CHD1-deficient MSC displayed less mineralization capacity (Figure 1G). Consistent with these findings, phenotype characterization data from the International Mouse Phenotype Consortium (54) revealed that heterozygous Chd1 knockout mice have lower bone mineral density and bone mineral content compared to wildtype mice (Supplementary Figure S2B). Together, these observations imply that CHD1 is required for osteogenesis by regulating the transcriptional program specifically at later stages of osteoblast differentiation.
Notably, the function of CHD1 in regulating differentiation-induced gene expression was not limited to the osteoblast lineage. CHD1 depletion also resulted in reduced induction of lineage-specific genes during adipocyte differentiation (Supplementary Figure S3A), suggesting a more general function of CHD1 in regulating differentiation-related transcriptional programs in MSC.
CHD1 occupancy increases near the TSS of differentiation-induced genes
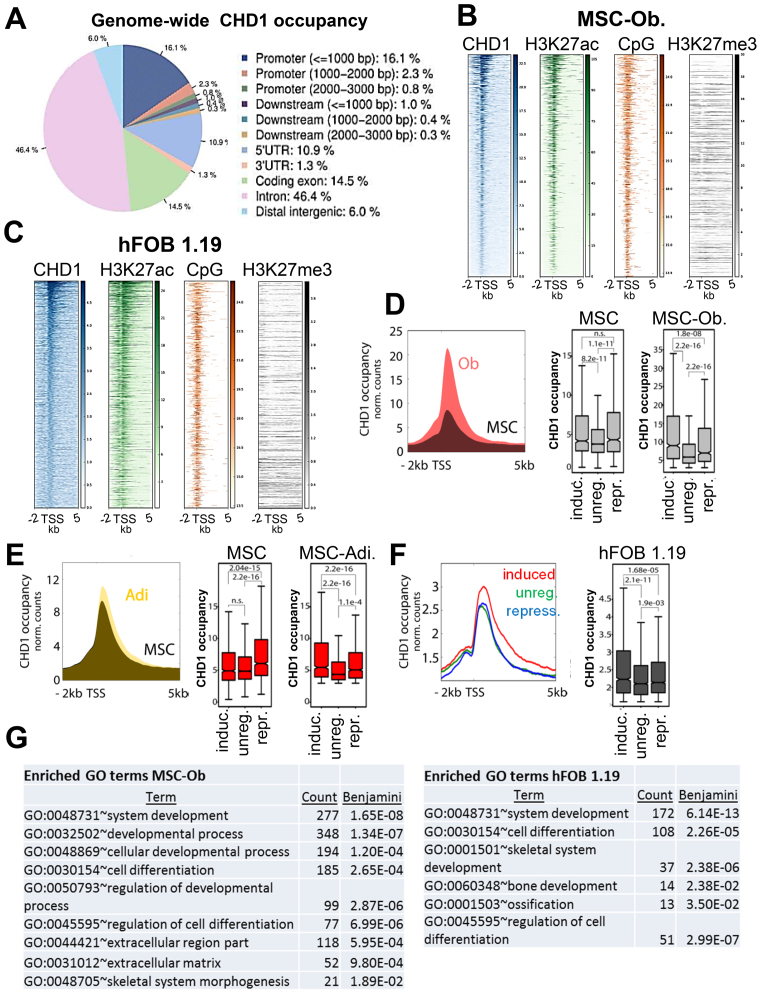
To understand the molecular mechanisms by which CHD1 functions during MSC differentiation, we studied its genome-wide occupancy during both adipogenesis and osteogenesis by chromatin immunoprecipitation followed by high throughput sequencing (ChIP-seq). Consistent with a function in controlling gene expression, most CHD1-enriched regions were located within genes, with the highest abundance in introns and promoter regions (Figure 2A). The strongest CHD1 occupancy was observed at the proximal region downstream of the TSS (Supplementary Figure S4A). The occupancy of CHD1 generally correlated with the intensity of the active histone modification H3K27ac and was co-localized with a higher CpG content near the TSS, but was not correlated with H3K27me3 in MSC and hFOB 1.19 (Figure 2B and C). Next, we tested if transcriptional changes occurring during differentiation are also accompanied by changes in CHD1 occupancy. Therefore, we focused on a region between 0 and 1 kb downstream of the TSS where CHD1 occupancy was highest and where CHD1 was previously shown to facilitate nucleosome turnover (16). Indeed, CHD1 occupancy significantly increased near the TSS of osteoblast-activated genes during differentiation (Figure 2D). Moreover, we observed a significant increase in CHD1 occupancy at induced genes during osteogenesis in comparison to genes which were repressed or unregulated during differentiation in MSC (Figure 2D). A similar pattern was also observed during adipogenesis (Figure 2E), suggesting that CHD1, like H2B monoubiquitination (30), may generally be required for MSC differentiation-associated gene expression changes. Together, these findings demonstrate that CHD1 occupancy near the TSS is strongly increased and accompanies regulatory changes during differentiation. To more precisely characterize the changes in CHD1 occupancy during osteoblast-differentiation we used DiffBind to identify differentially-associated regions and GREAT analysis for the functional categorization of these regions. Most of the differentially enriched regions were in close proximity to the TSS, but not intergenic regions (Supplementary Figure S4B). Notably, these genes were significantly enriched with genes associated with extracellular matrix components (Supplementary Figure S4C). Since differentiation of stem cells often involves transcriptional regulation of bivalent genes marked with both H3K4me3 and H3K27me3 (12), we examined whether CHD1 would similarly target these promoters. However, only a small fraction of bivalent genes which are activated upon osteoblast differentiation were dependent on CHD1 (Supplementary Figure S4D).
Figure 2.
CHD1 occupancy increases near the TSS of differentiation-induced genes. (A) Relative genome-wide CHD1 occupancy analysis at regulatory elements in osteoblast-differentiated MSC (MSC-Ob.) represented by a pie chart with the color code on the right. Analysis was performed via CEAS. (B, C) Occupancy of H3K27ac, CpG frequency and H3K27me3 relative to CHD1-occupied transcriptional start sites (TSS) ordered from high to low based on CHD1 occupancy in MSC-Ob. (B) and hFOB 1.19 (C). (D) Aggregate plot of CHD1 around the TSS of osteoblast differentiation-induced genes in undifferentiated (black) and differentiated (red) conditions. Quantification of CHD1 occupancy at induced (induc.), unregulated (unreg.) or repressed (repr.) gene groups in undifferentiated (MSC) and differentiated (MSC-Ob) conditions are shown on the right. P-values below 0.05 are shown unless indicated as not significant (n.s.). (E) Aggregate plot of CHD1 as described in D but for adipocyte differentiation-regulated gene groups (Adi.). (F) Aggregate plot of CHD1 around the TSS in hFOB 1.19 at genes induced (red), unregulated (green) or repressed (blue) during osteoblast differentiation. Boxplots on the right show quantitation of CHD1 occupancy with P-values indicated above boxplots. (G) The lists represent significantly enriched gene ontology (GO) terms at high CHD1-occupied genes around the TSS of osteoblast-differentiation induced genes in MSC-Ob. and hFOB 1.19. GO analysis was performed with the DAVID analysis tool with the adjusted P-value (Benjamini) and count of genes within the respective term.
To more precisely characterize the genes regulated during osteogenesis in hFOB 1.19 we classified these into two groups based on high and low CHD1 occupancy (Supplementary Figure S4E). Overall, we observed a significant enrichment of CHD1 at differentiation-induced genes in the highly expressed and highly CHD1-occupied fraction, but not in the lower expressed fraction (Supplementary Figure S4F, Figure 2F). This difference in CHD1 occupancy and its correlation with the induction of osteoblast-specific genes prompted us to perform GO analyses on the low and high occupied groups. The higher CHD1-occupied fraction of genes showed an enrichment of GO terms associated with cellular differentiation, developmental processes and the extracellular region, which were not enriched in the low bound fraction (Figure 2G, Supplementary Table S2). Interestingly, the top 20% of CHD1-enriched and differentiation-induced genes showed the highest association with cellular differentiation and skeletal terms compared to the others fractions (Supplementary Table S3), indicating that the highly CHD1-occupied genes are more closely associated with osteoblast-specific functions. Taken together, these results show that differentiation-induced genes display an increased recruitment of CHD1 specifically around the TSS of osteoblast differentiation-related genes.
CHD1-dependent genes display higher CHD1 occupancy near the TSS
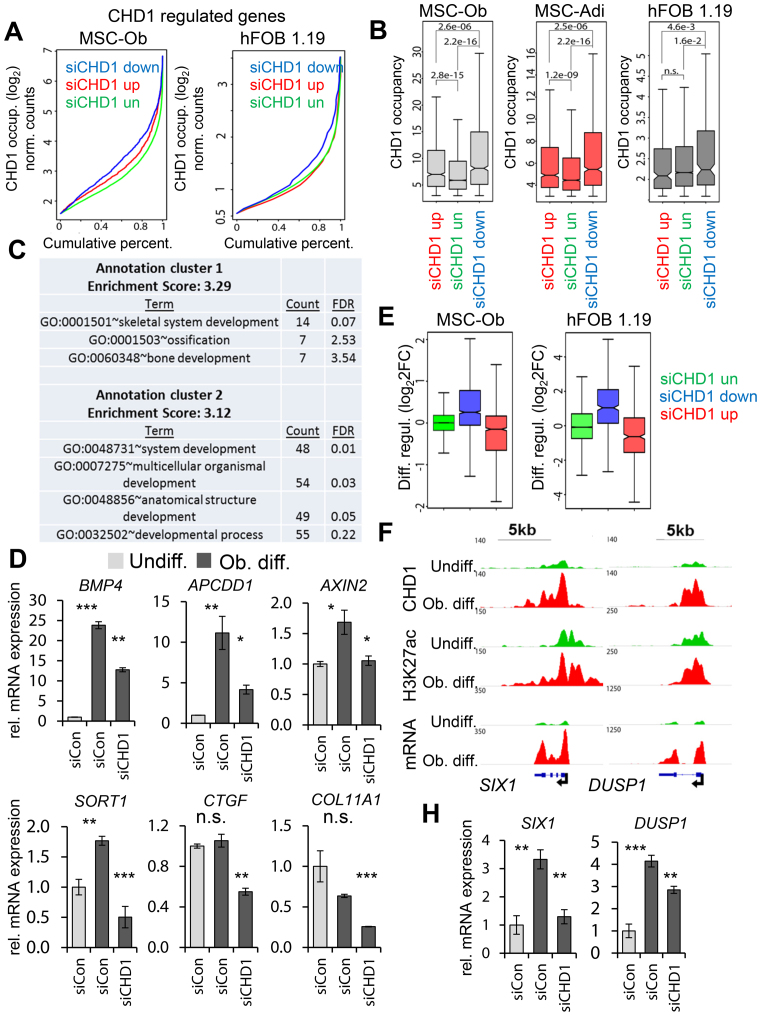
After observing high occupancy of CHD1 near the TSS of differentiation-induced genes and its requirement for transcriptional changes during differentiation, we next tested whether CHD1 occupancy is also associated with CHD1-dependent gene expression. Consistent with this notion, but in contrast with the enhancer-specific function of BRD4 (35), CHD1-dependent genes in both osteoblast and adipocyte-differentiated MSC as well as in hFOB 1.19 displayed significantly higher CHD1 occupancy near the TSS compared to down- or unregulated genes (Figure 3A, B). In hFOB 1.19, we next classified the CHD1-regulated genes into high and low CHD1-occupied fractions (Supplementary Figure S4G) according to the previously introduced threshold value (Supplementary Figure S4E). As expected, the low occupied fraction of CHD1-dependent genes was significantly less expressed compared to the highly occupied fraction (Supplementary Figure S4G), which was enriched in GO terms associated with skeletal system and developmental processes (Figure 3C). This was not the case for the low CHD1-occupied fraction (Supplementary Table S4). The dependency on CHD1 was further verified by qRT-PCR for genes from the high CHD1 fraction (Figure 3D, Supplementary Figure S5A). Notably, some of these candidate genes were previously shown to be highly predictive for ectopic bone formation (ELN, BMP4, COL11A1) (55,56). Moreover, consistent with a general importance for CHD1 in controlling osteoblast-specific transcriptional changes, we observed an induction of CHD1-dependent genes during osteoblast differentiation in both hFOB 1.19 and MSC (Figure 3E), which could be also verified for individual genes (Figure 3F and H). In contrast, genes upregulated after CHD1 depletion were mainly downregulated during differentiation and unregulated genes remained unchanged during differentiation. Altogether, these data indicate that osteoblast-specific genes highly occupied by CHD1 are more dependent on CHD1 function, supporting a direct function for CHD1 in the transcriptional regulation of these genes.
Figure 3.
CHD1-dependent genes are enriched for CHD1 occupancy near the TSS and are induced during differentiation. (A) Cumulative percentage plots represent CHD1 binding at CHD1-dependent (blue), -independent (green) or -upregulated (red) genes in osteoblast-differentiated MSC (MSC-Ob) and hFOB 1.19 after siRNA-mediated CHD1 depletion. Normalized log2 formatted occupancy values of CHD1 are plotted against the cumulative percentage. Significantly regulated (adjusted P-value < 0.05) genes were selected by a cutoff above or below ±0.5 log2-fold changes upon CHD1 depletion. (B) Boxplots of CHD1 occupancy at CHD1-regulated genes in osteoblast- (MSC-Ob) and adipocyte-differentiated (MSC-Adi) MSC as well as osteoblast-differentiated hFOB 1.19. P-values are shown above compared groups. (C) Gene Ontology (GO) terms of CHD1-dependent genes in hFOB 1.19 identified as in A. The two most enriched annotation clusters are listed with the adjusted P-value (Benjamini) and the count of genes within respective GO terms. (D) qRT-PCR gene expression analysis of CHD1-dependent genes enriched in the GO-terms shown in C in differentiated hFOB 1.19 (BMP4, APCDD1, AXIN2, SORT1, CTGF, COL11A1). The gene expression was calculated relative to the undifferentiated control samples and normalized to the housekeeping gene RPLP0. Values are mean ± SD (n = 3). (E) Differentiation-induced gene expression changes in MSC and hFOB 1.19 of CHD1-regulated gene groups as shown in A. The fold changes in gene expression are log2 transformed (log2FC) and represented by boxplots. (F) Genomic occupancy profiles of CHD1 and H3K27ac as well as mRNA expression profiles in undifferentiated (green) and osteoblast-differentiated conditions (red) at the differentiation-induced and CHD1-dependent genes SIX1 and DUSP1. ChIP- and RNA-seq values are normalized to RPGC and RPKM, respectively. The black arrow denotes the TSS and indicates the direction of transcription. (H) qRT-PCR analysis in MSC-Ob for DUSP1 and SIX1 in undifferentiated control (Undiff.) and osteoblast-differentiated conditions (Ob. diff.). Values are shown relative to the Undiff. control condition and are normalized to 18S rRNA. Mean ± SD (n = 3).
Given the pivotal role of transcription factors (TFs) in the regulation of gene expression we sought to analyze if CHD1-regulated genes may utilize a particular repertoire of TF during osteoblast cell fate commitment. Interestingly, none of the known TFs involved in osteoblast lineage commitment displayed a particular enrichment at the promoter regions of CHD1-dependent genes (Supplementary Figure S6A and S6B). This is not surprising given the fact that many lineage-specific TFs appear to function by binding to distal regulatory regions, rather than promoter-proximal regions. Consistent with general chromatin-associated events associated with transcriptional regulation, epigenetic regulators such as RBBP5, SIN3A, HDAC1 and EZH2 were among the factors most significantly enriched at the CHD1-occupied promoters. As expected, CHD1 was among the top hits as well. However, no particular differences in enrichment of potential factors were observed between the CHD1-dependent and CHD1-unregulated promoters, suggesting a complex interplay between distal lineage-specific enhancer regions and TSS-proximal CHD1 function during differentiation.
CHD1 regulates early RNAPII elongation and promotes H2A.Z occupancy around the TSS
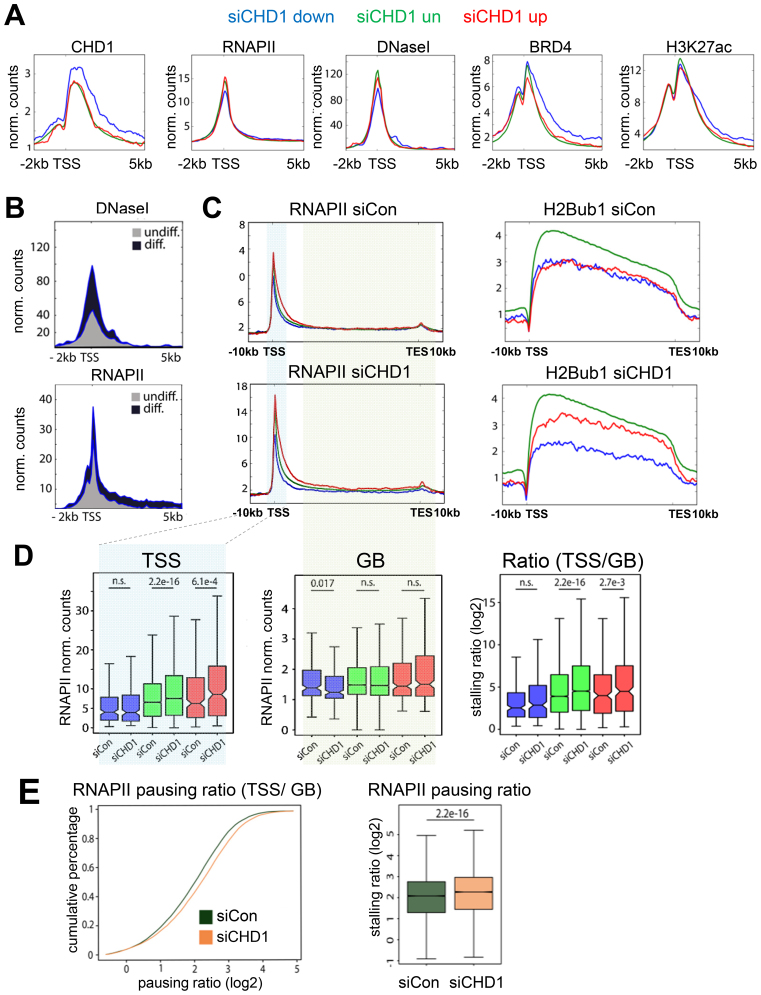
To further investigate the mechanisms by which CHD1 controls differentiation-induced gene expression, we analyzed the chromatin landscape around the TSS of CHD1-dependent and highly occupied genes (Figure 3A) by examining chromatin accessibility (DNase I hypersensitivity) as well as the occupancy of the histone modifications H3K27ac, H2Bub1, and RNAPII and Bromodomain-containing protein 4 (BRD4) in hFOB 1.19. BRD4 is a central regulator of lineage- and tumor-specific transcriptional progams and functions to increase transcriptional activity by binding to acetylated lysine residues at distal and proximal regulatory regions (30,34,35). CHD1-dependent genes showed a high occupancy of CHD1 which extended into the gene body, while displaying comparatively low RNAPII occupancy around the TSS (Figure 4A). Concordant with lower RNAPII levels, we also detected lower DNase I hypersensitivity but higher enrichment of BRD4 and H3K27ac at these sites (Figure 4A). Interestingly, during differentiation these regions displayed increased RNAPII occupancy as well as DNase I hypersensitivity, suggesting increased chromatin remodeling is associated with osteoblast-specific gene induction (Figure 4B). We next tested whether CHD1 is required for chromatin accessibility at the TSS of the dependent genes using Formaldehyde-Assisted Identification of Regulatory Elements (FAIRE) analyses. However, CHD1 depletion did not significantly affect chromatin accessibility at several CHD1-dependent osteoblast-specific genes (Supplemental Figure S7A). We further analyzed the underlying DNA sequence of CHD1-dependent genes by a nucleosome positioning prediction tool (57) and observed a higher probability of nucleosome occupancy downstream of the TSS of CHD1-dependent genes compared to repressed or unregulated genes (Supplementary Figure S7B). Thus, it is possible that the higher signals observed for CHD1, BRD4 and H3K27ac downstream of the TSS at these genes may be associated with more stably positioned nucleosomes.
Figure 4.
CHD1 is required for effective elongation of RNA Polymerase II. (A) Aggregate plots of CHD1, RNA Polymerase II (RNAPII), and DNaseI hypersensitivity as well as BRD4 and H3K27ac around the TSS of CHD1-dependent (blue), -repressed (red) and -unregulated (green) gene groups in hFOB 1.19. (B) DNaseI hypersensitivity and RNAPII occupancy are shown for CHD1-dependent genes in undifferentiated (grey) and differentiated (blue) conditions around the TSS in hFOB 1.19. (C) RNAPII and H2Bub1 aggregate plots over averaged CHD1-regulated gene groups (as in A) in hFOB 1.19 transfected with control (siCon) or CHD1 (siCHD1) siRNAs. (D) Quantification of RNAPII occupancy near the TSS, gene body (GB) and ratio of TSS and GB for CHD1-regulated gene groups (as in A) in hFOB 1.19 transfected with siCHD1 or siCon. P-values are shown above the respective comparison groups. (E) The pausing ratio of RNAPII in control (siCon) and CHD1-depleted (siCHD1) hFOB 1.19 is represented via a cumulative percentage plot. The quantification is shown in the boxplot on the right. Pausing ratios are shown as log2 transformed values. The P-values between the groups are indicated.
Since CHD1 was shown to increase nucleosome turnover immediately downstream of the TSS and subsequently support early elongation of RNAPII into the gene body (GB) (16,24), we analyzed the differential occupancy of RNAPII near the TSS and across the GB by using the previously described ‘pausing ratio’ estimation (51,52) as a proxy for release from promoter proximal pausing. Therefore, the ‘pausing ratio’ was defined as previously described (51,52) by dividing the occupancy of RNAPII near the TSS of each gene by its occupancy across the GB of the same gene. Furthermore, H2B monoubiquitination (H2Bub1), a histone modification previously described to be regulated by CHD1 (29) and positively correlated with the elongating RNAPII was measured as an additional read-out for transcriptional elongation (58). Similar to the changes in mRNA expression upon CHD1 depletion and consistent with decreased RNAPII elongation, we observed a decrease in H2Bub1 occupancy on the transcribed regions of CHD1-dependent, an increase at repressed genes, and no change at unregulated genes (Figure 4C). Consistently, RNAPII occupancy decreased at the GB of CHD1-dependent genes after CHD1 depletion, while unchanged near the TSS (Figure 4C and D), further supporting a specific role for CHD1 in the transition of RNAPII from initiation or early elongation into productive elongation during differentiation. Remarkably, we observed a global increase of RNAPII pausing after CHD1 depletion (Figure 4E), which was most prominent at unregulated and repressed genes due to a strong increase of RNAPII at the TSS-region, with less changes across the GB (Figure 4D). Despite this global increase of RNAPII pausing, no global effect on gene expression was detected, indicating differential regulatory mechanisms at highly CHD1-occupied, CHD1-dependent genes, which are mainly induced during differentiation, compared to CHD1-independent genes and genes upregulated in response to CHD1 depletion.
Previous studies showed that H2A.Z occupancy near the TSS decreases RNAPII pausing in Drosophila (18) through the formation of low salt-sensitive and instable nucleosomes together with H3.3, which may decrease the nucleosomal barrier for RNAPII (59,60). To further analyze the observed effects on RNAPII pausing and the regulation of CHD1-dependent genes in respect to H2A.Z and H3.3, we performed ChIP-seq for these histone variants after CHD1 depletion. Interestingly, whereas no significant change was observed in H3.3 occupancy, H2A.Z levels uniformly decreased near the TSS of all genes following CHD1 depletion (Supplementary Figures S7C and S8A). Notably, the reduction in H2A.Z occupancy was not due to altered protein levels (Supplementary Figure S7D). Furthermore, we tested whether lower H2A.Z levels were caused by a general decrease in H2A occupancy. However, H2A levels remained unaltered at the genes which displayed a reduction in H2A.Z occupancy following CHD1 knockdown (Supplementary Figure S8A), possibly suggesting a role for CHD1 in the formation of heterotypic nucleosomes containing canonical H2A and H2AZ. Since H2A.Z and H3.3 are also associated with enhancers (61,62), we analyzed the occupancy of the indicated histone variants at these sites as well (Supplementary Figure S8B). Surprisingly, whereas H2A.Z levels slightly increased at BRD4-bound osteoblast specific enhancers, H3.3 occupancy decreased substantially. Interestingly, RNAPII remained unaffected at these enhancer sites with CHD1 knockdown (Supplementary Figure S8B). Altogether, these findings indicate a differential function of CHD1 in affecting H3.3 and H2A.Z dynamics at TSS and enhancers, where decreased H2A.Z levels near the TSS following CHD1 depletion may result in increased RNAPII pausing and decreased transcriptional elongation of osteoblast-specific genes.
DISCUSSION
Differentiation requires an intricate, spatially and temporally orchestrated reprogramming of transcription which directs cell-fate decisions. These changes are coordinated by transcription factor binding at proximal and distal regulatory elements which promotes the recruitment of cofactors and the general transcription machinery, resulting in altered chromatin states and initiation of gene activity. Thus, chromatin remodeling, histone modification and histone exchange are central mechanisms in gene transcription, where they promote gene expression, in part, by enabling RNAPII elongation.
Although CHD1 is an intensively investigated chromatin remodeler, its genome-wide occupancy and importance for transcriptional regulation during differentiation had not been addressed in-depth to date. In this study, we uncover a previously unknown function of CHD1 in osteoblast differentiation. Importantly, we show a preferential pattern of CHD1-occupancy changes during osteoblast and adipocyte differentiation downstream of the TSS on differentiation-induced genes, where CHD1-dependence is tightly coupled to its occupancy. Consistent with our observations in human cells, an importance of CHD1 downstream of the TSS in facilitating productive elongation of RNAPII into the gene body by promoting nucleosome turnover and remodeling of the nucleosomal barrier was previously demonstrated in mouse embryonic fibroblasts (16).
Since a large fraction of CHD1-dependent genes in the investigated systems were induced during differentiation, CHD1 occupancy downstream of the TSS appears to play a particularly important role in the activation of differentiation-specific genes, while not necessarily being required for the expression of lineage-independent genes. This view is consistent with previous studies which revealed a requirement of CHD1 in spatiotemporal gene regulation, where it was required for gene activation in Drosophila and in human prostate cancer cells (20,25), as well as during reprogramming and differentiation in mouse cells (10,27). During gene activation, the chromatin landscape is remodeled from a transcriptionally silent and more condensed to an active and open chromatin state, which involves the exchange of histone variants, changes in post-translational histone modifications and increased nucleosome turnover (14,63–65). Surprisingly, although RNAPII occupancy was low at CHD1-dependent genes, we observed high levels of CHD1 occupancy around and downstream of the TSS, presumably coinciding with the first nucleosome (+1 nucleosome), which represents a barrier to the elongating RNAPII (66). We speculate that, as indicated in previous studies (16,24), increased CHD1 occupancy at these sites may be necessary for nucleosome turnover to facilitate the transition from transcriptional initiation to productive elongation. Nucleosome turnover can also alter the chromatin landscape by promoting the exchange of modified with unmodified or differentially modified histones (63,67). Furthermore, high CHD1 levels might be necessary to support the opening and remodeling of chromatin during gene activation at regions containing a particular epigenetic or chromatin signature or exhibiting a particular repertoire of chromatin-associated factors or transcription factors. In fact, the prominent BRD4 occupancy at these sites might support or be a direct consequence of such an event (68,69). Well positioned nucleosomes downstream of the +1 nucleosome may also represent additional barriers to RNAPII, which require the presence of chromatin remodelers such as CHD1 for efficient gene transcription.
In addition to and consistent with increased pausing of RNAPII at highly occupied CHD1-dependent genes, we observed decreased occupancy of the histone variant H2A.Z, but not canonical H2A, downstream of the TSS after CHD1 depletion. Interestingly, H2A.Z occupancy is negatively correlated with the +1 nucleosome barrier (18) and was further associated with nucleosome instability when in complex with the histone variant H3.3 (59,60,70). Our results support the notion that CHD1 modulates or promotes the exchange of the histone variant H2A.Z in a genome-wide manner near the TSS, thereby reducing nucleosome stability and promoting nucleosome turnover, resulting in a decreased barrier for RNAPII. Interestingly, we could not observe an effect of CHD1-depletion on H2A.Z occupancy at enhancer regions, pointing to another CHD1-independent mechanism at these sites. Although studies in Drosophila showed a CHD1-dependent incorporation of H3.3 into chromatin (71), we were unable to detect any significant change in H3.3 occupancy near the TSS in CHD1-depleted cells. Concordantly, it was shown that CHD1, unlike CHD2, had only minor effects on H3.3 turnover at single genes in human cells (72), whereas CHD1 rather maintained open DNA-regions. This further supports the idea that CHD1 contributes to establishing an active and open chromatin state at differentiation-induced genes.
Although CHD1-depletion generally impaired osteoblast differentiation-induced gene expression, it surprisingly resulted in enhanced transcription of the osteoblast marker genes ALPL and BGLAP in MSC. This is likely unrelated to the effects of CHD1 near the TSS as only low levels of CHD1 were detected at the TSS of the BGLAP or ALPL genes (data not shown). Interestingly, this inhibitory effect of CHD1 can be potentially explained by the recently discovered interaction of CHD1 with LSD1 (73), which was shown to repress osteoblast differentiation by silencing ALPL and BGLAP expression (74). In contrast to the higher expression of these early osteoblastic markers, CHD1 depletion led to impaired mineralization and decreased expression of extracellular matrix-associated genes. Interestingly, symptoms associated with osteomalacia also include high alkaline phosphatase levels accompanied by decreased bone mineralization. Although this disease is mainly described to be caused by malnutrition, heredity has also been implicated in osteomalacia (75,76). Thus, it is tempting to speculate whether mutation or altered expression of CHD1 in some patients could lead to osteomalacia.
Altogether, our results provide further mechanistic insight into the function of CHD1 during differentiation by analyzing its genome-wide occupancy and effects on gene expression. Remarkably, the significance of CHD1 for osteoblast biology is further supported by in vivo ectopic bone formation studies in mice. Furthermore, we provide evidence that CHD1 is required for efficient transcriptional regulation during lineage specification. Further studies will be required to determine the role of CHD1 in the context of different lineage-specifying transcription factors and transcriptional coregulatory complexes like Mediator. We hypothesize that the absence or decreased expression of CHD1, for example in prostate cancer where CHD1 is frequently mutated or deleted (77), may result in aberrant transcriptional changes which may affect therapeutic responsiveness. Analogous to the synthetic lethal effects we recently reported in response to PARP inhibitor treatment of CHD1-deficient cells following DNA damage (78), we speculate that the application of molecules such as BET inhibitors to CHD1-deficient cells may further increase RNAPII pausing and specifically target this subset of tumors. Together, these findings reveal an important role for CHD1 in lineage specification and provide a foundation for further analyses examining its function in various bone-related diseases as well as its tumor suppressor function and the targetability of CHD1-deficient tumors.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank G. Salinas-Riester, D. Indenbirken, and A. Grundhoff for support with next generation sequencing; V. Assmann for help with transduction experiments; and N. Molitor for excellent technical support.
Author contributions: The experiments were designed by S.A.J. and S.J.B. S.N. and W.X. contributed to bioinformatic analyses. S.J.B. and T.H. performed the in vitro experiments in hFOB 1.19 and hMSCs. S.J.B. generated all the ChIP-Seq and mRNA-Seq data. N.D. and M.K. conducted in vivo bone formation experiments. V.K. and Z.N. contributed intellectually to the planning of the project and data analysis. The manuscript was written by S.J.B., Z.N. and S.A.J. All authors read and corrected the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Academic Exchange Service (DAAD) (to S.N.); German Research Foundation [DFG; JO 815/3-1 to S.A.J.]; German Ministry for Science and Education (BMBF)-funded iBONE consortium [01KU1401A to S.A.J.]. Funding for open access charge: Internal funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kassem M., Bianco P.. Skeletal stem cells in space and time. Cell. 2015; 160:17–19. [DOI] [PubMed] [Google Scholar]
- 2. Méndez-Ferrer S., Michurina T.V., Ferraro F., Mazloom A.R., MacArthur B.D., Lira S.A., Scadden D.T., Ma’ayan A., Enikolopov G.N., Frenette P.S.. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010; 466:829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Owen M., Friedenstein A.J.. Stromal stem cells: marrow-derived osteogenic precursors. Ciba Found. Symp. 1988; 136:42–60. [DOI] [PubMed] [Google Scholar]
- 4. Sacchetti B., Funari A., Michienzi S., Cesare S.D., Piersanti S., Saggio I., Tagliafico E., Ferrari S., Robey P.G., Riminucci M. et al. . Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007; 131:324–336. [DOI] [PubMed] [Google Scholar]
- 5. Abdallah B.M., Kassem M.. New factors controlling the balance between osteoblastogenesis and adipogenesis. Bone. 2012; 50:540–545. [DOI] [PubMed] [Google Scholar]
- 6. Siersbæk R., Nielsen R., John S., Sung M.-H., Baek S., Loft A., Hager G.L., Mandrup S.. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J. 2011; 30:1459–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thompson B., Varticovski L., Baek S., Hager G.L.. Genome-wide chromatin landscape transitions identify novel pathways in early commitment to osteoblast differentiation. PLoS ONE. 2016; 11:e0148619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Venkatesh S., Workman J.L.. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015; 16:178–189. [DOI] [PubMed] [Google Scholar]
- 9. Chen T., Dent S.Y.R.. Chromatin modifiers: regulators of cellular differentiation. Nat. Rev. Genet. 2014; 15:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gaspar-Maia A., Alajem A., Polesso F., Sridharan R., Mason M.J., Heidersbach A., Ramalho-Santos J., McManus M.T., Plath K., Meshorer E. et al. . Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009; 460:863–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clapier C.R., Cairns B.R.. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009; 78:273–304. [DOI] [PubMed] [Google Scholar]
- 12. Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. et al. . A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006; 125:315–326. [DOI] [PubMed] [Google Scholar]
- 13. Cui K., Zang C., Roh T.-Y., Schones D.E., Childs R.W., Peng W., Zhao K.. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009; 4:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harada A., Okada S., Konno D., Odawara J., Yoshimi T., Yoshimura S., Kumamaru H., Saiwai H., Tsubota T., Kurumizaka H. et al. . Chd2 interacts with H3.3 to determine myogenic cell fate. EMBO J. 2012; 31:2994–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu G., Cui K., Northrup D., Liu C., Wang C., Tang Q., Ge K., Levens D., Crane-Robinson C., Zhao K.. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2013; 12:180–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Skene P.J., Hernandez A.E., Groudine M., Henikoff S.. The nucleosomal barrier to promoter escape by RNA polymerase II is overcome by the chromatin remodeler Chd1. eLife. 2014; 3:e02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stasevich T.J., Hayashi-Takanaka Y., Sato Y., Maehara K., Ohkawa Y., Sakata-Sogawa K., Tokunaga M., Nagase T., Nozaki N., McNally J.G. et al. . Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 2014; 516:272–275. [DOI] [PubMed] [Google Scholar]
- 18. Weber C.M., Ramachandran S., Henikoff S.. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol. Cell. 2014; 53:819–830. [DOI] [PubMed] [Google Scholar]
- 19. Zentner G.E., Henikoff S.. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 2013; 20:259–266. [DOI] [PubMed] [Google Scholar]
- 20. Morettini S., Tribus M., Zeilner A., Sebald J., Campo-Fernandez B., Scheran G., Wörle H., Podhraski V., Fyodorov D.V., Lusser A.. The chromodomains of CHD1 are critical for enzymatic activity but less important for chromatin localization. Nucleic Acids Res. 2011; 39:3103–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kelley D.E., Stokes D.G., Perry R.P.. CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma. 1999; 108:10–25. [DOI] [PubMed] [Google Scholar]
- 22. Lin J.J., Lehmann L.W., Bonora G., Sridharan R., Vashisht A.A., Tran N., Plath K., Wohlschlegel J.A., Carey M.. Mediator coordinates PIC assembly with recruitment of CHD1. Genes Dev. 2011; 25:2198–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sims R.J., Millhouse S., Chen C.-F., Lewis B.A., Erdjument-Bromage H., Tempst P., Manley J.L., Reinberg D.. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell. 2007; 28:665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Radman-Livaja M., Quan T.K., Valenzuela L., Armstrong J.A., van Welsem T., Kim T., Lee L.J., Buratowski S., van Leeuwen F., Rando O.J. et al. . A key role for Chd1 in histone H3 dynamics at the 3΄ ends of long genes in yeast. PLoS Genet. 2012; 8:e1002811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burkhardt L., Fuchs S., Krohn A., Masser S., Mader M., Kluth M., Bachmann F., Huland H., Steuber T., Graefen M. et al. . CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res. 2013; 73:2795–2805. [DOI] [PubMed] [Google Scholar]
- 26. Guzman-Ayala M., Sachs M., Koh F.M., Onodera C., Bulut-Karslioglu A., Lin C.-J., Wong P., Nitta R., Song J.S., Ramalho-Santos M.. Chd1 is essential for the high transcriptional output and rapid growth of the mouse epiblast. Development. 2015; 142:118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koh F.M., Lizama C.O., Wong P., Hawkins J.S., Zovein A.C., Ramalho-Santos M.. Emergence of hematopoietic stem and progenitor cells involves a Chd1-dependent increase in total nascent transcription. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E1734–E1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mao A.S., Mooney D.J.. Regenerative medicine: Current therapies and future directions. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:14452–14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee J.-S., Garrett A.S., Yen K., Takahashi Y.-H., Hu D., Jackson J., Seidel C., Pugh B.F., Shilatifard A.. Codependency of H2B monoubiquitination and nucleosome reassembly on Chd1. Genes Dev. 2012; 26:914–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karpiuk O., Najafova Z., Kramer F., Hennion M., Galonska C., König A., Snaidero N., Vogel T., Shchebet A., Begus-Nahrmann Y. et al. . The histone H2B monoubiquitination regulatory pathway is required for differentiation of multipotent stem cells. Mol. Cell. 2012; 46:705–713. [DOI] [PubMed] [Google Scholar]
- 31. Harris S.A., Enger R.J., Riggs B.L., Spelsberg T.C.. Development and characterization of a conditionally immortalized human fetal osteoblastic cell line. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 1995; 10:178–186. [DOI] [PubMed] [Google Scholar]
- 32. Simonsen J.L., Rosada C., Serakinci N., Justesen J., Stenderup K., Rattan S.I.S., Jensen T.G., Kassem M.. Telomerase expression extends the proliferative life-span and maintains the osteogenic potential of human bone marrow stromal cells. Nat. Biotechnol. 2002; 20:592–596. [DOI] [PubMed] [Google Scholar]
- 33. Chen L., Ditzel N.. In vivo heterotopic bone formation assay using isolated mouse and human mesenchymal stem cells. Bio-Protoc. 2015; 5:e1389. [Google Scholar]
- 34. Nagarajan S., Hossan T., Alawi M., Najafova Z., Indenbirken D., Bedi U., Taipaleenmäki H., Ben-Batalla I., Scheller M., Loges S. et al. . Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep. 2014; 8:460–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Najafova Z., Tirado-Magallanes R., Subramaniam M., Hossan T., Schmidt G., Nagarajan S., Baumgart S.J., Mishra V.K., Bedi U., Hesse E. et al. . BRD4 localization to lineage-specific enhancers is associated with a distinct transcription factor repertoire. Nucleic Acids Res. 2016; doi:10.1093/nar/gkw826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simon J.M., Giresi P.G., Davis I.J., Lieb J.D.. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat. Protoc. 2012; 7:256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Anders S., Huber W.. Differential expression analysis for sequence count data. Genome Biol. 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Warnes G.R., Bolker B., Bonebakker L., Gentleman R., Liaw W.H.A., Lumley T., Maechler M., Magnusson A., Moeller S., Schwartz M. et al. . gplots: various R programming tools for plotting data. 2016.
- 40. Huang D.W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 41. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; doi:10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. . Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shin H., Liu T., Manrai A.K., Liu X.S.. CEAS: cis-regulatory element annotation system. Bioinforma. Oxf. Engl. 2009; 25:2605–2606. [DOI] [PubMed] [Google Scholar]
- 46. Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J.. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004; 32:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R. et al. . Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012; 481:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G.. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010; 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Griffon A., Barbier Q., Dalino J., van Helden J., Spicuglia S., Ballester B.. Integrative analysis of public ChIP-seq experiments reveals a complex multi-cell regulatory landscape. Nucleic Acids Res. 2015; 43:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lin C.Y., Lovén J., Rahl P.B., Paranal R.M., Burge C.B., Bradner J.E., Lee T.I., Young R.A.. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012; 151:56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zeitlinger J., Stark A., Kellis M., Hong J.-W., Nechaev S., Adelman K., Levine M., Young R.A.. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007; 39:1512–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Alford A.I., Kozloff K.M., Hankenson K.D.. Extracellular matrix networks in bone remodeling. Int. J. Biochem. Cell Biol. 2015; 65:20–31. [DOI] [PubMed] [Google Scholar]
- 54. Brown S.D.M., Moore M.W.. The international mouse phenotyping consortium: past and future perspectives on mouse phenotyping. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2012; 23:632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Larsen K.H., Frederiksen C.M., Burns J.S., Abdallah B.M., Kassem M.. Identifying a molecular phenotype for bone marrow stromal cells with in vivo bone-forming capacity. J. Bone Miner. Res. 2010; 25:796–808. [DOI] [PubMed] [Google Scholar]
- 56. Twine N.A., Chen L., Pang C.N., Wilkins M.R., Kassem M.. Identification of differentiation-stage specific markers that define the ex vivo osteoblastic phenotype. Bone. 2014; 67:23–32. [DOI] [PubMed] [Google Scholar]
- 57. Kaplan N., Moore I.K., Fondufe-Mittendorf Y., Gossett A.J., Tillo D., Field Y., LeProust E.M., Hughes T.R., Lieb J.D., Widom J. et al. . The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009; 458:362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fuchs G., Hollander D., Voichek Y., Ast G., Oren M.. Cotranscriptional histone H2B monoubiquitylation is tightly coupled with RNA polymerase II elongation rate. Genome Res. 2014; 24:1572–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jin C., Felsenfeld G.. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007; 21:1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jin C., Zang C., Wei G., Cui K., Peng W., Zhao K., Felsenfeld G.. H3.3/H2A.Z double variant–containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet. 2009; 41:941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Brunelle M., Nordell Markovits A., Rodrigue S., Lupien M., Jacques P.-É., Gévry N.. The histone variant H2A.Z is an important regulator of enhancer activity. Nucleic Acids Res. 2015; 43:9742–9756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen P., Zhao J., Wang Y., Wang M., Long H., Liang D., Huang L., Wen Z., Li W., Li X. et al. . H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev. 2013; 27:2109–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Das C., Tyler J.K.. Histone exchange and histone modifications during transcription and aging. Biochim. Biophys. Acta. 2013; 1819:332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. West J.A., Cook A., Alver B.H., Stadtfeld M., Deaton A.M., Hochedlinger K., Park P.J., Tolstorukov M.Y., Kingston R.E.. Nucleosomal occupancy changes locally over key regulatory regions during cell differentiation and reprogramming. Nat. Commun. 2014; 5:4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Voss T.C., Hager G.L.. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet. 2014; 15:69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schones D.E., Cui K., Cuddapah S., Roh T.-Y., Barski A., Wang Z., Wei G., Zhao K.. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008; 132:887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Svensson J.P., Shukla M., Menendez-Benito V., Norman-Axelsson U., Audergon P., Sinha I., Tanny J.C., Allshire R.C., Ekwall K.. A nucleosome turnover map reveals that the stability of histone H4 Lys20 methylation depends on histone recycling in transcribed chromatin. Genome Res. 2015; 25:872–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Devaiah B.N., Case-Borden C., Gegonne A., Hsu C.H., Chen Q., Meerzaman D., Dey A., Ozato K., Singer D.S.. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat. Struct. Mol. Biol. 2016; 23:540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shi J., Whyte W.A., Zepeda-Mendoza C.J., Milazzo J.P., Shen C., Roe J.-S., Minder J.L., Mercan F., Wang E., Eckersley-Maslin M.A. et al. . Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013; 27:2648–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shu H., Nakamura M., Siretskiy A., Borghi L., Moraes I., Wildhaber T., Gruissem W., Hennig L.. Arabidopsisreplacement histone variant H3.3 occupies promoters of regulated genes. Genome Biol. 2014; 15:R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Konev A.Y., Tribus M., Park S.Y., Podhraski V., Lim C.Y., Emelyanov A.V., Vershilova E., Pirrotta V., Kadonaga J.T., Lusser A. et al. . The CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007; 317:1087–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Siggens L., Cordeddu L., Rönnerblad M., Lennartsson A., Ekwall K.. Transcription-coupled recruitment of human CHD1 and CHD2 influences chromatin accessibility and histone H3 and H3.3 occupancy at active chromatin regions. Epigenet. Chromatin. 2015; 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Metzger E., Willmann D., McMillan J., Forne I., Metzger P., Gerhardt S., Petroll K., von Maessenhausen A., Urban S., Schott A.-K. et al. . Assembly of methylated KDM1A and CHD1 drives androgen receptor-dependent transcription and translocation. Nat. Struct. Mol. Biol. 2016; 23:132–139. [DOI] [PubMed] [Google Scholar]
- 74. Ge W., Liu Y., Chen T., Zhang X., Lv L., Jin C., Jiang Y., Shi L., Zhou Y.. The epigenetic promotion of osteogenic differentiation of human adipose-derived stem cells by the genetic and chemical blockade of histone demethylase LSD1. Biomaterials. 2014; 35:6015–6025. [DOI] [PubMed] [Google Scholar]
- 75. Takeyari S., Yamamoto T., Kinoshita Y., Fukumoto S., Glorieux F.H., Michigami T., Hasegawa K., Kitaoka T., Kubota T., Imanishi Y. et al. . Hypophosphatemic osteomalacia and bone sclerosis caused by a novel homozygous mutation of the FAM20C gene in an elderly man with a mild variant of Raine syndrome. Bone. 2014; 67:56–62. [DOI] [PubMed] [Google Scholar]
- 76. Yue H., Yu J., He J., Zhang Z., Fu W., Zhang H., Wang C., Hu W., Gu J., Hu Y. et al. . Identification of two novel mutations in the PHEX gene in Chinese patients with hypophosphatemic rickets/osteomalacia. PLoS One. 2014; 9:e97830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Grasso C.S., Wu Y.-M., Robinson D.R., Cao X., Dhanasekaran S.M., Khan A.P., Quist M.J., Jing X., Lonigro R.J., Brenner J.C. et al. . The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012; 487:239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kari V., Mansour W.Y., Raul S.K., Baumgart S.J., Mund A., Grade M., Sirma H., Simon R., Will H., Dobbelstein M. et al. . Loss of CHD1 causes DNA repair defects and enhances prostate cancer therapeutic responsiveness. EMBO Rep. 2016; 17:1609–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.