Abstract
A unique feature of RNA polymerase II (RNA pol II) is its long C-terminal extension, called the carboxy-terminal domain (CTD). The well-studied eukaryotes possess a tandemly repeated 7-amino-acid sequence, called the canonical CTD, which orchestrates various steps in mRNA synthesis. Many eukaryotes possess a CTD devoid of repeats, appropriately called a non-canonical CTD, which performs completely unknown functions. Trypanosoma brucei, the etiologic agent of African Sleeping Sickness, deploys an RNA pol II that contains a non-canonical CTD to accomplish an unusual transcriptional program; all protein-coding genes are transcribed as part of a polygenic precursor mRNA (pre-mRNA) that is initiated within a several-kilobase-long region, called the transcription start site (TSS), which is upstream of the first protein-coding gene in the polygenic array. In this report, we show that the non-canonical CTD of T. brucei RNA pol II is important for normal protein-coding gene expression, likely directing RNA pol II to the TSSs within the genome. Our work reveals the presence of a primordial CTD code within eukarya and indicates that proper recognition of the chromatin landscape is a central function of this RNA pol II-distinguishing domain.
INTRODUCTION
Eukaryotic mRNA synthesis is orchestrated by the RNA polymerase II (RNA pol II) carboxy-terminal domain (CTD) (1–5). There are two types of CTDs: the canonical CTD, consisting of tandem repeats of the heptapeptide, Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 and the non-canonical CTD, devoid of any repeats. All well-studied model organisms contain a CTD of the canonical type within their RNA pol II (6,7). This CTD is known to undergo a multitude of post-translational modifications, which help signal the co-transcriptional activities that occur during the synthesis of monogenic mRNAs (8). In addition, the CTD modification codes communicate with the chromatin landscape during the transcription cycle (9).
While the myriad roles played by the canonical CTD have been illuminated over the past three decades, functions of non-canonical CTDs are largely unexplored (7,10). Trypanosoma brucei, the etiologic agent of African Sleeping Sickness, deploys an RNA pol II containing a non-canonical CTD to accomplish an unusual mode of transcription; all protein-coding genes are transcribed as polygenic precursor mRNAs (pre-mRNAs) that are initiated within several-kilobase-long regions, called transcription start sites (TSSs), upstream of the first protein-coding gene in arrays of genes organized head-to-tail within 11 large (1–5 Mb) chromosomes (11). This assertion is based on recent transcriptome analysis and 5΄ tri-phosphorylated RNA mapping data (12–14). TSSs appear to be devoid of conserved sequences and do not appear to be bound by normal transcription factors. These findings are consistent with our inability to detect conventional pre-mRNA gene promoter sequences. Our minimal understanding of trypanosome RNA pol II promoters derives from studies defining the spliced-leader (SL) RNA gene promoter, which appears to be an atypical promoter element (15).
In the case of the ∼167 head-to-head polygenic arrays present in the T. brucei genome, TSS regions are marked by histone variants H2Az and H2BV, as well as by a modified H4, H4K10ac (reviewed in (16)). It is possible that RNA pol II uses these histone marks to recognize TSS regions within the genome and ensure proper polygenic transcription. The resulting long, polycistronic pre-mRNAs are rapidly processed to produce stable mRNAs by 5΄ trans-splicing of a hypermethylated cap derived from a separately transcribed universal SL RNA and 3΄ polyadenylation (17).
The T. brucei RNA pol II non-canonical CTD is 284 amino-acids-long and contains an indispensible 90-residue central region (Figure 1A) (18). Like the canonical CTD, the non-canonical CTD is serine-rich (17%, compared to 7% for the entire polypeptide) and is post-translationally phosphorylated (18,19). However, the significance of these modifications in mRNA production is unknown. As protein-coding mRNAs are highly expressed by bacteriophage RNA polymerases driving their cognate promoters in transgenic trypanosomes, the non-canonical CTD is not crucial for pre-mRNA maturation. Thus, the essentiality of the non-canonical CTD likely lies in its role in TSS recognition.
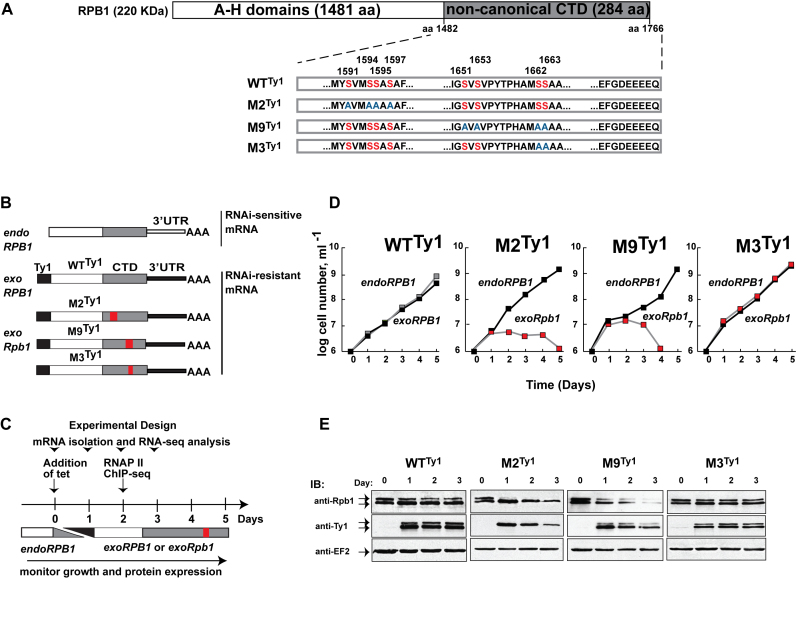
Figure 1.
Mutations in the CTD of T. brucei RPB1 cause varied phenotypes. (A) Schematic of T. brucei RPB1. The conserved A-H domains, characteristic of eukaryotic RNA pol II, make up the body of the polypeptide. They are followed by a 284-amino acid-long non-canonical CTD (gray) in place of the canonical CTD found in most model eukaryotes. Clusters of serine residues (red, numbered on top) in the WTTy1 CTD that were substituted with alanine residues (blue) in the cell lines M2Ty1, M9Ty1 and M3Ty1 are shown. Drawing is not to scale. (B) Schematic of genetic background in four transgenic cell lines. Endogenous RPB1 (endoRPB1) mRNAs, encoded by allelic pairs within chromosomes 4 and 8, contain similar 3΄UTRs (open bar) that are targeted for RNAi-mediated destruction in the presence of tetracycline. N-terminally tagged (black box) proteins are produced from RNAi-resistant mRNA containing a different 3΄UTR (closed bar), also in the presence of tetracycline. Red boxes mark regions containing the serine-to-alanine mutations. (C) Schematic of experimental design for growth and expression analyses. Stable transgenic cell lines were induced with tetracycline on Day 0. Time points indicate days after induction. The gray and black ramps indicate the corresponding decrease of endogenous RPB1 and increase of tagged proteins. (D) Growth curves of WTTy1, M2Ty1, M9Ty1 and M3Ty1 cells in the absence (black squares) and presence (gray/red squares) of tetracycline. Tetracycline addition caused endogenous RPB1 depletion and concomitant production of tagged proteins. (E) Immunoblot analyses of whole cell extracts prepared before (Day 0) and after (Days 1–3) tetracycline induction. Anti-RPB1 antibodies detect both endogenous RPB1 and exogenous wild-type, or mutant, Ty1-tagged RPB1 proteins. Anti-Ty1 antibodies detect only the tagged proteins. Robust expression of tagged protein in each of the cell lines is visible on Day 1. As RPB1 is predicted to have multiple and dynamic modifications, it often appears as a doublet. Anti-EF2 immunoblots are loading controls.
In this report, we used mRNA-seq and RNA pol II chromatin immunoprecipitation to observe the global effects of mutations to the non-canonical CTD on TSS recognition in vivo. Clusters of serine-to-alanine mutations in the CTD adversely affected cell growth and decreased mRNA production from TSS-proximal genes. Other RNA pol II-dependent transcripts were mostly unaffected, consistent with previously observed mRNA production initiated from internal sites within transcription units (12). Interestingly, RNA pol II occupancy in the CTD-mutant M2Ty1 and M9Ty1 parasites was shifted downstream from TSS regions, which was not the case for the WTTy1RNA pol II occupancy. These data indicate that the non-canonical CTD of T. brucei RNA pol II is essential for the proper positioning of polymerase to ensure normal mRNA production.
MATERIALS AND METHODS
Plasmid constructs, mutagenesis and T. brucei strain constructs
A plasmid containing a tetracycline-inducible RNAi gene fragment that targets the endogenous RPB1 3΄UTR was described previously (18). pAD76, a pAD74 derivative, was constructed by insertion of a DNA fragment (primer pair AD271/AD368) encoding the A-H domains of RPB1 (amino acid 1–1481) between the unique XbaI and BstZ 17I sites, followed by insertion of a DNA fragment (primer pair AD441/AD442), encoding the entire CTD (amino acids 1482–1766), between the unique AvrII and NsiI sites, of pAD74 (18). pAD76 derivatives that expressed mutant RPB1 proteins were constructed by swapping the wild-type CTD with an altered CTD between the AvrII and NsiI sites. CTD alterations were generated by site-directed mutagenesis of the wild-type CTD within pCR2.1-TOPO™ using a Change-IT™ Multiple Mutation Site Directed Mutagenesis kit (Stratagene) before they were introduced into pAD76. pAD76 and its derivatives were linearized using NotI to facilitate homologous recombination of the construct into an rRNA loci. All exogenous tagged-RPB1 proteins are expressed from a tetracycline-inducible ribosomal RNA gene promoter, which is transcribed by RNA pol I. A blasticidin-resistance gene within the constructs allowed for the selection of stable transformants. Molecular cloning otherwise was done using standard procedures and Escherichia coli strains.
Trypanosome culture and growth analysis
The T. brucei Lister 427 procyclic cell line 29-13 and all stable transgenic cell lines were grown in SDM-79 media supplemented with 10% fetal bovine serum (tetracycline-free) at 27°C in a humidified chamber containing 5% CO2 (30). AD-101 cells were produced by introducing a tetracycline-inducible construct containing the RNAi sequence that targets the 3΄UTR of RPB1. All other cell lines were derived from AD-101 by introducing a tetracycline-inducible exogenous RPB1 gene (that produces the tagged proteins) with an RNAi-resistant 3΄UTR. Clonal selections of all cell lines were done by serial dilution, using 2.5 μg/ml Phleomycin and 10 μg/ml Blasticidin. All cell lines were verified by genomic PCR and DNA sequence analyses.
Antibodies and western analysis
Anti-CTD antibody, produced in rabbit against a recombinant CTD peptide, detects endogenous and exogenous RPB1 (tagged proteins). Anti-Ty1 antibody (31), a mouse monoclonal antibody that detects the tagged proteins, was obtained from the Antibody and Bioresource Core Facility of The Rockefeller University and Memorial Sloan-Kettering Cancer Center. Anti-RPB4 antibody was described previously (18). Expression of RPB1 was determined by western blot analysis of whole cell extracts prepared from parasites, before and after one, two and three days of tetracycline induction. Protein extracts from 2 × 106 cells, prepared by boiling in SDS-sample buffer, were separated by 8% SDS-PAGE and analyzed by immunoblotting using the ECL™ kit from Pierce. Inclusion of the tagged proteins into the 12-subunit-RNA pol II enzyme was verified by western blot analysis of antibody-captured RNA pol II with anti-RPB4 antibody. Anti-EF2 antibody was from Santa Cruz (sc-13004). Anti-2,2,7-trimethylguanosine mouse mAb (K121) agarose conjugate was from Calbiochem (NA02A).
RNA isolation and semi-quantitative RT-PCR analysis
Total cellular RNA (from 107 cells) was isolated using TRIzol™ reagent (Invitrogen) and treated with RQ1 RNase-free DNase (Promega) to remove DNA contamination. Transcript levels of genes were compared by reverse transcription (RT) followed by semi-quantitative PCR analysis. 7SL RNA, a small nuclear RNA transcribed by RNA pol III, was used as an internal control. Briefly, cDNAs were synthesized from 1–2 μg of total RNA in a 20 μl RT reaction using 5 μM random hexamers and Superscript III™ (Invitrogen) enzyme. Following heat inactivation (75°C, 15 min) and RNA removal using RNase H, 1 μl RT reaction was used for PCR amplification. Each 25 μl PCR reaction contained 40 μM of dATP, dTTP, and dGTP, and 4 μM dCTP along with 0.02 μM [32P]-dCTP, 1U of LongAmp Taq DNA polymerase and 0.4 μM primer pairs for the target mRNA and 7SLRNA (Supplementary Table S4). An equimolar ratio of Competimer™ pair (3΄-end blocked by ddNTP) for 7SL RNA was used to prevent saturation amplification. PCR products, separated by 6% PAGE, were visualized by exposing to a PhosphoImager™ screen and quantified using Imagequant software.
mRNA-Seq
mRNA-Seq libraries were prepared following the Illumina™ small RNA library preparation method. Poly(A)+-containing RNA from 10 μg of total RNA was captured by two rounds of oligo-d(T)n-bead selection and fragmented by base treatment. Following adaptor ligation to fragmented RNA, cDNA libraries were prepared by reverse transcription and amplified by PCR. Purified cDNA libraries were sequenced on the Illumina HiSeq™ platform. Sequence reads were mapped to T. brucei 927 genome assembly version 5 using Tophat2 (v2.0.8b) set at the following parameters: no-coverage-search, no-novel-juncs, no-novel-indels. The mapped reads were then converted to gene expression values using Cuffdiff2 (v2.1.1) with the default parameters (32). The gene annotation consisted of the coding regions. Log2-fold changes were displayed on IGV browser. Volcano plots were generated using FPKM value (log2-fold change) versus P-value in R.
RNA pol II-ChIP and ChIP-Seq
Chromatin immunoprecipitation (ChIP) was performed on extracts from all four cell lines (WTTy1, M2Ty1, M3Ty1 and M9Ty1), prepared after two days of tetracycline addition, as described (33). Briefly, 5 × 108 cells were cross-linked with 1% formaldehyde, directly added into cell cultures, via mixing by constant shaking at 25°C for 20 min. Formaldehyde was quenched with glycine (0.125 M), and cell pellets were washed twice with ice-cold phosphate buffer saline and collected by centrifugation. To prepare for chromatin fragmentation, cell pellets were washed once each with Lysis Buffer 1 and 2 and chromatin was fragmented in Lysis Buffer 3 by sonication using a Bioruptor™ (Diagenode) at high setting for 15 min (30 s on, 30 s off). Sheared chromatin samples were clarified by centrifugation to remove particulate debris and cleared supernatant was subjected to immunoprecipitation by mixing with anti-Ty1 antibodies for 12–15 h at 4°C. Antibody-bound chromatin was captured using anti-mouse antibody-coated magnetic beads (Dynabeads™ M-280), reverse cross-linked to separate DNA from protein, and DNA was purified by extracting twice with phenol/chloroform/isoamyl alcohol solution and concentrated by ethanol precipitation.
ChIP-PCR of RNA pol II within the chromosome 3 and chromosome 7 regions was determined by 30 cycles of amplification of ChIP DNA using [α-32P] dCTP as tracer and primers listed in Supplementary Table S4. Input reactions contain 1/100th fraction of chromatin DNA used in the immunoprecipitation. PCR products were separated by gel electrophoresis (6% PAGE), and visualized by PhosphorImager scanning of dried gels.
ChIP-Seq libraries were constructed using NEBNext™ Ultra DNA Library Prep Kit for Illumina™ (NEB). Libraries were size selected for 200 bp DNA insert using AMPure™ XP beads (Beckman Coulter), PCR amplified and sequenced on HiSeq™ platform (Illumina). Raw reads were filtered and adapters were removed by FASTQC v0.10.1 using default parameters. The reads were mapped to the reference genome using bowtie v2 2.1.0. The non-unique reads were randomly distributed. Binding enrichment was called from the aligned reads using MACS2 2.0.10 and SPP (http://compbio.med.harvard.edu/Supplements/ChIP-Seq) using default parameters. All statistical analyses were performed using R.
SL RNA cap analysis
The 5΄ end hypermethylated spliced leader (SL) RNA Cap structure in T. brucei [m7G(5΄)ppp(5΄)m26 ApmApmCmpm3Ump] was detected using primer extension of a [γ-32P] 5΄-labeled oligonucleotide 5΄-CTGGGAGCTTCTCATACCCAATA-3΄ after hybridization to 2 μg total RNA from transgenic parasites before and after 2 days of tetracycline induction. Extension products, which are a mix of hypermethylated Cap1-4 species, were resolved by electrophoresis on a 10% polyacrylamide-7 M urea gel. Unmodified SL RNA (Cap0) was obtained from transgenic parasites treated with sinefungin for 3 h before isolation.
Nascent RNA synthesis analysis
T. brucei parasites (4 × 107 cells/assay), collected before and after 2 days of tetracycline induction, were permeabilized using lysolecithin. [α-32P]-UTP was added to actively transcribing cells and transcription reactions were performed for 10 min. Nascent radiolabeled RNA was isolated using a phenol–chloroform–isoamyl alcohol solution, resolved by electrophoresis on a 6% polyacrylamide–7 M urea gel and visualized by PhosphorImaging™.
RNA polymerase II activity assay
RNA pol II transcription assays were performed following the method described in (34). Briefly, nuclear extracts from all four cell lines (WTTy1, M2Ty1, M3Ty1 and M9Ty1), prepared after one day of tetracycline induction, were used to immune-capture the tagged-RNA pol II complexes using anti-Ty1 antibodies and anti-mouse antibody-coated magnetic beads (DynabeadsTM M-280). Bead-bound, immune-captured RNA pol II complexes were used in transcription assays on Calf Thymus DNA (Sigma) as template and [α-32P]-UTP as tracer. Mouse pre-immune IgG was used as control for immune-capture. The resulting RNAs were separated on a denaturing 6% polyacrylamide gel and detected by PhosphorImaging™.
RESULTS
Clusters of serine-to-alanine mutations in the essential 90-amino acid central region of the non-canonical CTD negatively impact both cell growth and long RNA abundance
To investigate how the T. brucei RNA pol II machinery uses its non-canonical CTD, we have set up a genetic system in which cell growth depends upon exogenously-expressed Ty1-tagged versions of RPB1 that contain an RNAi-resistant 3΄UTR (18) (Figure 1B and C). The endogenous RPB1 (endoRPB1) subunit of RNA pol II was depleted using tetracycline-induced RNA interference (RNAi) that targeted its RNAi-sensitive 3΄ UTR. After 1–2 days of RNAi induction, endoRPB1 was largely depleted in a control cell line, ultimately causing cell death (Supplementary Figure S1A). Derivative cell lines were produced that contained either an exogenous, tetracycline-inducible, Ty1-tagged wild-type CTD-containing-RPB1 protein (designated as tagged-wild-type CTD) or a mutant CTD-containing-Rpb1 protein (designated as tagged-mutant CTD) (Supplementary Table S1). We measured the growth of these cell lines to assess if mutant CTDs enable cell viability (Figure 1D).
Two cell lines (WTTy1, including the tagged-wild-type CTD, and M3Ty1, including the tagged-mutant CTD with S1662A and S1663A substitutions) grew normally, whereas two cell lines (M2Ty1, including the tagged-mutant CTD with S1591A, S1594A, S1595A and S1597A substitutions and M9Ty1, containing the tagged-mutant CTD with S1651A, S1653A, S1662A and S1663A substitutions) were unable to grow (Figure 1D). Each of these four cell lines expressed functional RNA pol II complexes (Supplementary Figures S1B and S2) with comparable amounts of tagged RPB1 (Figure 1E and Supplementary Table S2) that could transcribe the RNA pol II-dependent SL RNA genes (Supplementary Figure S3A and B). Nascent SL RNA production was consistent with this finding; only small decreases were observed when measured by [32P]-UTP incorporation in permeabilized cells (Supplementary Figure S3C and D). In contrast, we observed significant decreases in total [32P]-UTP incorporation and abundance of long RNAs in the growth-altered mutant cells, but not in the normally growing cells, which suggests a defect in polygenic transcription by M2Ty1 and M9Ty1 RNA pol II (Supplementary Figure S3C and D).
Mutations within the non-canonical CTD result in reduced expression of TSS-proximal genes
To determine the transcriptional effect of mutations within the M2Ty1 and M9Ty1 cell lines, we measured gene expression levels before and after their tagged-mutant CTDs were produced (Figure 1C). We also measured gene expression levels in the WTTy1 before and after its tagged-wild-type CTD was produced and M3Ty1 cell lines before and after its tagged-mutant CTD was produced. cDNA libraries, representing poly(A)+ RNA from 16 individual cell cultures, were deep sequenced to obtain quantitative mRNA measurements (mRNA-Seq). Heat maps were used to visualize gene expression changes on day 1, 2 and 3, in WTTy1 or M2Ty1 cells, compared to day 0, when no tagged protein was produced (Figure 2A and B, Supplementary Figure S5A and B). Reliance on the tagged proteins did not markedly alter overall mRNA production levels in any of the four cell lines. Specifically, less than 2% of protein-coding genes in WTTy1 and M3Ty1 cells, and ∼4% of protein-coding genes in M2Ty1 and M9Ty1 cells, showed ≥4-fold change in expression on day 2 compared to day 0. However, in both M2Ty1 and M9Ty1 cells, genes specifically situated immediately downstream of TSSs showed markedly reduced expression. This reduced expression of TSS-proximal genes became progressively more pronounced during the three-day experiment (Figure 2A and B, Supplementary Figure S5A and B).
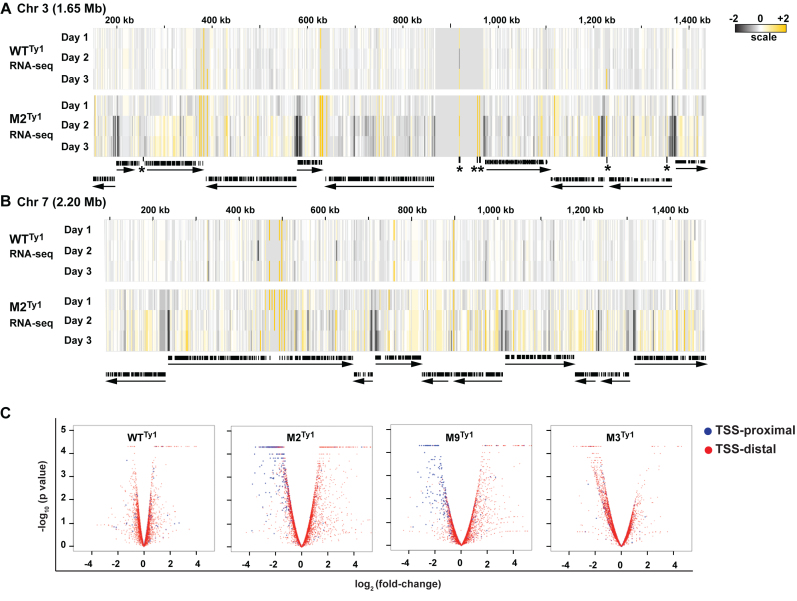
Figure 2.
mRNA-Seq shows reduced expression of TSS-proximal genes in M2Ty1 and M9Ty1 cells. (A and B) Heat map views of gene expression changes (Fragments Per Kilobase of transcript per Million mapped reads, log2-fold change) in WTTy1 and M2Ty1 cells on chromosomes 3 (A) and 7 (B) after 1 day (Day 1), 2 days (Day 2) and three days (Day 3) of tetracycline induction (compared to untreated cells). Data are shown using the IGV™ browser. Yellow and black show increases and decreases in expression, respectively. Black arrows indicate the length and direction of polycistronic transcription units. Black bars show coding sequences. Pre-ribosomal RNA gene clusters, tRNAs and snoRNAs are shown by asterisks (*). (C) Volcano plots for WTTy1, M2Ty1, M9Ty1 and M3Ty1 cells, showing gene expression (log2-fold changes) versus adjusted significance P-values (log10) for all protein coding genes, before and after two days of tetracycline addition. The set of 482 TSS-proximal genes is highlighted in blue; all other genes (∼8000) are shown in red.
To examine statistically whether the reduced expression of TSS-proximal genes is a general feature of M2Ty1 and M9Ty1 cells, we selected a representative TSS-proximal set of 482 genes, consisting of the first three genes from each of the 167 polygenic arrays (Supplementary Table S4; in a few cases, there are less than three proximal genes in an array) and determined their expression changes after 2 days of tagged-protein production. Expression of many TSS-proximal genes was significantly reduced (≥4-fold, P < 10−2) in the M2Ty1 and M9Ty1 mutants: 150 genes (31%) and 50 genes (10%), respectively (Figure 2C). In contrast, expression of no TSS-proximal genes in WTTy1, and only two TSS-proximal genes in M3Ty1, were reduced. Reduced expression of TSS-proximal genes in the M2Ty1 and M9Ty1 mutants is even evident after one day of tagged-protein production (Supplementary Figure S4).
We observed a striking spatial pattern between a gene's expression and its relative proximity to a TSS in the affected polygenic arrays in M2Ty1 and M9Ty1 cells. The first gene of each polygenic array showed the largest reduction in expression, whereas the downstream genes, in consecutive order, showed a less severe reduction (Figure 3A). We observed that the reduced expression correlated with published H2Az ChIP data (Figure 3B) (14). Additionally, expression of genes distal to a TSS, usually ∼8–10 kb downstream of the first gene, was unaltered. We confirmed these results for genes surrounding two divergently arranged TSSs through semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) using total RNA derived from cells before and after induction of mutant polymerase (Supplementary Figure S6A and B).
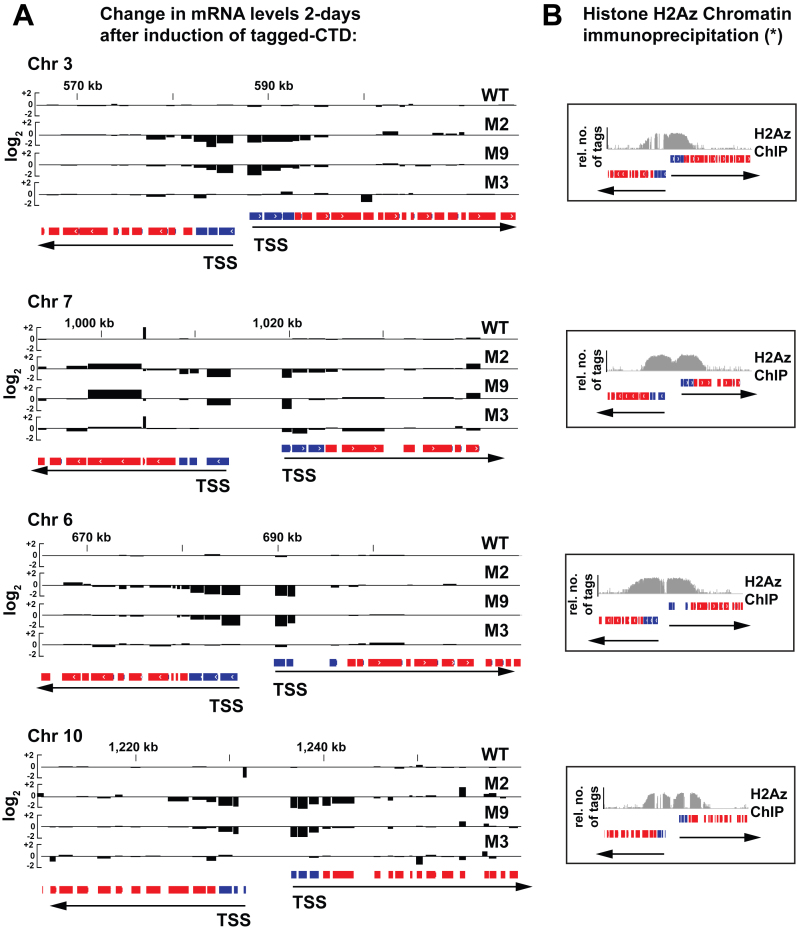
Figure 3.
Small clusters of mutations in the RNA Pol II CTD result in reduced RNA abundance within TSS-proximal genes. (A) Representative examples of transcript production at TSS-proximal and -distal regions on chromosomes 3, 7, 6 and 10 in cell lines WTTy1, M2Ty1, M9Ty1 and M3Ty1. Black bar plots show gene expression changes after 2 days of tetracycline-induced tagged RNA Pol II production relative to uninduced cells (log2-fold changes in Fragments Per Kilobase of transcript per Million mapped reads). The blue blocks denote TSS-proximal protein-coding genes. The red blocks denote TSS-distal protein-coding genes. The black arrows indicate the direction of transcription of polygenic units. The two sets of blue blocks in opposite orientations flank two divergently arranged TSSs. (B) Representative data showing that the ∼10 kb genomic loci, which include TSS regions, are marked by specific histone variants and histone modifications in wild-type T. brucei. Asterisk (*) indicates data is from Siegel et al. (2009). The variant histone H2Az occupancy (gray histograms) on regions shown corresponds to those in panel (A).
Mutations within the non-canonical CTD shift recruitment of RNA pol II downstream from TSS regions
In M2Ty1 and M9Ty1 cells, TSS-proximal genes were poorly expressed because they were either poorly transcribed into mRNAs or efficiently transcribed into highly unstable mRNAs. These two possibilities were distinguished by assessing RNA pol II engagement across the genome. Specifically, we performed chromatin immunoprecipitation (ChIP) assays in WTTy1 and M2Ty1 cells after 2 days of tagged-protein expression, and identified RNA pol II-associated DNA by deep sequencing (ChIP-Seq). RNA pol II ChIP-Seq of two biological replicate samples from both WTTy1 and M2Ty1 cells shows high correlations (Supplementary Figure S7 and S10). Analyses were done using combined replicate datasets.
Wild-type RNA pol II transcribes the SL RNA genes in T. brucei (15). As expected, RNA pol II in WTTy1 cells was highly associated (enrichment of ∼6-fold) with the transcribed region of the SL RNA gene clusters (Figure 4A). Similarly, RNA pol II in M2Ty1 cells was highly associated (enrichment of ∼8-fold) with the transcribed regions of the SL RNA gene clusters. Enrichment analyses in both cases were defined as the fold-change of RNA pol II ChIP DNA relative to total DNA (see Materials and Methods).
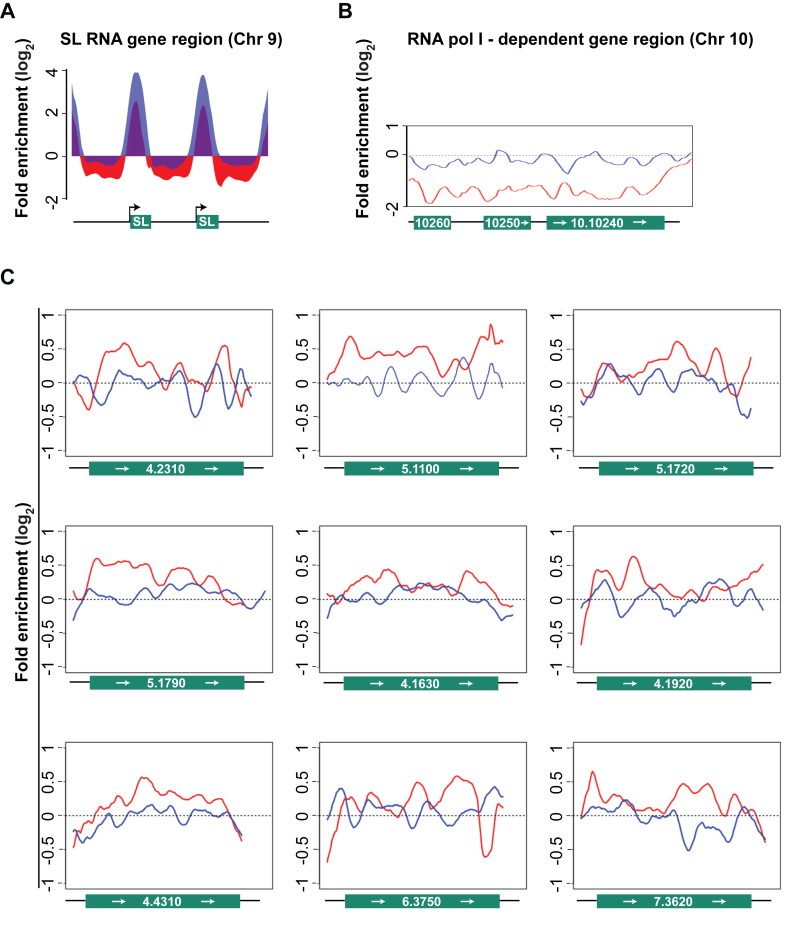
Figure 4.
ChIP-Seq analyses of RNA pol II engagement in WTTy1 and M2Ty1 cells. In all panels data from WTTy1 are red and data from M2Ty1 are blue. (A) Histograms of RNA pol II ChIP-enrichment relative to total DNA, on SL RNA genes in WTTy1 and M2Ty1 cells. A 4-kb region on chromosome 9, covering two SL RNA gene repeats (green boxes) plus flanking sequences (black line), is shown. Black arrows indicate direction of SL RNA transcription. The y-axis represents estimated ChIP enrichment (log2) relative to total DNA at each position based on smoothed tag density in each dataset. (B) Line plots show RNA pol II occupancy did not occur within RNAP I-dependent protein coding genes in either WTTy1 or M2Ty1 cells. An ∼10 kb region on chromosome 10, containing procyclins (EP1 and EP2; 10.10260 and 10.10250), procyclin-associated gene (PAG1; 10.10240) and flanking sequences (black line) is shown. The y-axis is as in (A). (C) Line plots show RNA pol II occupancy relative to total DNA, as in (A), on nine protein-coding genes that are expressed at high levels (above 90 percentile in RNA-Seq analysis; source is TriTrypDB) and are not within the TSS regions of any polygenic gene arrays. Gene IDs are shown as chromosome number followed by gene number. Each plot covers the entire coding region of the respective gene plus 200 base pair flanking sequences. Each of these genes are between 60 and 300 kb downstream from the first protein coding gene of the polygenic array in which they are embedded. These data show examples that WT Ty1 and M2Ty1 RNA pol II occupancy are often relatively the same at regions 60–300 kb downstream from the first protein-coding gene.
Wild-type RNA pol II does not transcribe the RNA pol I-dependent pre-rRNA gene cluster, nor does it transcribe the limited set of RNA pol I-dependent protein coding genes (20). As expected, RNA pol II in WTTy1 or M2Ty1 cells was not associated with the pre-rRNA gene cluster (data not shown) or the RNA pol I-dependent protein coding genes (Figure 4B), but does associate with RNA pol II-dependent protein coding genes (Figure 4C).
Wild-type RNA pol II's main function is to transcribe the polygenic protein-coding regions within T. brucei's 11 large chromosomes. RNA pol II occupancy, represented as positive log2-fold changes, is visible as orange peaks in the chromosome-wide line plots of WTTy1 and M2Ty1 ChIP-Seq analysis (Figure 5, Supplementary Figures S8 and S9). As expected, WTTy1 RNA pol II occupies the coding and intergenic regions of polygenic arrays, producing wide areas of relatively modest (∼1.2-fold, log2-fold changes), though clearly positive, ChIP-Seq enrichment distributed within the 167 polygenic arrays. In contrast to the RNA pol II patterns observed within the more typical monogenetic transcription units of other eukaryotes, we did not see enzyme enrichment at TSSs. This likely reflects the trypanosome-specific gene expression program that includes several-kilobase-long TSS regions, constitutively synthesized polygenic pre-mRNAs, and trans-spliced 7mG capped mRNAs (17).
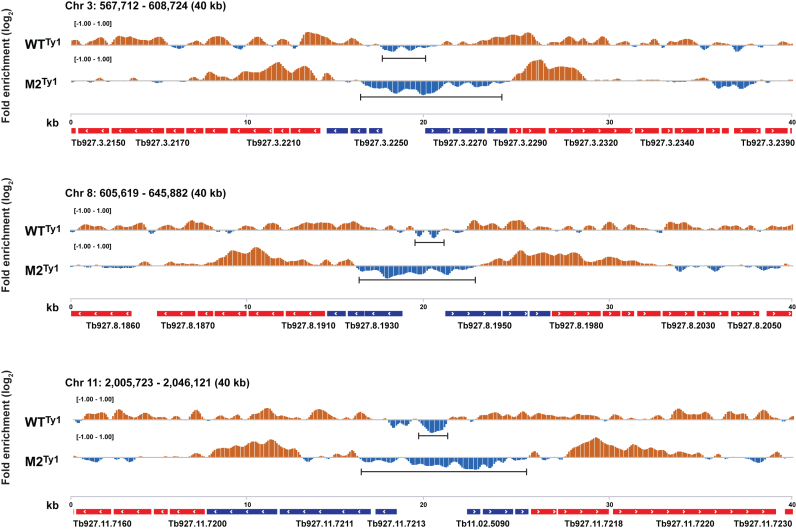
Figure 5.
ChIP-Seq shows altered recruitment of M2Ty1 RNA pol II to sites downstream from WTTy1 RNA pol II sites. Histograms, generated using the IGVTM browser, show RNA pol II ChIP enrichment relative to total DNA for WTTy1 (top) and M2Ty1 (bottom) cells in three representative TSSs, each encompassing a ∼40 kb region: chromosome 3 (top panel), chromosome 8 (middle panel) and chromosome 11 (bottom panel). The y-axes represent estimated ChIP enrichment (orange, positive; blue, negative). TSS-proximal and TSS-distal genes are shown in blue and red respectively. Arrows indicate orientation of gene transcription. Numbers above the gene arrays indicate the length (kb) of the chromosomal regions. The horizontal bar under each histogram marks the TSS regions that are deficient in RNA pol II occupancy. RNA pol II exclusions, represented as negative log2-fold changes, are visible as deep depressions in the chromosome-wide line plots of WTTy1 ChIP-Seq analysis. Notably, high RNA pol II enrichments were not found upstream of polygenic transcription units in T. brucei, in contrast to the high enzyme concentrations (ranging 2- to 8-fold) usually found upstream of the monogenic transcription units in most model systems.
There was a striking difference between the WTTy1RNA pol II and M2Ty1 RNA pol II occupancy within the chromosomal regions that define the probable promoters of polygenic pre-mRNA transcription. T. brucei's probable RNA pol II promoters are recognized by H2Az, H2BV and H4K10ac occupancy (12,13). These overlapping histone marks are broad; each one is ∼10 kb, covers a TSS region, and is upstream from, as well as within, the first several protein-coding regions of polygenic arrays. In WTTy1 cells, RNA pol II occupancy begins within these histone marks, consistent with its proper chromatin recognition of the TSS regions upstream for each polygenic gene array. In M2Ty1 cells, RNA pol II occupancy shows a downstream shift relative to the WTTy1 RNA pol II pattern. To probe the differences between the WTTy1 and M2Ty1 RNA pol II patterns within the TSS regions, we performed semi-quantitative PCR analyses on two representative TSS regions (Supplementary Figure S11). Consistent with the RNA pol II occupancy data shown in Figure 5 and Supplementary Figures S9 and S10, these data show that the TSS regions harbor much less mutant enzyme than wild-type enzyme.
Although it is established that co- and post-transcriptional processing of pre-mRNA plays a major role in linking RNA pol II activity to mRNA steady state populations, we assessed the relationship between the absence of mutant pol II within TSS regions and the levels of TSS-proximal genes determined in our RNA-seq data sets (Supplementary Figure S8). Although poor TSS recognition might be expected to lead to decreased TSS-proximal steady state mRNA levels, this will not always be the case as TSS-proximal genes are various distances from their cognate TSS regions, and post-transcriptional processes are known to play significant roles in mRNA production. Moreover, to provide an unbiased presentation of our RNA-seq data (Figure 5 and Supplementary Figures S9 and S10), we designated ‘TSS-proximal genes’ as the first three protein-coding regions of a polygenic unit, regardless of their position within a several-kilobase-long TSS region and regardless of their position relative to the H2Az, H2BV and H4K10ac marks. Thus, the first three mRNA-coding regions, color-coded as the ‘TSS-proximal gene set’ to describe our RNA-seq findings, map at various positions relative to the promoter-associated histone marks. The data analysis presented in Supplementary Figure S8 indicates that the downstream shift of mutant RNA pol II occupancy, relative to the WTTy1 RNA pol II pattern, only partially accounts for the alterations in steady state mRNA levels observed between the mutant and wild type cells.
An unexpected enrichment of RNA pol II occupancy was at the start of the downstream shift of the M2Ty1 RNA pol II pattern compared to the WTTy1 RNA pol II pattern (for example, compare ChIP-seq data corresponding to genes Tb927.3.2209 and Tb927.3.2210 in Figure 5 and all ‘zoomed in’ panels on Supplementary Figure S8). These 2–3 kb ChIP-enrichment regions in the M2Ty1 cells may contain non- processive enzyme, as they do not correspond to increased transcripts (mRNA-seq) from these regions.
DISCUSSION
Our study, the first detailed analysis of the global effects of mutating an RNA pol II non-canonical CTD, shows that the non-canonical CTD of T. brucei RNA pol II plays a critical role in navigating the genome. We have previously shown that the non-canonical CTD is essential for cell viability (18). We now demonstrate that minor alterations in the essential central region of the non-canonical CTD in T. brucei RNA pol II specifically affect the ability of RNA pol II to properly recognize the polygenic coding regions of the genome (Figure 2C). These same mutations do not affect the transcription of SL RNA genes. Thus, T. brucei RNA pol II likely uses its non-canonical CTD to discriminate polygenic mRNA gene arrays from SL RNA gene arrays in chromatin.
Having previously divided the CTD into thirds and studied the resulting truncations, we found that residues 1668–1766 were dispensable: Wild-type-level cell growth was maintained and the polymerase appeared intact (18). However, when residues 1577–1766 were removed, cells rapidly died. We therefore reasoned that a substitution mutation strategy, based on cluster serine-to-alanine mutagenesis (21–23) in the 1577–1668 region, would result in full-length RPB1-CTD mutants that would be altered in their transcriptional ability.
Experimental evidence and modeling suggest that canonical CTDs are flexible, disordered and conformationally mobile structures (5). Similarly, the T. brucei CTD appears to be highly disordered and mainly composed of ambiguous secondary structure/random coil, based on protein disorder algorithms and protein secondary structure prediction algorithms. Therefore, we do not expect our clustered serine-to-alanine mutagenesis strategy to result in gross CTD structural effects. Instead, it is likely that the local changes in the M2Ty1 and M9Ty1 mutants affect just a subset of CTD-mediated interactions and CTD-dependent modifications. Unfortunately, neither the CTD-mediated interactions nor CTD-dependent modifications have been extensively cataloged to date (24).
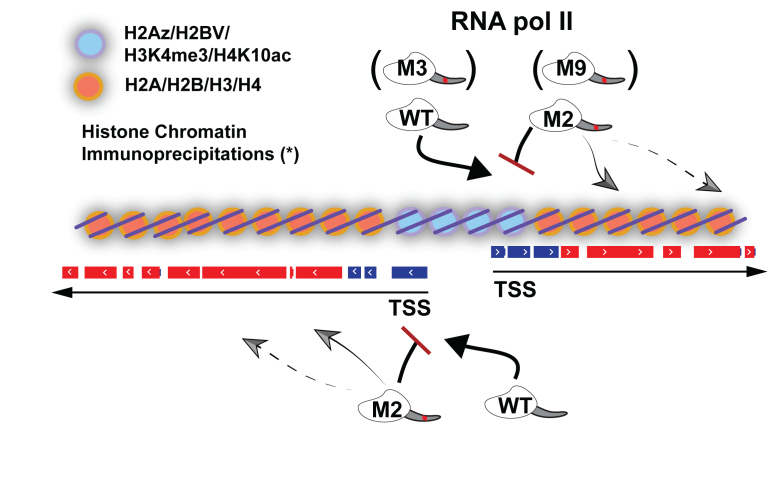
Although the production of polygenic pre-mRNAs and SL RNAs both require RNA pol II, their TSSs appear to have very different structures. The SL RNA gene TSS regions have well-defined specific promoter sequences and bind conventional RNA pol II transcription factors (25,26). The polygenic pre-mRNA TSS regions, while less well defined, are marked by specifically modified core histones and variant histones (13,14). It is likely that these chromatin marks govern RNA pol II recruitment in T. brucei as these organisms constitutively transcribe the clear majority of protein-coding genes, negating the need for a repertoire of factors to regulate the transcription of individual genes. Our model suggesting how RNA pol II reads the chromatin landscape is shown in Figure 6. The inability of the M2Ty1 and M9Ty1 RNA pol II to be recruited to TSSs likely prevents the subsequent CTD modifications (see Figure 1E) that are hallmarks of transcribing RNA pol II.
Figure 6.
Model showing WTTy1 (and M3Ty1) RNA pol II recruitment to chromatin regions at two divergently arranged TSSs, which have been shown to have a unique chromatin profile (blue circles). Specific clusters of mutations, in M2Ty1 (and M9Ty1) RNA pol II, prohibit recruitment to the TSS regions, and probably drive recruitment to internal chromatin regions (orange circles). Gene expression data (Figures 2 and 3) suggest M3Ty1 and M9Ty1 RNA pol II recruitments akin to WTTy1 and M2Ty1, respectively. TSS-proximal genes are in blue and TSS-distal genes are in red, following the convention established in Figure 3. Arrows indicate orientation of gene transcription. RNA pol II is drawn as an intact protein, containing the RPB1 markings established in Figure 1C. Asterisk (*) indicates chromatin profile based on Siegel et al. (2009).
It is unlikely that the histone marks or RNA pol II occupancy data reveal the precise dynamics of RNA pol II transcription within the T. brucei genome. For example, WTTy1 RNA pol II must transcribe the sequences upstream from the first protein-coding gene of a polygenic array as this upstream region contains essential SL RNA trans-splicing sites and a 5΄-untranslated region (5΄UTR). It appears that our ChIP assays do not capture the polymerase as it rapidly moves across non-coding gene regions after engaging chromatin within the TSS regions. Nevertheless, poor TSS-recognition by the mutant enzyme most likely accounts for the large downstream shift of enzyme engagement adjacent to the polygenic gene arrays within all chromosomes.
It is also unlikely that the lack of proper TSS recognition alone can fully explain the abnormal steady state mRNA pattern seen in the mutant parasites. For example, distance from TSS regions is known to influence gene expression levels in T. brucei (29). Our data show that the steady state levels of the genes far away from TSS regions appear to be increased (see Figure 2A and B) in mutant cells compared to wild type cells. If RNA pol II is blind to TSS regions, polymerase may be redirected to the internal regions of polygenic arrays. Internally initiated transcripts, recognized in T. brucei and the related parasite Leishmania major (12,27,28), may produce steady state mRNAs if they contain sufficient post-transcriptional signals. Additionally, a mRNA is more likely to be produced if it is far away from a TSS region, and thus more likely to benefit from an internal initiation event. Finally, a lack of proper TSS recognition by RNA pol II is expected to generate abnormal pre-mRNA transcripts which are unrecognizable to the cell's co and post-transcriptional machinery. These compromised machineries would produce the abnormal mRNA pattern seen in the M2Ty1 and M9Ty1 mutants and the loss of parasite viability.
In conclusion, the T. brucei RNA pol II non-canonical CTD permits polymerase to be properly recruited to TSS regions within chromatin. We speculate that in primitive organisms the CTD encodes information that enables RNA pol II to read the chromatin landscape present in the nucleus of growing parasites. Molecular genetic studies on non-canonical CTD-containing RNA pol II machinery will illuminate our speculation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank James F. Theis, George A.M. Cross and F. Nina Papavasiliou for critical reading of the manuscript.
Footnotes
Present address: Mahrukh Banday, Becton Dickinson, 7 Loveton Circle, MC, Sparks, MD 21152, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [AI108290, AI111453]. Funding for open access charge: NIH-NIAID [111453].
Conflict of interest statement. None declared.
REFERENCES
- 1. Cramer P., Armache K.J., Baumli S., Benkert S., Brueckner F., Buchen C., Damsma G.E., Dengl S., Geiger S.R., Jasiak A.J. et al. Structure of eukaryotic RNA polymerases. Annu. Rev. Biophys. 2008; 37:337–352. [DOI] [PubMed] [Google Scholar]
- 2. Darnell J.E., Jr Reflections on the history of pre-mRNA processing and highlights of current knowledge: a unified picture. RNA. 2013; 19:443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Egloff S., Dienstbier M., Murphy S.. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends Genet. 2012; 28:333–341. [DOI] [PubMed] [Google Scholar]
- 4. Eick D., Geyer M.. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev. 2013; 113:8456–8490. [DOI] [PubMed] [Google Scholar]
- 5. Corden J.L. RNA polymerase II C-terminal domain: Tethering transcription to transcript and template. Chem. Rev. 2013; 113:8423–8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chapman R.D., Heidemann M., Hintermair C., Eick D.. Molecular evolution of the RNA polymerase II CTD. Trends Genet. 2008; 24:289–296. [DOI] [PubMed] [Google Scholar]
- 7. Yang C., Stiller J.W.. Evolutionary diversity and taxon-specific modifications of the RNA polymerase II C-terminal domain. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:5920–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol. Cell. 2009; 36:541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lenstra T.L., Benschop J.J., Kim T., Schulze J.M., Brabers N.A., Margaritis T., van de Pasch L.A., van Heesch S.A., Brok M.O., Groot Koerkamp M.J. et al. The specificity and topology of chromatin interaction pathways in yeast. Mol. Cell. 2011; 42:536–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu P., Kenney J.M., Stiller J.W., Greenleaf A.L.. Genetic organization, length conservation, and evolution of RNA polymerase II carboxyl-terminal domain. Mol. Biol. Evol. 2010; 27:2628–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aslett M., Aurrecoechea C., Berriman M., Brestelli J., Brunk B.P., Carrington M., Depledge D.P., Fischer S., Gajria B., Gao X. et al. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010; 38:D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kolev N.G., Franklin J.B., Carmi S., Shi H., Michaeli S., Tschudi C.. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010; 6:e1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thomas S., Green A., Sturm N.R., Campbell D.A., Myler P.J.. Histone acetylations mark origins of polycistronic transcription in Leishmania major. BMC Genomics. 2009; 10:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Siegel T.N., Hekstra D.R., Kemp L.E., Figueiredo L.M., Lowell J.E., Fenyo D., Wang X., Dewell S., Cross G.A.. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009; 23:1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gilinger G., Bellofatto V.. Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res. 2001; 29:1556–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Croken M.M., Nardelli S.C., Kim K.. Chromatin modifications, epigenetics, and how protozoan parasites regulate their lives. Trends Parasitol. 2012; 28:202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liang X.H., Haritan A., Uliel S., Michaeli S.. Trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell. 2003; 2:830–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Das A., Bellofatto V.. The non-canonical CTD of RNAP-II is essential for productive RNA synthesis in Trypanosoma brucei. PLoS One. 2009; 4:e6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Urbaniak M.D., Martin D.M., Ferguson M.A.. Global quantitative SILAC phosphoproteomics reveals differential phosphorylation is widespread between the procyclic and bloodstream form lifecycle stages of Trypanosoma brucei. J. Proteome Res. 2013; 12:2233–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roditi I., Clayton C.. An unambiguous nomenclature for the major surface glycoproteins of the procyclic form of Trypanosoma brucei. Mol. Biochem. Parasitol. 1999; 103:99–100. [DOI] [PubMed] [Google Scholar]
- 21. Charbon G., Breunig K.D., Wattiez R., Vandenhaute J., Noel-Georis I.. Key role of Ser562/661 in Snf1-dependent regulation of Cat8p in Saccharomyces cerevisiae and Kluyveromyces lactis. Mol. Cell. Biol. 2004; 24:4083–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen C., Agnes F., Gelinas C.. Mapping of a serine-rich domain essential for the transcriptional, antiapoptotic, and transforming activities of the v-Rel oncoprotein. Mol. Cell. Biol. 1999; 19:307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sebastian S., Grutter C., Strambio de Castillia C., Pertel T., Olivari S., Grutter M.G., Luban J.. An invariant surface patch on the TRIM5alpha PRYSPRY domain is required for retroviral restriction but dispensable for capsid binding. J. Virol. 2009; 83:3365–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Badjatia N., Ambrosio D.L., Lee J.H., Gunzl A.. Trypanosome cdc2-related kinase 9 controls spliced leader RNA cap4 methylation and phosphorylation of RNA polymerase II subunit RPB1. Mol. Cell. Biol. 2013; 33:1965–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Palenchar J.B., Bellofatto V.. Gene transcription in trypanosomes. Mol. Biochem. Parasitol. 2006; 146:135–141. [DOI] [PubMed] [Google Scholar]
- 26. Lee J.H., Nguyen T.N., Schimanski B., Gunzl A.. Spliced leader RNA gene transcription in Trypanosoma brucei requires transcription factor TFIIH. Eukaryot. Cell. 2007; 6:641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martinez-Calvillo S., Yan S., Nguyen D., Fox M., Stuart K., Myler P.J.. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell. 2003; 11:1291–1299. [DOI] [PubMed] [Google Scholar]
- 28. Clark M.B., Amaral P.P., Schlesinger F.J., Dinger M.E., Taft R.J., Rinn J.L., Ponting C.P., Stadler P.F., Morris K.V., Morillon A. et al. The reality of pervasive transcription. PLoS Biol. 2011; 9:e1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kelly S., Kramer S., Schwede A., Maini P.K., Gull K., Carrington M.. Genome organization is a major component of gene expression control in response to stress and during the cell division cycle in trypanosomes. Open Biol. 2012; 2:120033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wirtz E., Leal S., Ochatt C., Cross G.A.. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999; 99:89–101. [DOI] [PubMed] [Google Scholar]
- 31. Bastin P., Bagherzadeh Z., Matthews K.R., Gull K.. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 1996; 77:235–239. [DOI] [PubMed] [Google Scholar]
- 32. Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L.. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013; 31:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee T.I., Johnstone S.E., Young R.A.. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 2006; 1:729–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Das A., Li H., Liu T., Bellofatto V.. Biochemical characterization of Trypanosoma brucei RNA polymerase II. Mol. Biochem. Parasitol. 2006; 150:201–210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.