Abstract
Adaptive prediction is a capability of diverse organisms, including microbes, to sense a cue and prepare in advance to deal with a future environmental challenge. Here, we investigated the timeframe over which adaptive prediction emerges when an organism encounters an environment with novel structure. We subjected yeast to laboratory evolution in a novel environment with repetitive, coupled exposures to a neutral chemical cue (caffeine), followed by a sublethal dose of a toxin (5-FOA), with an interspersed requirement for uracil prototrophy to counter-select mutants that gained constitutive 5-FOA resistance. We demonstrate the remarkable ability of yeast to internalize a novel environmental pattern within 50–150 generations by adaptively predicting 5-FOA stress upon sensing caffeine. We also demonstrate how novel environmental structure can be internalized by coupling two unrelated response networks, such as the response to caffeine and signaling-mediated conditional peroxisomal localization of proteins.
Keywords: adaptive prediction, structured environments, conditioned fitness, peroxisomal translocation, variant analysis
Introduction
Diverse organisms including microbes have evolved mechanisms to gain fitness advantage by sensing an environmental cue to anticipate and prepare in advance for a future selective pressure, a strategy known as adaptive prediction (AP) (Tagkopoulos et al. 2008; Mitchell et al. 2009). AP is generally beneficial to microbes as it can confer fitness advantage over competitors, facilitate competition for resources, and help with evasion of predators or host-defense systems (Woelfle et al. 2004; Johnson et al. 2008; Calhoun and Kwon 2010; Whitaker et al. 2010). However, AP can become disadvantageous in poorly structured environments or those with unpredictable patterns of change, especially when the advanced preparedness is maladaptive (Mitchell and Pilpel 2011). In fact, it takes just a few hundred generations in a novel environment for Escherichia coli to lose its capability to predict a downshift in oxygen upon sensing an upshift in temperature (Tagkopoulos et al. 2008). While prior studies have demonstrated the existence of AP and the rapidity with which it is lost (Tagkopoulos et al. 2008; Mitchell et al. 2009), emergence of this behavior has not previously been reported under laboratory controlled conditions, making it challenging to elucidate its evolutionary and mechanistic underpinnings.
Results and Discussion
Experimental Design of a Structured Environment to Probe AP Emergence
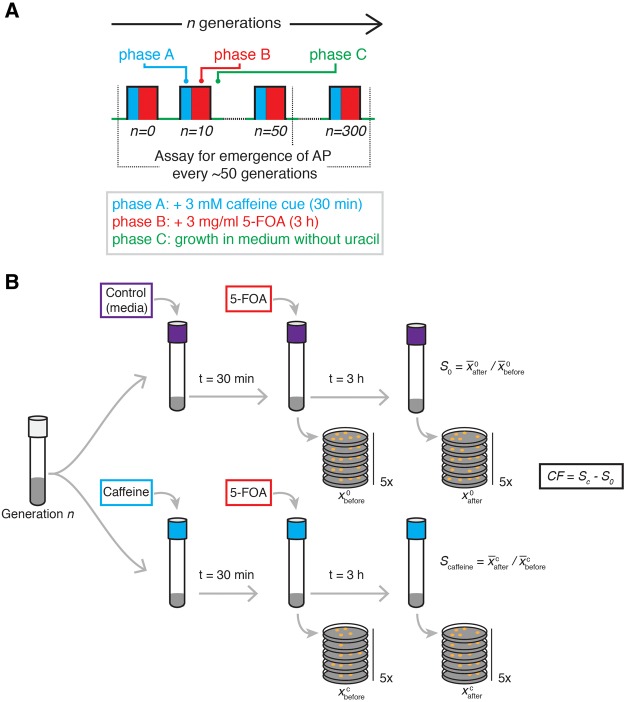
We sought to investigate the time scale over which AP can emerge in a microbial population subjected to laboratory evolution in a novel structured environment. In line with Pavlov’s classical conditioning experimental design (Pavlov 1927), we subjected yeast over every ∼10 generations to a 30-min exposure to a low innocuous dose of 3 mM caffeine (phase A) followed by a 3-h exposure to a sublethal dose (3 mg ml−1) of 5-fluoroorotic acid (5-FOA, phase B) (fig. 1a). Analogous to neutral stimuli used in classical conditioning experiments (Pavlov 1927), while caffeine triggers a global, pleiotropic response differentially regulating hundreds of genes in yeast (Kuranda et al. 2006; Reinke et al. 2006; Wanke et al. 2008; Rallis et al. 2013), it has no observable growth effect at the concentration used in our experiments (supplementary fig. S1, Supplementary Material online). The use of a neutral environmental cue (here caffeine) is important as it helps to differentiate AP from other evolutionary adaptations influenced by cross-protection (Dhar et al. 2013), where resistance to one stress has components that confer increased resistance to a second unrelated stress. 5-FOA is converted by orotidine 5′-phosphate decarboxylase (Ura3) into 5-fluorouracil, which is toxic to the cell (Boeke et al. 1984) (supplementary fig. S2, Supplementary Material online). Exposure to 5-FOA subsequent to caffeine represents a novel pair of stimuli that has not been previously associated by yeast. The conditional selection for 5-FOA resistance and counter-selection against uracil auxotrophy (phase C) is an important feature that we exploited to weed out cells that might have acquired constitutive resistance through mechanisms such as loss-of-function mutations in URA3. At intervals of ∼50 generations, we performed survival assays to assess whether conditioning with caffeine cued the population to better withstand a sub-lethal dose of 5-FOA. Specifically, the assays quantified changes in 5-FOA survival upon conditioning with caffeine, relative to survival without caffeine pre-treatment, defined as conditioned fitness (CF) (fig. 1b); hence CF > 0 is indicative of AP as it reflects survival increase because of pre-treatment with caffeine. Importantly, we also ascertained that CF was time dependent on caffeine pre-treatment for 30 min, and not present if caffeine and 5-FOA were added simultaneously in the survival assay (supplementary fig. S3, Supplementary Material online).
Fig. 1.
—Experimental design for investigating laboratory evolution of AP. (a) Yeast cultures were subjected to 30 cycles of laboratory evolution in a novel structured environment, with 10 generations between cycles. Each cycle had three phases: in phase A, cultures were exposed to 3 mM caffeine for 30 min; followed by 3-h exposure to 3 mg/ml 5-FOA in phase B; thereafter, an aliquot of the culture was transferred to fresh medium for overnight growth without uracil (phase C). To prevent temporal conditioning, short random intervals (< 2 h) were introduced between cycles. (b) Conditioned fitness (CF), defined as relative change in survival to 5-FOA given caffeine as a cue, was calculated using a culture aliquot that was exposed to caffeine or just growth medium (control), prior to treatment with 5-FOA. Survival was assessed by sampling immediately before or after adding 5-FOA, by counting colony formation units (CFUs) on five replicate plates (see Materials and Methods).
Synthetic Engineered Circuit to Prime Emergence of AP
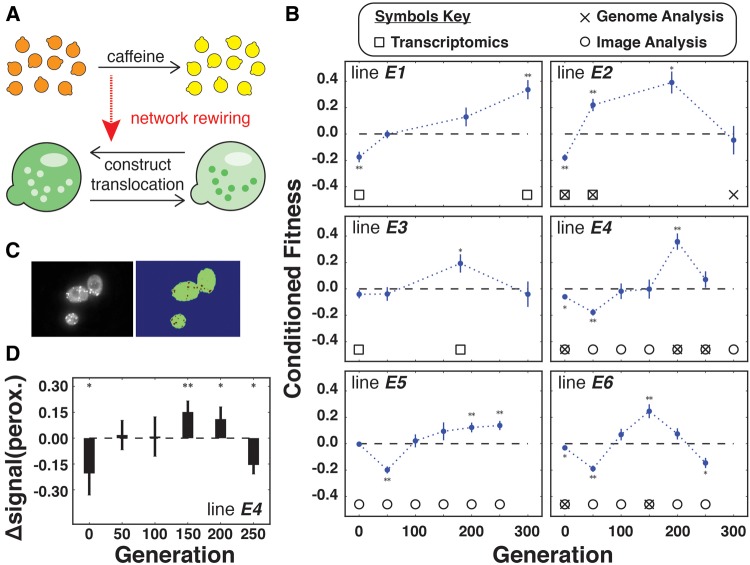
We first investigated whether AP could emerge through natural selection of mutations that internalize environmental structure by rewiring the gene regulatory network of yeast, linking the caffeine response to an unrelated process that reduced 5-FOA toxicity. Our experimental strategy was motivated by the observation that caffeine triggers a global pleiotropic response that modulates, among other pathways, signaling by the target of rapamycin complex 1, TORC1 (Wanke et al. 2008; Rallis et al. 2013). TORC1 is a serine/threonine kinase with a role in globally coordinating cell growth and homeostasis in response to a diverse array of environmental cues (Zhang et al. 2011; Laplante and Sabatini 2012; Hughes Hallett et al. 2014). It is activated by changes in oxygen levels, amino acid concentrations, energy levels and growth factors and negatively regulated by stress and certain chemicals including rapamycin and caffeine (Rallis et al. 2013). Its homolog, TORC2, functions independently of TORC1 to regulate metabolism, cytoskeletal organization, cell survival, and protein translocation, and is insensitive to caffeine (Reinke et al. 2006; Jung et al. 2010). We engineered a yeast strain to have the potential to acquire AP by exploiting crosstalk between TORC1 and TORC2 to induce peroxisomal translocation of Ura3 in response to caffeine exposure. This translocation would provide a conditional increase of 5-FOA resistance by virtue of the change in Ura3 compartmentalization from the cytosol to peroxisomes. Specifically, we fused URA3 (and GFP) to GPD1, which encodes a NAD+-dependent glycerol 3-phosphate dehydrogenase with an N-terminal type 2 peroxisomal targeting signal (PTS2). Phosphorylation of Gpd1 by the TORC2-pathway (Lee et al. 2012) triggers translocation of Gpd1 to the peroxisome (Jung et al. 2010) independently of caffeine treatment. In this regard, the engineered strain can be considered to be “one step removed” from acquiring AP, which could emerge through selection of a mutation(s) that linked the caffeine-responsive TORC1 network to TORC2-mediated adaptive translocation of Gpd1-EGFP-Ura3 to peroxisomes (fig. 2a). Importantly, this approach enabled us to determine if AP emerged by quantifying the effect of caffeine pre-treatment on the sub-cellular distribution of GFP fluorescence. To facilitate evolution of AP, we generated a mixed population of genotypes by introducing genetic variation through UV mutagenesis of the engineered yeast strain (Holland et al. 2014) (see Materials and Methods).
Fig. 2.
—Rapid emergence of AP in an engineered yeast strain. (a) Schematic diagram for mechanistic model: caffeine induces a global pleiotropic response in yeast bringing cells into a different state. A mutation(s) causing a network rewiring linking the caffeine-induced response and the signaling network for peroxisomal localization of the engineered Gpd1-EGFP-Ura3 construct could potentially generate AP. (b) Laboratory evolution of AP in six mutagenized, engineered yeast lines. Significant CF (Mann–Whitney U test, P value < 0.05, indicated by asterisks) was observed in at least one time point in each evolution cell line, reaching a maximum of CF = 0.390 (cell line E2, n = 190), which indicates that caffeine pre-treatment resulted in 39% increase in 5-FOA survival. Error bars indicate standard deviation. Symbols mark when transcriptomics, whole genome re-sequencing and image analysis were performed (see key at the top for details). (c) Fluorescence in cytoplasm and peroxisomes was quantified from at least 50 cells in each condition. Examples of representative segmented cells are shown (left) and each microscopy image was segmented and quantified (right, see supplementary methods, Supplementary Material online, for details). (d) Bars represent changes in relative peroxisomal fluorescence signal before and after caffeine exposure, asterisks indicate significance. Specifically to cell line E4, exposure to caffeine shifted Gpd1-EGFP-Ura3 to the peroxisome at generations n = 150 and n = 200. Significant CF was observed also at n = 200. Both caffeine-conditioned phenotypes, increase in 5-FOA survival and peroxisomal translocation of Gpd1-EGFP-Ura3, disappeared at n = 250.
Six independent lines of the mutagenized, engineered strain populations (heretofore referred as E# lines, where # indicates identity of the replicate) were subjected to laboratory evolution, as described earlier. Remarkably, AP emerged across all lines, appearing in one case within 50 generations (E2, CF = 0.39, Mann–Whitney U test, P value < 0.05; fig. 2b). A key question was whether any of the three lines had acquired AP through selection of mutants that conditionally re-localized Gpd1-EGFP-Ura3 to peroxisomes, upon sensing caffeine. At 50-generation intervals, we performed fluorescence microscopy on three lines (E4, E5, and E6) before and after 30 min of caffeine exposure, and used image analysis to quantify cytoplasmic and peroxisomal abundance of Gpd1-EGFP-Ura3 (fig. 2c). Strikingly, we observed that emergence of AP in one of the three lines (E4) correlated with caffeine-dependent re-localization of Gpd1-EGFP-Ura3 to peroxisomes. Interestingly, the ability of E4 to conditionally translocate Gpd1-EGFP-Ura3 eventually disappeared by 250 generations, also correlating with loss of AP (fig. 2d).
To evaluate cue-specificity and whether AP can emerge with other environmental cues, we repeated the laboratory evolution experiment using identical design but with menadione in place of caffeine. We chose menadione as an alternative chemical stimulus specifically because global gene expression patterns are distinct when cells are treated with menadione and caffeine, and menadione does not significantly influence GDP1 expression (Jung et al. 2010). Again, AP emerged over a similar time scale (50–200 generations), confirming that laboratory evolution of AP is a generalizable phenomenon, not restricted to a particular environmental cue (supplementary fig. S4, Supplementary Material online). Conclusively, our results demonstrate that AP emerges in remarkably short time frames through laboratory evolution of yeast when they experience an environment with novel temporal and structured changes in two stimuli.
Emergence of AP from Natural Genotypes
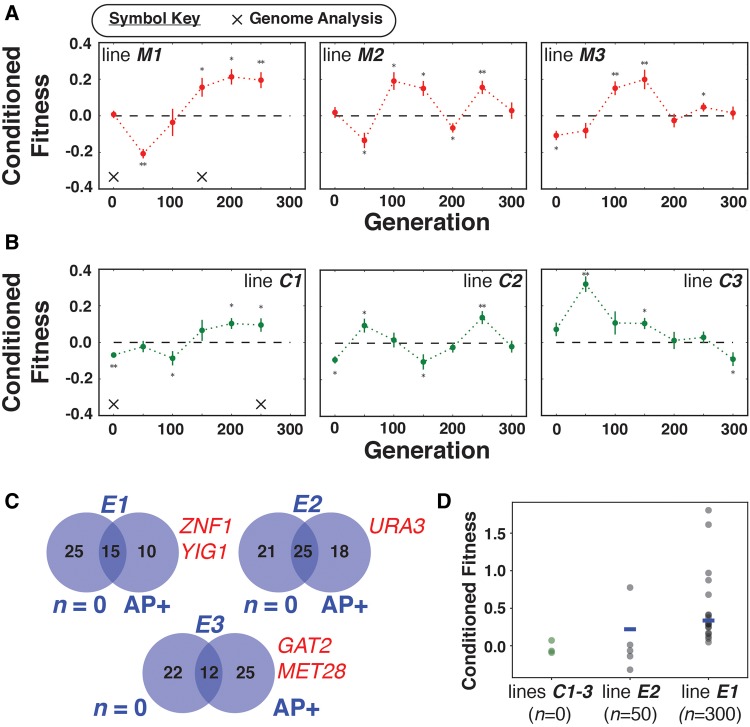
The observation that only one of the engineered lines linked caffeine-response with Gpd1-EGFP-Ura3 translocation to peroxisomes suggested that the mechanism for AP in the other evolution lines were independent of Gpd1-EGFP-Ura3 translocation. To explore this, we repeated the laboratory evolution experiment without the engineered GPD1-EGFP-URA3 construct using 3 independent cell lines (M# lines) of a UV-mutagenized S. cerevisiae BY4741 URA3 strain. Strikingly, AP rapidly emerged across all M lines within 100–150 generations (CF = 0.090–0.213, Mann–Whitney U test, P value < 0.05; fig. 3a). The consistency of this and our previous results motivated us to investigate whether genetic variation via mutagenesis was necessary for the rapid emergence of AP. We performed laboratory evolution with three lines (C# lines), each derived from a single colony of an S. cerevisiae BY4741 URA3 strain. We observed that AP emerged transiently across two out of three replicate C lines and was sustained over multiple generations in only one line (C1, CF = 0.103, Mann–Whitney U test, P value < 0.05; fig. 3b).
Fig. 3.
—Rapid emergence of AP in mutagenized and clonal lines of yeast. (a) Laboratory evolution of AP in M cell lines. AP emerged within 100–150 generations, reaching a maximum CF of 0.213 (M1 at n = 200). AP was sustained over at least 50–100 generations and it consistently emerged in other cell lines. (b) Sustained AP emerged only in C1 cell line and appeared in at least one time point in all cell lines. Asterisks indicate significant CF (Mann–Whitney U test, P value < 0.05), error bars indicate standard deviation. (c) Unique and shared caffeine-induced transcript changes before and after AP emerged in three evolution lines. Novel caffeine-responsive transcript changes included downregulation of URA3 in line E2, and also downregulation of several transcription factors in lines E1 and E3 (indicated in red font). (d) Distribution of CF values for clonal isolates before and after emergence of AP; lines C1–3 are isogenic cultures derived from single colonies of BY4741 URA3; blue lines represent CF values from BY4741 GPD1-EGFP-URA3 mixed populations (as in fig. 2b).
Mechanistic Insights from Sequence and Transcriptome Analysis
To better understand the mechanism for AP, we performed whole genome re-sequencing on three engineered lines (E2, E4 and E6), a mutagenized line (M1), and a clonal line (C1) across different time points of the laboratory evolution (i.e., at the start of the experiment, after AP emerged, and, in some lines, after it was lost). After applying conservative filters (see supplementary methods, Supplementary Material online), we discovered that 305 mutations had accumulated at 114 loci across the evolved lines, demonstrating that AP had emerged through selection of mutations during laboratory evolution, even in a C line that originated from single colony. Importantly, the only consistent pattern across all lines was the accumulation of mutations in two genes associated with de novo biosynthesis of pyrimidines, URA2 and URA6. Ura2 catalyzes the first two enzymatic reactions in the formation of the pyrimidine ring, while Ura6 catalyzes the phosphorylation of pyrimidine nucleoside monophosphates at a later step of the pathway. Notably, every mutation discovered in the two genes resulted in nonsynonymous substitutions: E2, R1100L (ν = 0.40); E4, N930D (ν = 1.0); E6, D2186N (ν = 1.0); M1, R2051C (ν = 0.66) and R1100C (ν = 0.28), all in Ura2; and C1, R2051H in Ura2 (ν = 1.0) and L22R in Ura6 (ν = 0.26) (supplementary file S1, Supplementary Material online). While none of these mutations mapped to amino acid residues within active sites of the two enzymes, PROVEAN (Choi et al. 2012; Choi and Chan 2015) predicted that mutations in the ATCase domain (R2051H, R2051C and D2186N), and the CPSase domain (R1100C, R1100L and N930D) have deleterious effects on the function of Ura2. The deleterious mutations in Ura2 and Ura6 could potentially decrease levels of the downstream product dihydroorotic acid (DHO), which has a role in the transcriptional upregulation of URA3 during uracil starvation (Flynn and Reece 1999). While these mutations do not abolish uracil biosynthesis, they could lower DHO levels, which in turn would downregulate URA3 expression to increase 5-FOA resistance. However, decreased flux through the de novo pyrimidine biosynthetic pathway by itself does not explain AP, since CF is represented by the increase in 5-FOA survival because of caffeine pre-treatment. Lower basal levels of URA3 resulting from Ura2 mutations could in principle set a primed state for caffeine-induced downregulation of URA3. Alternatively, we hypothesize that mutations at other genomic loci act in combination with the URA2 and/or URA6 mutations to conditionally increase 5-FOA resistance. Notably, within each cell line, 41–100 mutations mapped to genic and intergenic regulatory elements, affecting 25–55 genes including signal transduction genes, suggesting that alterations in the underlying gene regulatory network architecture might have contributed to linking caffeine response to increased capacity for dealing with 5-FOA (McGregor et al. 2012; Sorek et al. 2013). We performed transcriptome profiling (see supplementary methods, Supplementary Material online) on lines E1, E2 and E3 to investigate whether AP had emerged from rewiring of the caffeine response regulatory network (square symbols in fig. 2b). Comparative analysis of caffeine response of each evolution line at n = 0 generations and after AP had emerged demonstrated that this response had changed in distinct ways across all lines, and in at least one line it was rewired to a known mechanism for increasing 5-FOA resistance. Specifically, when AP emerged at generation n = 50, line E2 downregulated URA3 upon sensing caffeine (log2 fold-change: −2.09; P value < 0.01, supplementary file S2, Supplementary Material online) (fig. 3c). Together with the caffeine-induced re-localization of GFP to peroxisomes in line E4, the conditional down-regulation of URA3 in line E2 provides evidence that AP had emerged in these two lines via rewiring of two unrelated networks, one for sensing caffeine and one for increasing 5-FOA resistance.
Clonal Isolates Recapitulate Population Phenotypes
In some evolved lines, AP appeared transiently and was correlated to an overall increase in 5-FOA resistance (supplementary fig. S5, Supplementary Material online). While it took longer for a constitutive strategy to emerge (possibly due to phase C counter-selection of simple loss-of-function mutations in uracil biosynthesis, such as in URA3), it is not surprising that once it emerged, this strategy quickly displaced AP. In other words, AP appears to be a phenotypic trait attributable to few cells in the population that were subsequently outcompeted by cells that gained constitutively higher resistance to 5-FOA. While these cells might have acquired a novel strategy for higher 5-FOA resistance without losing their uracil biosynthesis capability, they could also be cheaters that are uracil auxotrophic mutants (ura-) exploiting other cells to complement their uracil need. In either case, the transience appears to be a consequence of selection of constitutively resistant or cheater mutants from a heterogeneous population of cells with and without AP. We investigated whether there was evidence for such population heterogeneity by quantifying CF of 24 clonal isolates from mixed populations of two lines—5 CFUs from line E2 at n = 50 generations, and 19 CFUs from line E1 at n = 300 generations. The distribution of CF values of clonal isolates recapitulated well the overall phenotype of each of the two lines, and demonstrated that few clones had significantly higher CF while others had little to no CF (fig. 3d). The high level of heterogeneity also suggested that AP might emerge through interactions among different variants within a mixed culture, with each variant playing specialized roles vis-à-vis uracil biosynthesis and 5-FOA resistance. This mixed population behavior is reminiscent of variegation in yeast populations mediated by epigenetic phenomena (Allshire and Ekwall 2015; Norman et al. 2015). Regardless of the mechanism, transience of AP might ultimately be a consequence of a change in population structure due to high frequency of coupled caffeine-5-FOA exposures in our experimental design; future experiments could investigate if longer periods of phase C or lower cell densities might prevent displacement of strains that possess AP as an individual or group-level trait.
Conclusions and Future Directions
We have demonstrated that laboratory evolution in a novel structured environment can consistently generate AP in yeast within a remarkably short timeframe. The whole genome sequence analysis suggested that reduced flux through the uracil biosynthesis pathway in conjunction with a more complex repertoire of mutations in regulatory genes contributes to adaptively predicting 5-FOA toxicity upon sensing caffeine. Additional experimentation will be required to characterize which regulatory mutations are genomically linked and how they mechanistically couple caffeine-sensing and conditionally increasing 5-FOA resistance. Furthermore, the transience of AP observed in some evolution lines may result from the instability of the underlying mechanism or the emergence of cheater sub-populations (King and Masel 2007; Beaumont et al. 2009; Levy et al. 2012). We predict that periodicity, strength and duration of stimulus, types of coupled environmental changes and use of counter-selection all likely influence the dynamics of both emergence and duration of AP. For all these reasons, a systems approach that cuts across molecular, genome-wide, single-cell, population and temporal scales will be necessary to fully characterize the underlying mechanisms for AP, how they emerge, and how they are lost. The capability to evolve novel AP in the laboratory makes it possible to conduct such multiscale systems analysis for dissecting and characterizing its molecular and mechanistic underpinnings. This laboratory evolution framework also permits investigation into ecological implications of AP with regard to its role in enabling adaptation of an organism to new environmental conditions.
Materials and Methods
Yeast Strains and Plasmids
Yeast strains used in this study were derived from the parental strain Saccharomyces cerevisiae BY4741. BY4741 GPD1-EGFP-URA3 strain was constructed by genomic integration of a PCR fragment containing EGFP (enhanced green fluorescent protein) and URA3 (orotidine-5′-phosphate decarboxylase gene) in-frame, amplified using the pYM27 and pRS426 plasmids, respectively. This PCR cassette contained 40 bp of homology on each side to facilitate the integration into the target downstream region of GPD1 open reading frame. The PCR cassette was sequenced after genomic insertion and no mutation was found from original sequences. For BY4741 URA3 strain, a PCR fragment containing URA3 gene was amplified from S. cerevisiae S288C with primers ∼200 bp from upstream and downstream regions of URA3 ORF, and it was transformed into the BY4741 strain.
Culture Conditions
Complete synthetic media (CSM) or CSM-uracil (ura) media with 2% glucose were used as media for cell cultures. Cells were grown at 30 °C on a rotator in a 2-ml volume. Cell densities of liquid cultures were determined from absorbance measurements at 600 nm using a BioPhotometer (Eppendorf). Serial dilution was performed by transferring an aliquot of cultures into fresh media and adjusting optical density (OD) to a final value of A600 nm = 1.5 in 1 ml. Subsequently cells were exposed to caffeine (phase A) and 5-FOA (phase B). An aliquot of 200 μl of culture was taken at the end of phase B, washed twice and transferred into 2 ml of CSM-ura for phase C. Mutagenesis was performed with UV light using Stratalinker UV Crosslinker Model 2,400 at 9,300 mJ cm−2 such that we observed 20% cell survival.
Conditioned Fitness Measurements
Every ∼50 generations along the evolutionary experiment, a culture aliquot was transferred into two different tubes and diluted to a cell density of A600 nm = 1.5 in 1 ml volume. Each sample was subsequently exposed to either phase A (with the cue) or phase Acontrol (using media instead of the cue), and subsequently both samples were subjected to 5-FOA in phase B. Cell survival was assessed by sampling the cultures immediately before and after phase B. Culture aliquots were adequately diluted, plated onto five CSM agar plates, incubated at 30 °C, and colony formation units (CFUs) were counted. We defined conditional fitness (CF) as the cue-specific effect on survival, calculated as survival difference to phase B prior exposure to caffeine () with respect to control () as follows:
| (Eq. 1) |
Survival () was then calculated as relative change of CFUs after phase B as,
| (Eq. 2) |
where and are average CFU counts before and after phase B, respectively. Significance of survival differences were assessed with Mann–Whitney U hypothesis tests.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Author Contributions
A.L.G.D.L., A.K., K.D.B., J.J.S., J.D.A., and N.S.B. designed the study. A.L.G.D.L. and A.K. performed the experiments. A.L.G.D.L. performed the quantitative analysis including image and sequencing analysis. F.D.M. assisted with analysis and study design. A.L.G.D.L. and N.S.B. drafted the manuscript. All authors wrote the manuscript.
Supplementary Material
Acknowledgments
We thank M. Pan, K. Johnson, J. Timberlake, Z. Simon, and D. O’Dell for technical assistance. J. Bletz, G. Glusman, M. Robinson, and C. Lausted for comments and feedback. We also thank members of the Baliga and Aitchison labs for helpful discussions. F.D.M. is a postdoctoral fellow of the Canadian Institutes of Health Research. This work was supported by National Institutes of Health (P50 GM076547 [J.D.A. and N.S.B.] and P41 GM109824 [J.D.A.]).
Literature Cited
- Allshire RC, Ekwall K.. 2015. Epigenetic regulation of chromatin states in Schizosaccharomyces pombe. Cold Spring Harb Perspect Biol. 7:a018770.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont HJE, Gallie J, Kost C, Ferguson GC, Rainey PB.. 2009. Experimental evolution of bet hedging. Nature 462:90–93. [DOI] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR.. 1984. A positive selection for mutants lacking orotidine-5’-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 197:345–346. [DOI] [PubMed] [Google Scholar]
- Calhoun LN, Kwon YM.. 2010. The effect of long-term propionate adaptation on the stress resistance of Salmonella enteritidis. J Appl Microbiol. 109:1294–1300. [DOI] [PubMed] [Google Scholar]
- Choi Y, Chan AP.. 2015. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics btv195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP.. 2012. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7(10):e46688.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar R, Sägesser R, Weikert C, Wagner A.. 2013. Yeast adapts to a changing stressful environment by evolving cross-protection and anticipatory gene regulation. Mol Biol Evol. 30:573–588. [DOI] [PubMed] [Google Scholar]
- Flynn PJ, Reece RJ.. 1999. Activation of transcription by metabolic intermediates of the pyrimidine biosynthetic pathway. Mol Cell Biol. 19:882–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland SL, Reader T, Dyer PS, Avery SV.. 2014. Phenotypic heterogeneity is a selected trait in natural yeast populations subject to environmental stress. Environ Microbiol. 16:1729–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes Hallett JE, Luo X, Capaldi AP.. 2014. State transitions in the TORC1 signaling pathway and information processing in Saccharomyces cerevisiae. Genetics 198:773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CH, Egli M, Stewart PL.. 2008. Structural insights into a circadian oscillator. Science 322:697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Marelli M, Rachubinski RA, Goodlett DR, Aitchison JD.. 2010. Dynamic changes in the subcellular distribution of Gpd1p in response to cell stress. J Biol Chem. 285:6739–6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, Masel J.. 2007. The evolution of bet-hedging adaptations to rare scenarios. Theor Popul Biol. 72:560–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuranda K, Leberre V, Sokol S, Palamarczyk G, Francois J.. 2006. Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol Microbiol. 61:1147–1166. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM.. 2012. mTOR signaling in growth control and disease. Cell 149:274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Jeschke GR, Roelants FM, Thorner J, Turk BE.. 2012. Reciprocal phosphorylation of yeast glycerol-3-phosphate dehydrogenases in adaptation to distinct types of stress. Mol Cell Biol. 32:4705–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Ziv N, Siegal ML.. 2012. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol. 10:e1001325.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor S, Vasas V, Husbands P, Fernando C.. 2012. Evolution of associative learning in chemical networks. PLoS Comput Biol. 8:e1002739.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A, et al. 2009. Adaptive prediction of environmental changes by microorganisms. Nature 460:220–224. [DOI] [PubMed] [Google Scholar]
- Mitchell A, Pilpel Y.. 2011. A mathematical model for adaptive prediction of environmental changes by microorganisms. Proc Natl Acad Sci U S A. 108:7271–7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman TM, Lord ND, Paulsson J, Losick R.. 2015. Stochastic switching of cell fate in microbes. Annu Rev Microbiol. 69:381–403. [DOI] [PubMed] [Google Scholar]
- Pavlov IP. 1927. Conditioned reflexes: an investigation of the physiological activity of the cerebral cortex. London: Oxford University Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallis C, Codlin S, Bähler J.. 2013. TORC1 signaling inhibition by rapamycin and caffeine affect lifespan, global gene expression, and cell proliferation of fission yeast. Aging Cell 12:563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke A, Chen JCY, Aronova S, Powers T.. 2006. Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J Biol Chem. 281:31616–31626. [DOI] [PubMed] [Google Scholar]
- Sorek M, Balaban NQ, Loewenstein Y.. 2013. Stochasticity, bistability and the wisdom of crowds: a model for associative learning in genetic regulatory networks. PLoS Comput Biol. 9:e1003179.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagkopoulos I, Liu Y-C, Tavazoie S.. 2008. Predictive behavior within microbial genetic networks. Science 320:1313–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanke V, et al. 2008. Caffeine extends yeast lifespan by targeting TORC1. Mol Microbiol. 69:277–285. [DOI] [PubMed] [Google Scholar]
- Whitaker WB, et al. 2010. Modulation of responses of Vibrio parahaemolyticus O3:K6 to pH and temperature stresses by growth at different salt concentrations. Appl Environ Microbiol. 76:4720–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH.. 2004. The adaptive value of Circadian clocks: an experimental assessment in Cyanobacteria. Curr Biol. 14:1481–1486. [DOI] [PubMed] [Google Scholar]
- Zhang J, et al. 2011. Mapping the interaction of Snf1 with TORC1 in Saccharomyces cerevisiae. Mol Syst Biol. 7:545.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.