Abstract
Nuclear factor TDP-43 is known to play an important role in several neurodegenerative pathologies. In general, TDP-43 is an abundant protein within the eukaryotic nucleus that binds to many coding and non-coding RNAs and influence their processing. Using Drosophila, we have performed a functional screening to establish the ability of major hnRNP proteins to affect TDP-43 overexpression/depletion phenotypes. Interestingly, we observed that lowering hnRNP and TDP-43 expression has a generally harmful effect on flies locomotor abilities. In parallel, our study has also identified a distinct set of hnRNPs that is capable of powerfully rescuing TDP-43 toxicity in the fly eye (Hrb27c, CG42458, Glo and Syp). Most importantly, removing the human orthologs of Hrb27c (DAZAP1) in human neuronal cell lines can correct several pre-mRNA splicing events altered by TDP-43 depletion. Moreover, using RNA sequencing analysis we show that DAZAP1 and TDP-43 can co-regulate an extensive number of biological processes and molecular functions potentially important for the neuron/motor neuron pathophysiology. Our results suggest that changes in hnRNP expression levels can significantly modulate TDP-43 functions and affect pathological outcomes.
INTRODUCTION
Recent experimental advances have highlighted that alterations in RNA metabolism are very common in neurodegenerative diseases (1–4). From a mechanistic point of view, at the base of these alterations there is often the dysfunction of one or several RNA binding proteins that play an important role in regulating the functioning of neurons, especially at the synaptic level (5,6).
For this reason, the identification of proteins and events potentially altered during the course of disease might represent a critical step for outlining novel therapeutic strategies to delay disease onset and/or progression. Unfortunately, this is not an easy task because nuclear factors seldom work alone in determining the processing and ultimate fate of a transcribed RNAs. In fact, most of them work via a complex network of interactions with other factors which can contribute to modify their functional properties in a cell- or developmental-specific manner (7).
In this respect, TDP-43, as one of the major hnRNP protein involved in Amyotrophic Lateral Sclerosis (ALS) and Frontotemporal Dementia (FTD) (8,9), is no exception to this rule. In recent years, several high-throughput studies performed on samples from human ALS/FTD patients or in TDP-43 animal and cellular disease models have uncovered a huge number of RNA processing events possibly dysregulated during disease (10–16).
In parallel with this huge number of potential targets, proteomic analyses have identified more than 100 factors that can bind to TDP-43 and potentially modify its actions (17–20). Even a more selected estimate obtained on the basis of functional co-immunoprecipitation studies still yields more than 20 interactions that can modify the biological properties of TDP-43 (21).
In particular, hnRNP A/B family members represent one of the key functional interactors of TDP-43 (22) and can play an autonomous role in neurodegeneration. Mutations in these proteins have been found to be independently involved in causing ALS and multisystem proteinopathy (23), whilst ALS-linked mutations in the Ubiquilin-2 gene have been shown to abolish interaction of this factor with hnRNP A1, A3 and U (24). Moreover, a cholinergic-associated loss of hnRNP A/B expression has been found associated with Alzheimer Disease (25) and a loss in hnRNP A1 expression has been reported in ALS spinal cord motoneurons carrying TDP-43 inclusions (26). Regarding hnRNP proteins and ALS, it is also important to mention the possible role played by hnRNP A3 and especially H/F proteins binding to the G4C2 expansions of the C9orf72 region (27–31). Of interest, there is also the observation that the interaction between TDP-43 and hnRNP proteins is highly conserved throughout evolution. For example, the Drosophila homolog of TDP-43 (TBPH) being capable of recognizing human hnRNP A/B proteins and vice versa (32). Furthermore, the importance of this interaction for the pathology has also been recently highlighted by reports showing that overexpression of hnRNP U and hnRNP A1/A2 can inhibit TDP-43-induced neuronal cell death in NSC34 cells (33). Similarly, in Drosophila, the ability of TDP-43 to suppress CGG RNA- Fragile X-associated tremor/ataxia syndrome associated phenotypes relies on the interplay of this factor with two fly homologs of hnRNP A2/B1, whose overexpression could attenuate TDP-43-toxicity (32–34). Also in Drosophila, it has been found that the fly ortholog of human DAZAP1 can act as a dominant modifier of a Valosin-containing protein (VCP) mutation causing neurodegeneration, at least partly, because of a toxic gain-of-function of TDP-43 redistribution from the nucleus to the cytoplasm (35). Finally, we have also recently reported that elevated human hnRNP levels in the brain of FTLD-TDP patients may represent a defense mechanism in repressing the inclusion of a TDP43-controlled, toxic exon, within the Sort1 gene (36).
Therefore, a growing body of evidence suggests that variation in expression of different hnRNP proteins may represent a general response against TDP-43 gain- and loss-of-function pathological effects, either by binding directly to TDP-43 or by acting through common targets. To test this hypothesis in a systematic manner, we have utilized the evolutionary conserved hnRNPs present in Drosophila melanogaster that may be considered one of the best model systems nowadays available for the study of TDP-43 biology and ALS pathology (37,38). Our results suggest that the majority of hnRNP proteins within the nucleus can affect TDP-43 functionality both in loss-of-function and gain-of-function disease models. Importantly, the ability by these hnRNPs to modify fly phenotypes can also be observed in their human homologs with regards to TDP43-controlled events, especially at the pre-mRNA splicing level.
MATERIALS AND METHODS
Fly strains and maintenance
The complete genotype of the fly stocks are indicated below:
W1118, w; GMR-Gal4, w; GMR-Gal4, UAS-TBPH, yw; UAS-mCD8::GFP, w; Elav-Gal4; UAS-Dicer-2, Elav-Gal4, tbphΔ23; UAS-TBPHRNAi/UAS-Dicer-2. The RNA interference (RNAi) strains were obtained from Vienna Drosophila Resource Center (VDRC) Drosophila stock centre and Bloomington Stock Center. All stocks and crosses were maintained at 25°C on a 12:12 h light:dark cycle, at constant humidity on standard cornmeal medium.
Eye phenotype and examination
Eyes morphology of 1 day post-eclosion flies were examined and given points were scored for the presence of loss of pigmentation, presence of neuronal death (black spot), retinal collapse and ommatidial fusion. Points were assigned on the following scale: one point was given each phenotype present, two points were given if the affected area was more than 5%, three points were given if the compromised area was more than 30% and four points were given if the affected area was more than 65%. Additional two points could be given for the presence of a high number of black spots. For each genotype over 100 eyes were examined.
Climbing assay
To assess the negative geotaxis movement in adult flies, we followed the previously established protocol (32). Shortly, groups of 20 aged flies were transferred to the bottom of a 50-ml cylinder without anaesthesia. After 30 s of adaptation, climbing ability was measured by counting the flies that reached the top of the cylinder (10 cm) in 15 s. The experiments were performed at 25°C.
Western blotting in flies
Total proteins extract were obtained from adult heads. The material has been squeezed in lysis buffer 1× (Lysis buffer composition 1.5×: 225 mm NaCl, 15 mm Tris, 7.5 mm ethylenediaminetetraacetic acid (EDTA), 15% glycerol, 7.5 mm ethylene glycol-bis(β-aminoethyl ether)-N,N,N',N'-tetraacetic acid (EGTA), 75 mm NaF, 6 M urea, 7.5 mm Dithiothreitol (DTT) and protease inhibitor) and then clarified by a short centrifugation at 0.5 × g. The proteins were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted on 0.2 μm nitrocellulose membrane (Whatman Protran). Membranes were blocked overnight in 5% non-fat dried milk in Tris-buffered saline (TBS)-0.01% Tween 20 and probed with anti-Flag M5 (Sigma, 1:10 000). Anti-Actin (Sigma1:5000) was used as a total protein loading control. Proteins were detected with Femto SuperSignal substrate (Thermo Scientific).
Gene knockdown
Human neuroblastoma SH-SY-5Y cells were cultured in Dulbecco's modified Eagle's medium–Glutamax-I (Gibco-BRL, Life Technologies Inc., Frederick, MD, USA) supplemented with 10% fetal bovine serum (Gibco-BRL, Life Technologies Inc., Frederick, MD, USA) and Antibiotic-Antimycotic-stabilized suspension (SigmaAldrich, St Louis, MO, USA) at 37°C incubator with humidified atmosphere of 5% CO2. The siRNA sense sequences used for silencing the different target proteins were the following: luciferase (control): uaaggcuaugaagagauac, TDP-43: gcaaagccaagaugagccu, DAZAP1: gagacucugcgcagcuacu, hnRNP Q/Syncrip: agacagugaucucucucau, hnRNP R cauuugggaucuacgucuu. After twice gene silencing by Hyperfectamine (Qiagen Inc, Gaithersburg, MD, USA), cells were collected and divided into two aliquots; one-half for western blot and the other half for reverse transcriptase-polymerase chain reaction (RT-PCR) analysis.
Western blotting
Total protein extracts were obtained by cell sonication in lysis buffer composed of 1× phosphate buffered saline (PBS) and 1× Complete Protease Inhibitor Cocktail (Roche Diagnostics, Mannheim, Germany). Proteins extract (20 μg) from each sample was loaded on a 10% SDS-PAGE. The gel was then electroblotted onto a Nitrocellulose blotting membrane according to standard protocols (Amersham Biosciences, Uppsala, Sweden) and blocked with 5% skimmed milk (non-fat dry milk in 1× PBS and 0.1% Tween-20). Proteins were probed with antibodies in order to confirm the gene knockdown efficacy and detected with enhanced chemiluminescence (ECL) Western Blotting Substrate (Thermo Scientific, Rockford, IL, USA). Tubulin, available in-house, was used as total protein loading control.
RNA extraction and RT-PCR analysis
RNA was obtained using Eurogold Trifast (Euroclone, Milan, Italy), following the manufacturer's instructions. One microgram of total RNA was used in the retrotranscription reaction with random primers and Moloney murine leukemia virus (M-MLV) Reverse Transcriptase (Gibco-BRL, Life Technologies Inc., Frederick, MD, USA). The primers for the target genes are including: POLDIP3 Fw 5΄-gcttaatgccagaccgggagttg-3΄; POLDIP3 Rv 5΄-tcatcttcatccaggtcatataaatt-3΄; TNIK Fw 5΄-caaaggcgagagaaggagctg-3΄; TNIK Rv 5΄-ctgatgctgaagggaaactaag-3΄; STAG2 Fw 5΄-gtatgtttacttggaaaagttcatg-3΄; STAG2 Rv 5΄-tgattcatccataattgaagctgga-3΄; MADD Fw 5΄-gacctgaattgggtggcgagttccct-3΄; MADD Rv 5΄-cattggtgtcttgtacttgtggctc-3΄. PCR conditions for POLDIP3 and TNIK: 94°C for 2 min, 94°C for 30 s, 56°C for 1 min and 72°C for 45 s for 35 cycles; and 72°C for 10 min for the final extension. PCR conditions for STAG2 and MADD: 94°C for 2 min, 94°C for 30 s, 50°C for 30s and 72°C for 1 min for 35 cycles; and 72°C for 10 min for the final extension. PCRs were optimized to be in the exponential phase of amplification and products were routinely fractionated in 1.5% (wt/vol) agarose gels. After quantifications by densitometric analysis using ImageJ software, the statistical significance was calculated using unpaired t-test. P < 0.05 was considered significant (n = 3) (*P < 0.05, **P < 0.01 and ***P < 0.001).
TDP-43 and DAZAP1 co-immunoprecipitations
HeLa cells (70% of confluence) were transfected with 3 μg of pFLAG-TDP-43 wild-type using the Effectene reagent. After 24 h, cell culture medium was removed and cells were washed with cold PBS and harvested. Cells were lysed in 500 μl of IP buffer (20 mM Tris pH 7.5, 110 mM NaCl, 0.5% Triton-X, 1× Complete Protease Inhibitor Cocktail) by sonication (3 min, mid power), in ice-cooled sonicating bath (BioRuptor, Diagenode, Belgium). The cell lysate was pre-cleared by incubation with 30 μl Protein A/G PLUS agarose beads (Santa Cruz Biotechnology Inc., Dallas, Texas, USA) in IP buffer for 1.5 h at 4°C. The pellets were discarded and the supernatants were used for immunoprecipitation: the cell lysates were incubated with 2 μg of mouse monoclonal anti-FLAG M2 antibody (Sigma-Aldrich) on a rotating device for an hour at 4°C. Then, 30 μl of Protein A/G PLUS agarose beads were added to each sample and incubated overnight at 4°C. The pellet was then washed three times in ice-cold IP buffer. The supernatants was discarded, and the pellet was re-suspended in 30 μl of 3× sample loading dye. The samples were fractionated by SDS-PAGE (10%) and analyzed by immunoblotting 1:2000 rabbit polyclonal anti-TDP-43 antibody (ProteinTech), with 1:500 rabbit polyclonal anti-DAZAP1 antibody and 1:500 rabbit polyclonal anti-hnRNP H antibody previously described (39,40).
RNA immunoprecipitation and RT-PCR analysis
Twenty-four hours after transfectin of 3 μg flag-DAZAP1 by Effectene, HeLa cells were collected using HEGN buffer (20 mM Hepes pH 7.7, 150 mM NaCl, 0.5 mM EDTA, 10% Glycerol, 0.1% Triton X-100, 1 mM DTT) and sonicated after adding protease inhibitory cocktail (Roche). HeLa lysate (40 μg) was incubated for 1 h at 4°C in HEGN buffer together with Protein A/G Agarose beads (Santa Cruz Biotechnology Inc., Dallas, TX, USA), pre-coated with 5 μg of anti-Flag antibody from Sigma, (IP-Flag) or with uncoated beads as controls (IP-Beads). After washes with HEGN + DOC 0.2% + Urea 0.5M, mRNA was phenol–chloroform extracted from immunoprecipitated RNPs. The abundance of possible DAZAP1 target genes was measured by quantitative real-time PCR, using a Biorad Real-Time PCR System and SYBR Green I Master (Roche), as described below.
RNA sequencing and functional analysis of differentially expressed genes
Total RNA was extracted from TDP-43 and DAZAP1 depleted SH-SY-5Y cells as described. As control, we used total RNA extracted from SH-SY-5Y cells treated with siRNA against luciferase. RNA sequencing was performed at Eurofins (www.eurofins.com) using an Illumina HiSeq 2500 machine.
Data processing was carried out with HiSeq Control Software v2.0.12.0, Basecalls performed with RTA v1.17.21.3. Reads were aligned to the human reference (GRCh38) using BWA-MEM (bwa-0.7.12) with standard parameters. Only uniquely aligned reads were used for expression profiling. Gene expression was measured based on annotation GRCh38.ensembl_genes_77_all_exons using feature Counts (Subread package 1.4.6).
Raw read counts were converted to Counts per million (CPM) values using Trimmed mean of M-values (TMM) normalization (edgeR package). Differential expression analysis was performed using the edgeR package. Features had to have a counts-per-million value of more than one in at least three samples or were removed. The interpretation of the differential expression genes data and pathway mapping were performed by using the PANTHER (Protein ANalysis THrough Evolutionary Relationships) Classification System (41), the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (42) and UniProt (43) tools.
The statistical significance of the overlap between genes regulated by TDP-43 (siTDP-43) and those regulated by DAZAP1 (siDAZAP1) was evaluated by calculating the representation factor and associated probability (hypergeometric distribution test) as implemented in http://nemates.org/MA/progs/overlap_stats.html (44). The overlap was visualized with Venn diagrams using http://www.pangloss.com/seidel/Protocols/venn.cgi.
Accession numbers
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (45) and are accessible through GEO Series accession number GSE97262 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97262).
Quantitative real-time PCR analysis
Total RNA was extracted from SH-SY5Y cells transfected with siRNA against TDP-43, DAZAP1, hnRNP Q and hnRNP R or co-transfected with siRNA against TDP-43 in the presence of siRNA against each of the three tested hnRNPs. RNA obtained from SH-SY5Y cells treated with siRNA against luciferase was used as negative control. For each RNA samples RT-PCR was performed in order to synthetize cDNA. The quantification of gene expression levels was carried out by real-time PCR, using SYBR green technology. The gene-specific PCR primer pairs used are as follows: MADD, forward 5΄-ttgagaccaactctgccaca-3΄, reverse 5΄-agactcgctggctcacatct-3΄; BRD8, forward 5΄-gcagcctgttacagatgac-3΄, reverse 5΄-aatagttgacaaatccataggc-3΄; TNIK, forward 5΄-tggaacatacgggcaagttt-3΄, reverse 5΄-tcctcttcatcccctgtgac-3΄. To normalize the results, the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and sometimes hSDHA were used (GAPDH, forward 5΄-cgctctctgctcctcctgtt-3΄, reverse 5΄-ccatggtgtctgagcgatgt-3΄; SDHA, forward 5΄-tgggaacaagagggcatctg-3΄, reverse 5΄-ccaccactgcatcaaattcatg-3΄). Furthermore, qPCR analysis was performed to validate RNAseq data obtained from SH-SY5Y cells silenced against luciferase (control), TDP-43 and DAZAP1. The gene-specific PCR primer pairs used are as follows: ELAVL3, forward 5΄-cctcaaattacagacgaagacca-3΄, reverse 5΄-gctgacgtacaggttagcatc-3΄; NOVA2 forward 5΄-aaggcgaatacttcctgaaggt-3΄, reverse 5΄-tactaggcatacccgctctgt-3΄; RELN forward 5΄-ttggaggttccagtgctttc-3΄, reverse 5΄-aaactgaggttggttgtggg-3΄; STX3 forward 5΄-tcggcagaccttcggattc-3΄, reverse 5΄-tcctcatcggttgtctttttgc-3΄; CELF5 forward 5΄-aaactcttcgtgggccagat-3΄, reverse 5΄-ggcacagtaggtgaggaagg-3΄; ACHE forward 5΄-agaaagcgtcttccggttct-3΄, reverse 5΄-tacgagccctcatccttcac-3΄; TNF forward 5΄-cctctctctaatcagccctctg-3΄, reverse 5΄-gaggacctgggagtagatgag-3΄; TNFRSF9 forward 5΄-ttggatggaaagtctgtgcttg-3΄, reverse 5΄-aggagatgatctgcggagagt-3΄; ICAM1 forward 5΄-ggccggccagcttatacac-3΄, reverse 5΄-tagacacttgagctcgggca-3΄; YPEL4 forward 5΄-cggagatctctggagcagac-3΄, reverse 5΄-gtgtttcagggcaggagaag-3΄;. Real-time PCRs were performed on a MiniOpticon real-time PCR and on a CFX96 real-time systems (Bio-Rad, Hercules, CA, USA). The expression levels were determined using the 2−ΔΔCT method (Schmittgen and Livak, 2008). The mean of relative expression levels ± standard deviation of three independent experiments are reported. Statistical significance was calculated using unpaired t-test (indicated as ** for P ≤ 0.01 and as * for P ≤ 0.05).
Flp-In HEK293 cell line expressing TDP-43 aggregates and RNA silencing of DAZAP1, hnRNP Q and hnRNP R
Flp-In HEK293 cells (Invitrogen) with inducible siRNA resistant FLAG-tagged wild-type TDP-43 (75 × 104) were seeded in 6-well plates in 1.5 ml of culture medium containing serum and antibiotics. A total of 4 μl of 40 μM siRNA against DAZAP1 (Sigma), hnRNP Q (Sigma) and hnRNPR (Sigma) was diluted in 90 μl of Opti-MEM (Life Technologies) and 6 μl of HiPerFect Transfection Reagent (Qiagen) was added to the diluted siRNA. A siRNA against fire-fly luciferase (Sigma) was used as a control. The same procedure of RNA silencing was repeated the day after (second silencing) and the third day the cells were seeded in a new 6-well plates containing microscope slides treated with 0.1 mg/ml Poly-L-Lysine (Sigma) (for immunofluorescence analysis) and silencing was repeated against each hnRNPs for the last time (third silencing). After 24 h the culture medium was changed and the TDP-43 12XQ/N aggregation was induced by addition of 1 μg/ml tetracycline (Sigma). After 48 h the cells were prepared for immunofluorescence analysis or western blot analysis.
Immunofluorescence analysis of Flp-In HEK293 cell line expressing TDP-43 aggregates
Flp-In HEK293 cells with inducible siRNA resistant FLAG-tagged wild-type TDP-43 were plated and treated as described above. For immunofluorescence analysis the cells were washed three times with PBS, fixed in 3.2% paraformaldehyde in PBS for 1 h at room temperature and permeabilized by using 0.3% Triton in PBS for 5 min on ice. Cells were then blocked with 2% BSA/PBS for 20 min at room temperature and immunolabeled with 1:200 rabbit polyclonal antibody anti-TDP-43 (ProteinTech Group) and 1:200 mouse monoclonal antibody anti-FLAG (Sigma) overnight at 4°C. The day after the cells were washed three times with PBS, incubated with 1:500 anti-mouse AlexaFluor 594 (Invitrogen) and 1:500 anti-rabbit AlexaFluor 488 (Invitrogen) for 1 h at room temperature and coverslipped with Vectashield-DAPI mounting medium (Vector Laboratories, Burlingame, CA, USA). Each slide was analyzed using Nikon Ti-E confocal microscope with a 60× oil objective and a pinhole size of 1.2 AU.
RESULTS
Suppression of hnRNPs expression can modify the neurotoxic phenotypes induced by TBPH overexpression in Drosophila eyes
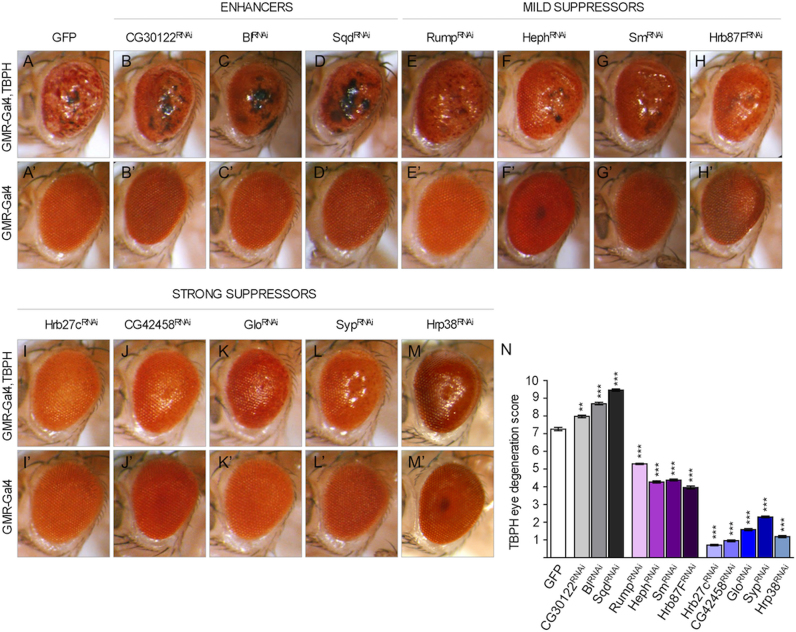
To identify physiological partners of TDP-43 function among the hnRNP family members, we analyzed first how many human hnRNP proteins presented orthologs in Drosophila. We focused attention on 12 human hnRNPs having a counterpart conserved in the fly with a significant level of confidence (Table 1) as calculated using the DRSC Integrative Ortholog Prediction Tool (DIOPT, http://www.flyrnai.org/diopt) (46). In order to determine which of these genes were able to modulate TDP-43 activity in vivo, we completed a systematic gene suppression screening by crossing flies overexpressing, specifically in eye compartment, the TDP-43 ortholog in Drosophila (TBPH), with a collection of transgenic lines carrying RNAi sequences against these 12 evolutionary conserved hnRNP proteins (Table 1). In this study we included Hrp38/hnRNP A1 previously investigated (32) as a reference for this experiment. Thus, for the hnRNPs screening, 23 independents RNAi strains were crossed against GMR-Gal4 flies expressing TBPH in their eyes (Figure 1A–M and Table 2). Moreover, to evaluate the specificity of the genetic modifications, the RNAi lines were alike crossed against GMR-Gal4 flies without expressing TBPH in the genetic background (Figure 1A’–M’ and Table 2).
Table 1. Selected human hnRNPs with their corresponding fly orthologs.
| Human gene | Fly gene | % identity (protein) | % similarity (protein) | ID RNAi |
|---|---|---|---|---|
| hnRNPA1 | Hrp38 | 48 | 63 | 31303 BSC |
| hnRNP A3 | Hrb87F/CG12749 | 50 | 63 | 100732 KK |
| hnRNP C | -/CG42458 | 30 | 47 | 47828 GD |
| 108072 KK | ||||
| hnRNP D | Sqd/CG16901 | 41 | 58 | 32395 GD |
| hnRNP F/H | Glo/CG6946 | 28 | 40 | 27752 GD |
| 110653 KK | ||||
| hnRNP I (PTBP1) | Heph/CG31000 | 54 | 66 | 33735 GD |
| 110749 KK | ||||
| hnRNP K | HnRNP-K/CG13425 | 35 | 45 | 2912 GD |
| 105271 KK | ||||
| hnRNP L | Sm/CG9218 | 35 | 49 | 28117 GD |
| 108351 KK | ||||
| 44658 GD | ||||
| hnRNP M | Rump/CG9373 | 28 | 42 | 44659 GD |
| 1000001KK | ||||
| 33011 GD | ||||
| hnRNP Q/R | Syncrip/CG17838 | 44 | 56 | 33012 GD |
| 110542 KK | ||||
| hnRNP U | -/CG30122 | 25 | 40 | 106984 KK |
| 16040 GD | ||||
| DAZAP1 | Hrb27c/CG10377 | 35 | 47 | 16041 GD |
| 101555KK |
The % identity and % similarity for each ortholog protein are shown as reported by the DRSC Integrative Ortholog Prediction Tool (DIOPT, http://www.flyrnai.org/diopt). The used fly transgenic lines carrying RNA interference sequences against these 12 evolutionary conserved hnRNP proteins are also indicated. All RNAis belong to VDRC Stock Center (GD and KK libraries) except for hnRNPA1/Hrp38 RNAi (#31303) obtained from Bloomington Stock Center (BSC).
Figure 1.
RNAi-mediated disruption of hnRNP candidate genes in Drosophila eyes. (A–M) RNAi-mediated knockdown of hnRNPs alters phenotype of TBPH gain-of-function eyes. Compared the control fly expressing TBPH (A) GMR-Gal4, TBPH/GFP with the following genotypes (B) GMR-Gal4, TBPH /CG30122RNAi, (C) GMR-Gal4, TBPH;Bl RNAi, (D) GMR-Gal4, TBPH/SqdRNAi, described as enhancers of TBPH phenotype; (E) GMR-Gal4, TBPH;RumpRNAi, (F) GMR-Gal4, TBPH/HephRNAi, (G) GMR-Gal4, TBPH; Sm RNAi, (H) GMR-Gal4, TBPH/Hrb87FRNAi described as mild suppressors; (I) GMR-Gal4, TBPH; Hrb27c RNAi, (J) GMR-Gal4, TBPH/CG42458 RNAi, (K) GMR-Gal4, TBPH/Glo RNAi, (L) GMR-Gal4, TBPH; Syp RNAi, (M) GMR-Gal4, TBPH; Hrp38 RNAi indicated as strong suppressors. (A’–M’) RNAi-mediated knockdown of hnRNPs does not alter phenotype of wild-type eye. Compared the control fly (A’) GMR-Gal4/GFP with the follow genotypes (B’) GMR-Gal4/CG30122RNAi, (C’) GMR-Gal4;BlRNAi, (D’) GMR-Gal4/SqdRNAi, (E’) GMR-Gal4;RumpRNAi, (F’) GMR-Gal4/HephRNAi, (G’) GMR-Gal4;SmRNAi, (H’) GMR-Gal4; Hrb87F RNAi (I’) GMR-Gal4; Hrb27c RNAi, (J’) GMR-Gal4/CG42458 RNAi, (K’) GMR-Gal4/GloRNAi, (L’) GMR-Gal4; SypRNAi , (M') GMR-Gal4; Hrp38RNAi. (N) Quantitative analyses of TBPH eye phenotype degeneration. Data show mean phenotype score ± SEM. ***P < 0.001 calculated by non-parametric analysis Mann–Whitney U-test.
Table 2. Summary of the functional genetic screening silencing hnRNPs in Drosophila eyes.
| hnRNP RNAi | ||||
|---|---|---|---|---|
| Human gene | Fly gene symbol | ID RNAi | TBPH GOF | Wild-type |
| Enhancers | ||||
| hnRNP U | -/CG30122 | 106984 KK | ** | ns |
| 2912 GD | *** | ns | ||
| hnRNP K | Bl/CG13425 | 105271 KK | ns | ns |
| hnRNP D | Sqd/CG16901 | 32395 GD | *** | ns |
| Mild Suppressors | ||||
| 44658 GD | *** | ns | ||
| hnRNP M | Rump/CG9373 | 44659 GD | *** | ns |
| 1000001 KK | ns | ns | ||
| hnRNP I | Heph/CG31000 | 33735 GD | *** | ns |
| 110749KK | *** | ns | ||
| hnRNP L | Sm/CG9218 | 28117 GD | *** | ns |
| 108351 KK | *** | ns | ||
| hnRNP A3 | Hrb87F/CG12749 | 100732 KK | *** | mp |
| Strong Suppressors | ||||
| 16040 GD | *** | ns | ||
| DAZAP1 | Hrb27c/CG10377 | 16041 GD | *** | ns |
| 101555 KK | *** | ns | ||
| hnRNP C | -/CG42458 | 47828 GD | *** | ns |
| 108072 KK | *** | ns | ||
| hnRNP F/H | Glo/CG6946 | 27752 GD | *** | ns |
| 110653 KK | *** | ns | ||
| 33011 GD | *** | ns | ||
| hnRNP R/Q | Syp/CG17838 | 33012 GD | *** | ns |
| 110542 KK | *** | mp | ||
| hnRNP A1 | Hrp38/CG9883 | 31303BSC | *** | ns |
Results obtained after the RNAi silencing of 12 genes in eyes (GMR-Gal4 driver) of TBPH gain-of-function flies (TBPH GOF) or in eyes of wild-type flies. The TBPH GOF column collected the results of phenotypic interactions between hnRNPs silenced in TBPH GOF background, describing the hnRNP RNAis as enhancers, strong suppressors or mild suppressors of the TBPH phenotype. Legend of symbols in table: (***) P-value < 0.001; (**) P-value < 0.01. The wild-type column collected the results of hnRNPs silencing in wild-type background; (ns) indicated that no significant phenotype modification was revealed; (mp) indicated the presence of a mild rough phenotype.
TBPH transgene expression (Figure 1A), if compared to Green Fluorescent Proteins (GFP) expression alone (Figure 1A’), induced high eye degeneration with loss of pigmentation, neuronal death with presence of black spot and ommatidial fusion, in coincidence with previous studies (35,47). Interestingly, we found that only few silenced genes (CG30122, Bl and Sqd) enhanced the neurodegenerative phenotypes induced by TBPH gain-of-function (Table 2). In fact, the disorganization of the retinal structures with significant reduction in the organ size and the presence of extensive necrotic areas were observed in these three cases (Figure 1B–D and N). On the contrary, using 19 RNAi lines 9 genes were able to ameliorate (mildly or strongly) the structural defects in the external organization of the retinas and prevent the necrotic degeneration of the photoreceptors due to TBPH overexpression (Table 2). In particular, we were able to find a group of strong suppressors (Hrb27c, CG42458, Glo, Syncrip/Syp, and Hrp38) whose silencing rescued almost completely the TBPH-gain-of-function phenotype (Figure 1I–M and N) and a group of mild suppressors (Hrb87F, Sm, Heph and Rump) that rescued only partially the TBPH-overexpression-dependent phenotype (Figure 1E–H and N). Interestingly, we also observed that the expression of the majority of these genetic modifiers alone in the Drosophila eyes did not produce apparent defects in the organization of these tissues, indicating that these genes were capable to rather specifically modulate TBPH function in vivo (Figure 1B’–M’ and Table 2).
Interference of hnRNPs activity in TBPH loss-of-function background dramatically modifies flies locomotor abilities
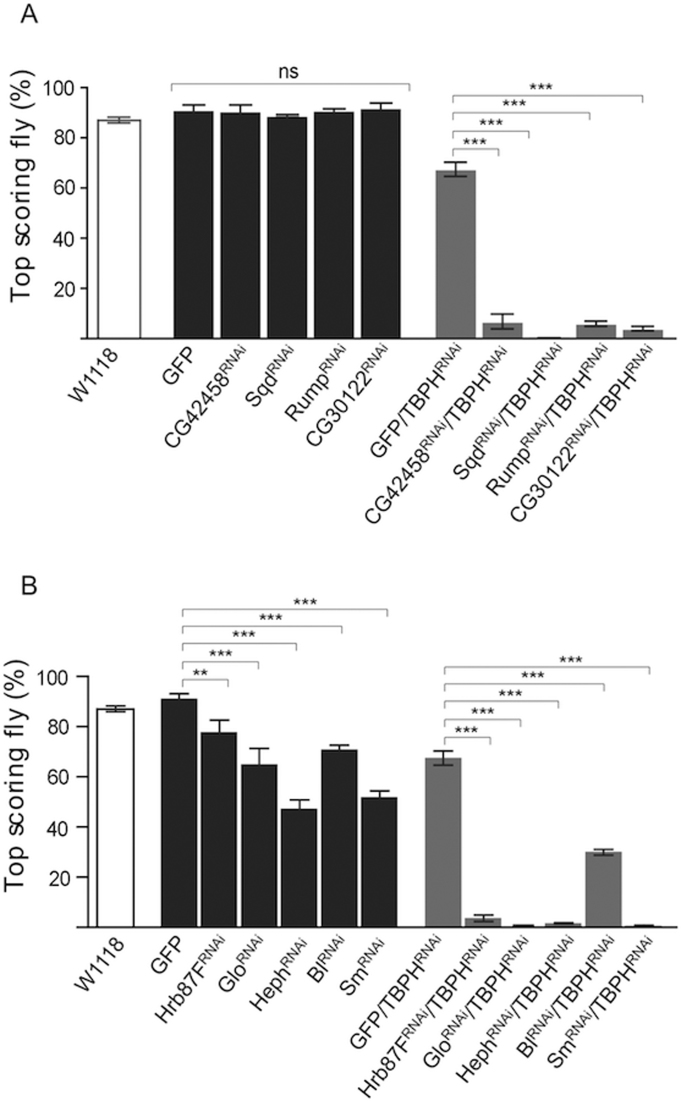
In a previous work, we demonstrated that the hnRNP protein Hrp38 was able to enhance the neurodegenerative phenotype induced by the reduction of TBPH activity in Drosophila neurons (Romano et al., (32)). Therefore, to complete the genetic analysis described above and further explore whether others hnRNPs presented similar phenotypic interactions with TBPH in central nervous system, we generated hypomorphic alleles in this locus. To this aim, RNAi against TBPH with the pan-neuronal driver Elav-Gal4 in TBPH heterozygous backgrounds (Elav-Gal4, tbphΔ23/+; UAS-TBPHRNAi, UAS-Dicer-2) was induced in flies also expressing the Dicer-2 enzyme (to potentiate gene silencing efficiency)(Figure 2A–B light grey columns). This model was previously used successfully and presents locomotive defects in specific climbing assays (Romano et al., (32)). In addition to establish if TBPH and hnRNPs were functionally related in the context of metabolic pathways, we induced pan-neuronal suppression of hnRNP proteins alone (Elav-Gal4/UAS-hnRNPRNAi, Figure 2A–B dark grey columns). Interestingly, we found that the ELAV-driven silencing of CG42458, Sqd, Rump and CG30122 did not affect climbing abilities of wild-type background, control, flies (Figure 2A, dark grey column, and Table 3) substantially confirming what reported after silencing these genes in the eye compartment (Figure 1). On the contrary, the neuronal silencing of CG42458, Sqd, Rump and CG30122 in TBPH reduced background (TBPH hypomorphic allele) produced a strong enhancement in the climbing phenotypes (Figure 2A, light grey columns). The suppression of a second group of hnRNPs (Hrb87F, Glo, Heph, Bl and Sm) provoked significantly locomotion impairment in flies, thus overlapping even alone the neurological defects triggered by the TBPH loss-of-function (Figure 2B and Table 3). In addition the silencing of Syp and Hrb27c caused respectively the complete paralysis and lethality in wild-type as well as in Elav-Gal4, tbphΔ23/+; UAS-TBPHRNAi/+ flies (see Table 3). Thus, the silencing of Hrb87F, Glo, Heph, Bl, Sm, Syp, and Hrb27c in neurons, produced already climbing defects by their own, making difficult to establish whether the genetic interactions that we observed in TBPH hypomorphic backgrounds were due to direct gene dose effects (Figure 2B, Table 3). In this respect, it is also important to note that silencing of Syncrip/hnRNP R/Q, Hrb27c/DAZAP1 and Hrp38/hnRNP A1 did not alter TBPH expression levels directly as determined by Western blot (Supplementary Figure S1).
Figure 2.
RNAi-mediated disruption of hnRNP candidate genes in Drosophila central nervous system. (A and B) The climbing ability analysis of hnRNPs silenced flies in wild-type background (Elav-Gal4; UAS-Dicer-2) are depicted in dark gray and in TBPH hypomorphic alleles (Elav-Gal4, tbphΔ23/+; UAS-TBPHRNAi, UAS-Dicer-2) are depicted in light gray. Additional W1118 control is reported in white. ns = not significant, **P < 0.01, ***P < 0.001 calculated by one-way ANOVA. Error bars SEM.
Table 3. Functional genetic interactions screening in the Drosophila central nervous system.
| hnRNP RNAi | ||||
|---|---|---|---|---|
| Human gene | Fly gene symbol | ID RNAi | TBPH-RNAi enhancers | Wild-type |
| hnRNP C | -/CG42458 | 47828 GD | *** | ns |
| 108072 KK | *** | ns | ||
| hnRNP D | Sqd/CG16901 | 32395 GD | *** | ns |
| 44658 GD | *** | ns | ||
| hnRNP M | Rump/CG9373 | 44659 GD | *** | ns |
| 1000001KK | *** | ns | ||
| hnRNP U | -/CG30122 | 106984 KK | *** | ns |
| hnRNP A3 | Hrb87F/CG12749 | 100732 KK | *** | * |
| hnRNP F/H | Glo/CG6946 | 27752 GD | *** | *** |
| 110653 KK | *** | *** | ||
| hnRNP I | Heph/CG31000 | 33735 GD | *** | *** |
| 110749 KK | *** | *** | ||
| hnRNP K | Bl/CG13425 | 2912 GD | *** | *** |
| 105271 KK | *** | *** | ||
| hnRNP L | Sm/ CG9218 | 28117 GD | *** | *** |
| 108351 KK | *** | *** | ||
| 33011 GD | paralized | paralized | ||
| hnRNP Q/R | Syp/CG17838 | 33012 GD | paralized | paralized |
| 110542 KK | paralized | paralized | ||
| 16040 GD | lethal | lethal | ||
| DAZAP1 | Hrb27c/CG10377 | 16041 GD | lethal | lethal |
| 101555KK | lethal | lethal | ||
The results of climbing assay obtained after the RNAi silencing of 11 hnRNP genes in central nervous system (Elav-Gal4) in TBPH hypomorphic and wild-type backgrounds. Legend of symbols in table: (***) P-value < 0.001 (*) P-value < 0.05, (ns) not significant. In the case of Syp/hnRNP R/Q and Hrb27c/DAZAP1 silencing, the climbing abilities were not measured because the flies appeared completely paralyzed or did not born, respectively.
The human orthologs of hnRNPs Hrb27c (DAZAP1) and Syncrip/Syp (hnRNP Q and R) can contribute to regulate TDP43-controlled mRNA splicing and gene expression events
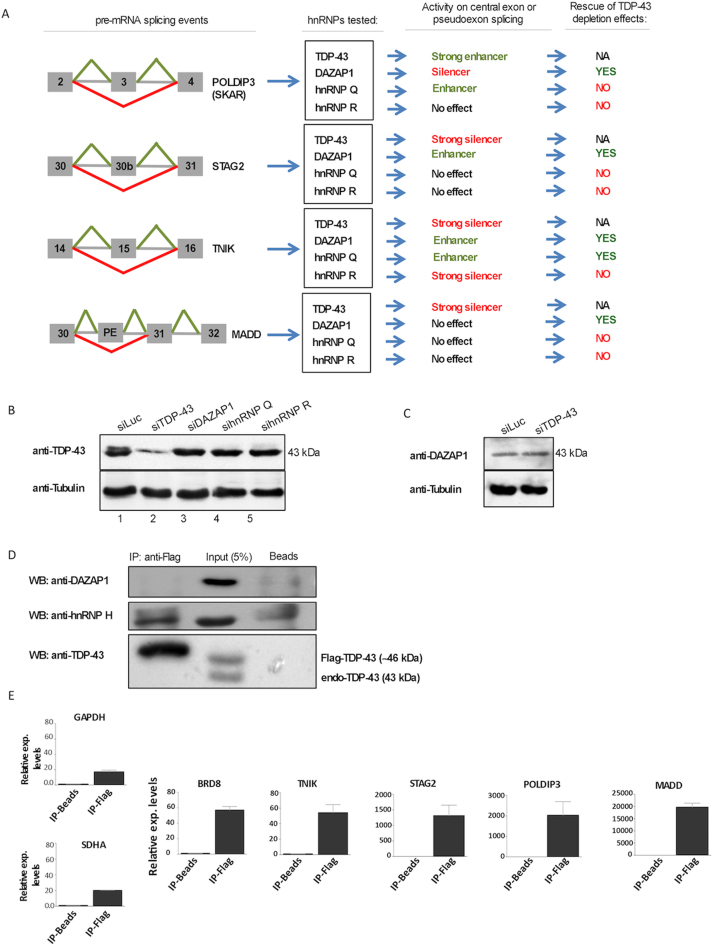
The results obtained in the fly model suggested that different hnRNPs might be directly and differentially involved in the molecular regulation of TBPH/TDP-43 activity. We decided to concentrate on the action of DAZAP1 and hnRNP Q/R because these proteins had an extremely pronounced negative effect when knocked out in combination with TDP-43 (Table 3) but, at the same time, seemed to be protective in the gain-of-function expression model (Figure 1I, L and Table 2).
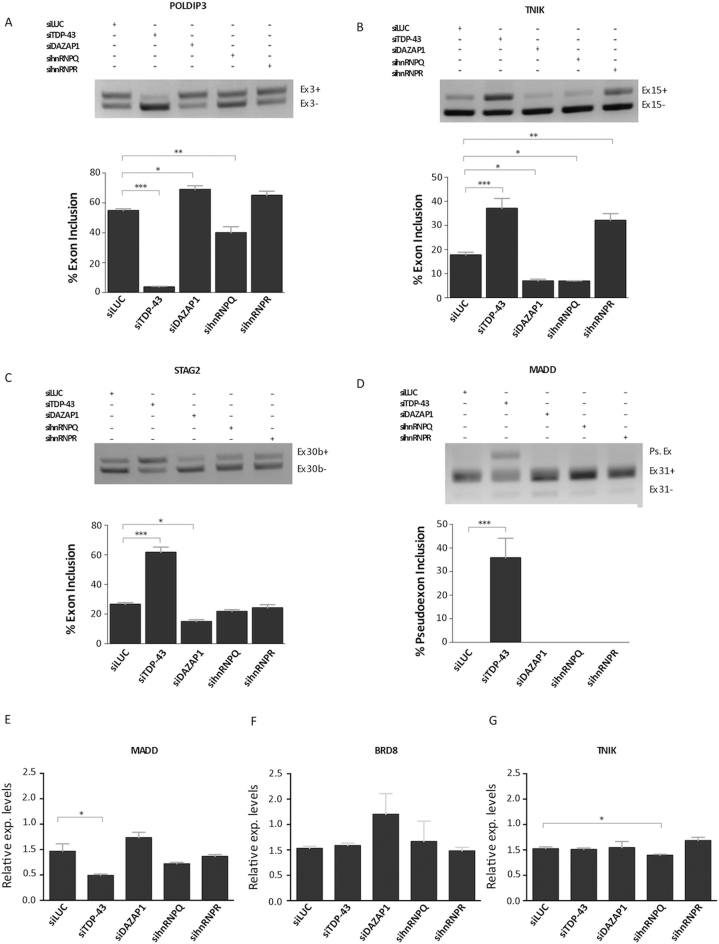
Therefore, in order to determine the influence of these hnRNPs on TDP-43-regulated events, we tested their functions in a series of pre-mRNA splicing processes known to be consistently altered by TDP-43 depletion (the efficiency of hnRNP depletion in the SH-SY-5Y cell line used for this experiment is reported in Supplementary Figure S2). Thus, the analyzed events were the splicing of SKAR/POLDIP3 exon 3 (48); TNIK exon 15 (14); STAG2 exon 30b (49); and MADD exon 31 (49).
As shown in Figure 3A–D, the removal of TDP-43 caused a drop in the inclusion of POLDIP3 exon 3 (Figure 3A), an increase in TNIK exon 15 inclusion (Figure 3B), an increase in STAG2 exon 30b inclusion (Figure 3C) and loss of exon 31 inclusion plus activation of a pseudoexon in the MADD gene (Figure 3D). Interestingly, removal of DAZAP1, hnRNP Q and hnRNP R also had a noticeable and consistent effect on the recognition of these exons. In particular, removal of DAZAP1 could increase POLDIP3 exon 3 inclusion even above normal inclusion levels (Figure 3A), and decrease TNIK exon 15 (Figure 3B) and STAG2 (Figure 3C) exon 30b inclusion below normal levels, in a specular manner with respect to TDP-43. Similarly to DAZAP1, also silencing of hnRNP Q could decrease TNIK exon 15 inclusion below normal levels (Figure 3B). In the case of hnRNP R, an effect on splicing could be detected on TNIK exon 15 inclusion that could also be increased above normal levels in a manner similar to TDP-43 (Figure 3B). Finally, DAZAP1, hnRNP Q and R do not seem to affect MADD splicing profile like TDP-43, which induces exon skipping and pseudoexon inclusion (Figure 3D).
Figure 3.
Effects of DAZAP1 and hnRNP Q/R depletion on TDP-43 controlled events. Differentially treated SH-SY-5Y cells were used to validate the effects of various hnRNPs on TDP-43 controlled genes. RT-PCR analysis was performed for the following splicing events: POLDIP3/SKAR exon 3 (A), TNIK exon 15 (B), STAG2 exon 30b (C) and MADD exon 31 (D). The agarose gel was loaded with the following samples: control siRNA Luciferase transfected cells (lane 1, siLuc), depleted of TDP-43 (lane 2, siTDP-43), depleted of DAZAP1 (lane 3, siDAZAP1), depleted of hnRNP Q (lane 4, sihnRNPQ) and depleted of hnRNP R (lane 5, sihnRNPR). The identity of the various transcripts is reported on the right. For the MADD gene, the appearance of a pseudoexon is also reported (Ps.Ex.). *P < 0.05, **P < 0.01, ***P < 0.001 (n = 3), calculated by student's t-test. The effects of DAZAP1 and hnRNP Q/R depletion were also tested on gene expression events controlled by TDP-43. Real-time PCR quantification analysis of MADD (E), BRD8 (F) and TNIK (G) endogenous transcript levels following siRNA transfection in SH-SY-5Y cells from three independent experiments. Each bar reports the mean ± standard deviation of three independent experiments. The single asterisks indicate significant differences (P ≤ 0.05) between the indicated measurements.
In parallel to pre-mRNA splicing, another TDP-43 controlled event that can also be tested using this approach is the effects on gene expression levels of TDP-43 targets previously published by our laboratory. To this aim, we examined the levels of MADD and BRD8 expression levels as these were previously reported to be reduced following the TDP-43 RNAi (49). TNIK was also added as a control, since its splicing levels were affected by most of these hnRNPs. As shown in Figure 3E–G, however, in SH-SY-5Y cells TDP-43 seemed to have a statistically significant effect only on the expression levels of the MADD gene. None of the other hnRNPs was able to affect expression independently with the only exception of hnRNP Q that downregulated TNIK expression for a small but statistically significant effect.
Rescue of TDP43-controlled mRNA splicing and gene expression events by DAZAP1 and hnRNP Q and R
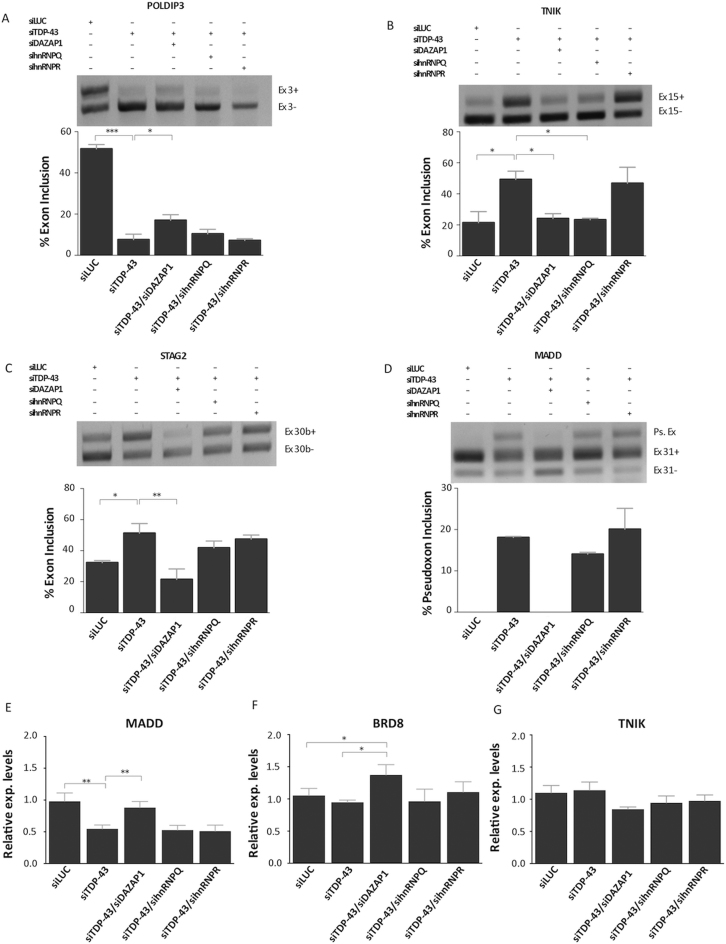
Considering these changes, it was then interesting to take into account whether the effects mediated by the single hnRNPs could be used to rescue splicing events disrupted by the absence of TDP-43. We then proceeded with the removal of DAZAP1, hnRNP Q and hnRNP R in parallel with TDP-43 silencing, to determine whether and how these proteins could further affect these splicing events.
As shown in these figures (Figure 4A–D), DAZAP1 removal significantly rescued the inclusion of POLDIP3 exon 3, the skipping of TNIK exon 15 and the skipping of STAG2 exon 30b. With regards to the MADD gene, although removal of DAZAP1 did not significantly rescue exon 31 inclusion, it did prevent pseudoexon inclusion with a very high efficiency. With regards to hnRNP Q, its removal was effective to rescue exon skipping of TNIK exon 15 but had no effect in all other cases. Finally, removal of hnRNP R had no effect to modify the changes induced by TDP-43 knock down in any of these four genes. As shown in Figure 3E–G, it was interesting to note that also for gene expression profiles the knockout of DAZAP1 could significantly rescue the gene expression levels of the MADD and BRD8 genes. No effect could be observed, however, for the knockout of either hnRNP Q or R.
Figure 4.
Rescue of TDP-43 controlled pre-mRNA splicing and gene expression events. RT-PCR of SH-SY-5Y cell lines were used to validate the potential effects of hnRNP depletion on the splicing profile of various TDP-43 controlled genes: POLDIP3/SKAR exon 3 (A), TNIK exon 15 (B), STAG2 exon 30b (C) and MADD exon 31 (D). The agarose gel was loaded with the following samples: control siRNA Luciferase transfected cells (lane 1, siLUC), depleted of TDP-43 (lane 2, siTDP-43), depleted of TDP-43 and DAZAP1 (lane 3, siTDP-43/siDAZAP1), depleted of TDP-43 and hnRNP Q (lane 4, siTDP-43/sihnRNPQ), and depleted of TDP-43 and hnRNP R (lane 5, siTDP-43/sihnRNPR). The identity of the various transcripts is reported on the right. For the MADD gene, the appearance of a pseudoexon is also reported (Ps.Ex.). *P < 0.05, **P < 0.01, ***P < 0.001 (n = 3), calculated by one-way ANOVA. The effects of DAZAP1 and hnRNP Q/R depletion were also tested on gene expression events controlled by TDP-43. Real-time PCR quantification analysis of MADD (E), BRD8 (F) and TNIK (G) transcript levels following siRNA transfection in SH-SY5Y cells. Each bar reports the mean ± standard deviation of three independent experiments. The double and single asterisks indicate significant differences (P ≤ 0.01 and P ≤ 0.05, respectively) between the indicated measurements.
The results of these rescue experiments are summarized in Figure 5A and from this figure it is clear that, out of all these three hnRNPs tested, DAZAP1 is the most consistent modifier of the tested TDP-43 targets and that can also effectively rescue the effects of TDP-43 depletion on RNA splicing and MADD expression.
Figure 5.
DAZAP1 connections with TDP-43 expression and regulated events. (A) schematic diagram reporting the effects of TDP-43, DAZAP-1, hnRNP Q and hnRNP R effects of the four TDP43-controlled splicing events in SH-SY-5Y cells. (B) TDP-43 expression levels measured by western blot following siRNA silencing in SH-SY-5Y cells of DAZAP1, hnRNP Q and hnRNP R. Silencing of TDP-43 is also reported as a control. (C) Effects of TDP-43 silencing in SH-SY-5Y cells on DAZAP1 protein expression levels in siRNA TDP-43 treated versus untreated cells. (D) Co-immunoprecipitation experiments using flag-TDP-43 to check for binding to DAZAP1 (upper panel). The presence of hnRNP H1 in the immunoprecipitated sample is used as a positive control (middle panel). The lower panel shows the levels of flagged and endogenous TDP-43 in the Input and immunprecipitated sample. (E) RNA immunoprecipitation experiments to control for DAZAP1 binding to the BRD8, TNIK, STAG2, POLDIP3, MADD transcripts and also two housekeeping genes, GAPDH and SDHA (used as controls).
DAZAP1 does not affect TDP-43 expression but can bind in vivo to TDP43-controlled mRNAs
Therefore, in order to further characterize the role of these hnRNPs in terms of TDP-43 expression and targets we first tested whether these hnRNPs could directly affect TDP-43 expression, especially DAZAP1. However, a western blot for TDP-43 following the silencing of DAZAP1, hnRNP Q and hnRNP R showed that TDP-43 expression was not appreciably affected by silencing of these proteins (Figure 5B). Also of note, TDP-43 silencing was not capable of affecting significantly the expression of DAZAP1 (Figure 5C) and that of hnRNP Q and R (Supplementary Figure S3A).
We then tested whether silencing of these three factors could alter the nuclear localization of TDP-43 leaving its relative expression levels unchanged. However, immunohistochemical analysis DAZAP1, and hnRNP Q and R silenced cells showed that TDP-43 retained its mostly nuclear localization (Supplementary Figure S3B–E).
Another possible connection between TDP-43 and these hnRNPs could be represented by their ability to bind directly TDP-43 and affect its functional properties. This is certainly a possibility for hnRNP Q, as previous co-IP experiments have shown a direct interaction between this protein and TDP-43 (18). In parallel, several proteomic studies on TDP-43 have also confirmed the presence of hnRNP R in complexes that contain TDP-43 (18,20).
No previous experimental reports, however, have uncovered a possible interaction between TDP-43 and DAZAP1. In order to address this issue, therefore, we have performed co-immunoprecipitation experiments by transfecting flagged-TDP-43 in HeLa cells and testedthe presence of DAZAP1 by western blot. The results of this experiment, presented in Figure 5D, show that TDP-43 and DAZAP1 do not seem to co-IP together (hnRNP H co-immunoprecipitation with TDP-43 is reported as positive control of experimental conditions).
It was then decided to test whether DAZAP1 was capable of binding to the TDP-43 pre-mRNA targets analyzed in Figures3 and 4. To do this, we performed RNA-IP analysis by transfecting a flag-DAZAP1 protein in SH-SY-5Y cells, immunoprecipitating and testing by RT-qPCR of the binding of this protein to the POLDIP3, TNIK, MADD and STAG2 transcripts. As shown Figure 5E, all these transcripts were highly enriched (contrary to what was observed for two control genes, GAPDH and SDHA).
TDP-43 together with DAZAP1 can alter the expression of a high number of neuronal and synaptic mRNAs in SH-SY5Y cells
Based on the results of these RNA-IP experiments, we next sought to identify all the common target genes whose expression may be co-regulated by TDP-43 and DAZAP1 in a more neuronal setting. To achieve this, we then performed RNA sequencing analysis of the human neuroblastoma SH-SY5Y cell line depleted by TDP-43 or DAZAP1, by RNAi and compared results.
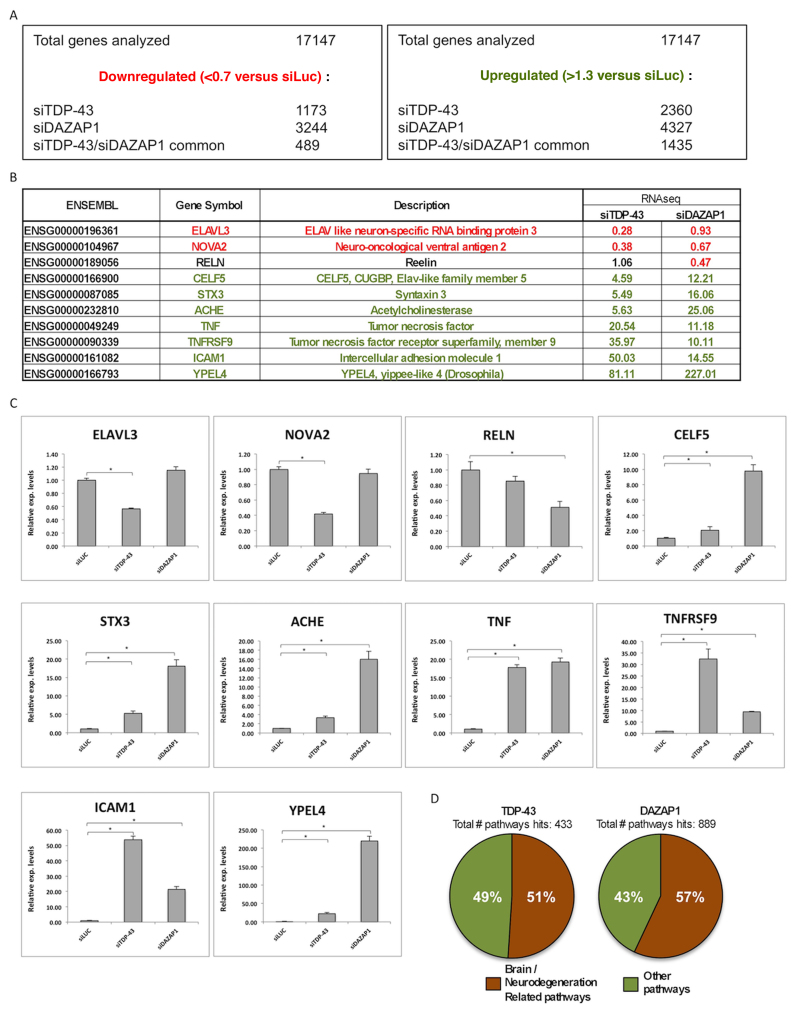
The putative differentially expressed genes generated by RNA-Seq following silencing were identified by comparing to a control sample treated with anti-luciferase siRNA (siLuc) (upregulation cut-off: >1.3; downregulation cut-off: <0.7-fold change). Using these criteria, among the total 17147 genes analyzed by RNA-seq for TDP-43, there were a total of 3533 genes differentially expressed between siTDP-43 and siLUC treated cells. Of the 3533 differentially expressed genes, 1173 (33%) genes were downregulated while 2360 (67%) genes were upregulated (Figure 6A). Conversely, for DAZAP1, there were a total of 7571 genes differentially expressed between siDAZAP1 and siLUC treated cells. Of the 7571 differentially expressed genes, 3244 (43%) genes were downregulated while 4327 (57%) genes were upregulated. The top 100 hits for TDP-43 and their relative changes in expression levels are reported in Supplementary Table S1 whilst the top 250 hits for DAZAP1 are reported in Supplementary Table S2 (with neuronal pathway genes highlighted).
Figure 6.
Validation of the TDP-43 or DAZAP1 silencing and comparison between RNA-seq and qRT-PCR results. (A) Summary of downregulated (<0.7× versus siLUC) and upregulated (>1.3× versus siLUC) genes after siTDP-43 or siDAZAP1 treatments. The number of common (between siTDP-43 and siDAZAP1) downregulated and upregulated genes is also shown. (B) List of genes associated with brain functions (ELAV3, NOVA2, RELN, STX3, ACHE, YPEL4, CELF5) or inflammation (TNF, TNFRSF9, ICAM1) selected for validation of the RNA-seq analysis. The expression levels of genes following siTDP-43 or siDAZAP1 treatments versus the control condition (siLUC) is indicated. (C) Validation of RNA-seq by real time PCR of the ten selected transcripts. The results are represented as relative expression compared with the control (siLUC). (D) Pathways analysis of differentially expressed genes following TDP-43 and DAZAP1 depletion as determined by PANTHER, DAVID and UniProt analyses.
Most importantly, the RNA-seq analysis identified 489 co-downregulated and 1435 co-upregulated genes in siTDP-43 and siDAZAP1 treated cells which represent a notable target overlap (Figure 6A). According to the DAVID database (50), many of these genes are involved in brain metabolism or potentially related with neurodegeneration.
In particular, from this common list of hits, we then selected for qRT-PCR validation ten genes that have been described to possess a relation with brain/inflammation, according to the GEO functional annotations of the listed genes (Figure 6B). The fold changes obtained from qRT-PCR were compared with RNA-seq expression analysis results (Figure 6C).
Initially, we gave priority to the genes top ranked in the siTDP-43 RNA-seq data and then looked for the common targets, potentially related with neurodegeneration, regulated by DAZAP1. Among the differentially expressed genes identified, the coregulation was particularly evident for seven gene, CELF5 (CUGBP, Elav-Like Family Member 5), Syntaxin 3 (STX3), Acetylcholinesterase (ACHE) Tumor necrosis factor (TNF), Tumor necrosis factor receptor superfamily, member 9 (TNFRSF9), Intercellular adhesion molecule 1 (ICAM1) and YPEL4 (yippee like 4) that were highly upregulated both by TDP-43 and DAZAP1 RNAi (Figure 6B and C). On the other hand, we found genes such as ELAV like neuron-specific RNA binding protein 3 (ELAV3) and Neuro-oncological ventral antigen 2 (NOVA2) that were prominently downregulated by TDP-43 RNAi (and NOVA2 also by DAZAP1 RNAi). Reelin (RELN), shown to be regulated only by siDAZAP1 in the RNA-seq analysis, was selected as a ‘neutral’ (for TDP-43 RNAi treatment) and qPCRs confirmed these data.
We searched for enriched Gene Ontology (GO) annotations in the set of genes that were commonly up- or downregulated by TDP-43 and DAZAP1. To this aim, the list of significantly TDP-43 and DAZAP1 regulated transcripts were sorted into GO term categories for ‘molecular function’ and ‘biological process’ by using the UniProtKB Retrieve/ID mapping tool (http://www.uniprot.org/uploadlists/). Regarding TDP-43, 2269 out of the 3535 statistically significantly regulated Ensembl identifiers were successfully mapped to 8481 UniProtKB IDs. Regarding DAZAP1, 5137 out of the 7571 statistically significantly regulated Ensembl identifiers were successfully mapped to 19541 UniProtKB IDs.
For both TDP-43 and DAZAP1, the molecular function of GO terms associated with regulated transcripts include a large percentage of transcription factor activity, transcription factor binding, nucleic acid binding transcription factor activity, signal transducer activity, transporter activity, molecular function regulator and receptor activity (Supplementary Figure S4A).
On the other hand, in the biological process category, similar percentage in genes involved in ‘cellular process’, biological regulation, single-organism process and ‘metabolic processes’ groups were notably represented for both siTDP-43 and siDAZAP1 (Supplementary Figure S4A). It is also interesting to note that, among the subcategories, the molecular events most represented were signal transduction, autophagy, protein folding/unfoding, immune system process, biological adhesion, locomotion, presynaptic process involved in chemical synaptic transmission and behavior (data not shown).
In addition, the statistical significance of the overlap between the lists of genes regulated by the two splicing factors was also evaluated (Supplementary Figure S4B). The overlap between both the downregulated and upregulated gene sets was significantly higher than expected for independent sets of genes (Supplementary Figure S4B).
As expected, high enrichment scores for both the up- and downregulated gene sets were found, for clusters related to neurodegenerative disease and nervous system-related genes (Supplementary Figure S4C and Table S4). In particular, a consistent number of genes was also found to be related with inflammatory responses, supporting a potentially important impact of TDP-43 and DAZAP1 for neuroinflammation and motor neuron pathophysiology, as recently reviewed (51).
Finally, another important feature of this analyses was the observation that a number of transcripts potentially related with the activity of the cholinergic system have been found to be altered (Supplementary Table S3). Even taking in consideration the glutamatergic (and not cholinergic) nature of Drosophila motoneurons, this finding might be particularly interesting from the point of view of human ALS disease that is an example of cholinergic neurodegeneration.
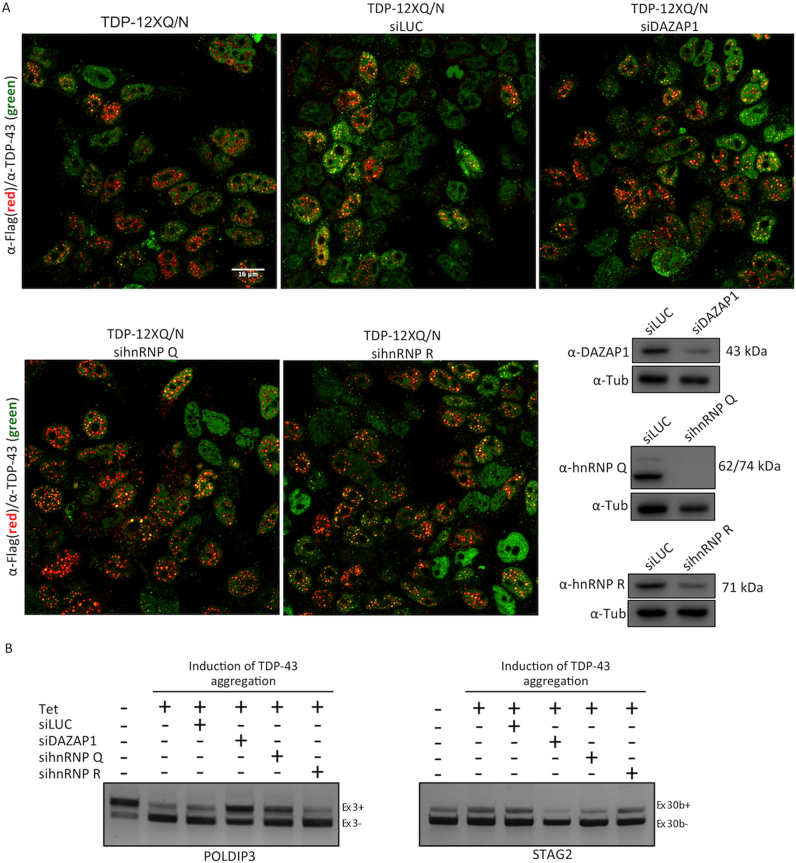
Effects of depleting DAZAP1 and hnRNPQ/R in a loss-of-function model of TDP-43 pathology
In order to further support these results in a more realistic scenario of TDP-43 pathology, we have taken advantage of a new aggregation system that is based on the Tet-dependent expression in HEK-293 cells of repetitions of the prion-like, Q/N-repeated sequence at the C-terminus of TDP-43 (52,53). The advantage of this system over previous methodologies is principally based on its ability to induce endogenous TDP-43 aggregation in cells without altering the expression levels of the TARDBP gene or introducing mutations in its sequence. Of interest, the effects of this induced aggregation have been recently shown to be comparable to TDP-43 knockdown in SH-SY-5Y cells (54). Therefore, using this system we have tested whether knockdown of DAZAP1, hnRNP Q and hnRNP R can affect the aggregation of endogenous TDP-43. As shown in Figure 7A, knockdown of these proteins, in fact, cannot reduce the accumulation of TDP-43 aggregates in the nucleus and cytoplasm of cells. Nonetheless, Figure 7B shows that removal of DAZAP1 and hnRNP Q can restore the inclusion of POLDIP3 exon 3 and the skipping of STAG2 exon 30b which was induced by the formation of the aggregates. As in the knockdown experiments performed in SH-SY-5Y cells, hnRNP R had no ability to rescue either effect. In this system, the splicing profiles of the TNIK and MADD genes could not be tested as it occurs differently in HEK293 cells compared to the SH-SY-5Y cell lines.
Figure 7.
Immunofluorescence assay of Flp-In HEK293 FLAG-tagged wild-type TDP-43 cells. (A) The merged image shows the localization of the endogenous TDP-43 (green) and aggregated (red) TDP43-F4L-12XQ/N after RNA silencing of DAZAP1, hnRNP Q and hnRNP R. A siRNA against fire-fly Luciferase (Luc) was used as a control. Scale bars: 16 μm. (B) RT-PCR of Flp-In HEK293 cell lines expressing TDP-43 aggregates were used to validate the potential effects of hnRNP depletion on the splicing profile of POLDIP3 exon 3 and STAG2 econ 30b. Western blot analysis of RNA silencing against DAZAP1, hnRNP Q and hnRNP R. Antibody anti-Tubulin (α-Tub) was used as a loading control.
Alterations in the expression of hnRNP proteins in ALS patients
Finally, it was of interest to find existing supporting evidence for these findings in a disease scenario. Therefore, as a preliminary approach, we explored GEO deposited levels of transcripts expressed in human cells and/or tissues from ALS and Spinal Muscolar Atrophy (SMA) patients where TDP-43 levels are modified. As a result, we found two connected studies (55,56) where global gene expression profiling in 13 muscle disease groups of ALS patients showed the simultaneous alteration in the mRNA levels of the hnRNPs highlighted by our experiments: TDP-43 (+121% versus controls) DAZAP1 (+118% versus controls), hnRNP Q/Syncrip (+120% versus controls) and hnRNPR (+141% versus controls). The observation that alterations in ALS patients may not simply be limited to TDP-43 but can eventually also occur in the expression of these hnRNPs opens the way for further indepth analyses of the global importance of hnRNP variations in ALS disease.
DISCUSSION
In general, all RBPs present in the eukaryotic nucleus jointly collaborate to regulate all the aspects of an mRNA life cycle, from transcription to translation (57). How they achieve this with remarkable accuracy is still the subject of numerous studies. First, because each of these proteins generally binds hundreds or thousands of transcripts and as a result, determining specific versus non-specific interactions is still a challenging question (58). Second, even if we knew exactly where each hnRNP was binding in the eukaryotic transcriptome we still lack a comprehensive knowledge of the functional properties of each RNA binding protein. Moreover, both RNA-binding properties and function can be deeply affected by post-translational modifications which are very common and can potentially affect pathology especially in ALS and FTD (59–61). In conclusion, even for well known hnRNP proteins that have been studied for several years it is still difficult to predict exactly which transcripts or cellular pathways could be affected by their presence or absence. Most importantly, it is still very difficult to predict what kind of influence they may have on each other, either by affecting their expression/function or binding to similar sets of targets (62).
For this reason, a better understanding of this issue may have considerable importance for our understanding of neurodegenerative processes. The working hypothesis is that differences in expression levels of hnRNP proteins within a neuron in the presence of the same disease-causing mutation or aberrant aggregation of key RBP proteins identified, such as TDP-43 and FUS/TLS may result in profound consequences.
In this work, therefore, we have addressed in a comprehensive manner the possible effect of most major nuclear hnRNP proteins on TDP-43 pathology using well characterized fly models: analyzing both loss-of-function and gain-of-function scenarios. To do this, we have exploited the fact that hnRNPs represent a very ‘old’ and conserved class of proteins. As a result, we have been able to focus our attention on 12 human hnRNPs (Table 1) for which fly strains expressing shRNA are available.
Depletion of almost all these hnRNPs on a reduced TDP-43 expression background was observed to generally have a deleterious effect on the locomotor abilities of flies, although to different extent. This result may not be particularly surprising, as many of these proteins are already known to participate in alternative splicing processes that play a central role in brain development and functioning (32,63).
As a central conclusion, the picture that emerges from this study is that the majority of all major hnRNP proteins have some potential to modify TDP-43 functional consequences both in a loss-of-function and gain-of-function scenarios, meaning similarly regulated target genes, metabolic pathways of sub-cellular mechanisms.
In particular, out of all the tested hnRNPs, we have decided for several reasons to further concentrate our efforts on three hnRNPs: DAZAP1, hnRNP Q and hnRNP R. First of all, because they displayed a rather interesting behavior with respect to the TDP-43 disease models. In the loss-of-function scenarios, in fact, knockout of these hnRNPs caused paralysis or lethality of flies. In a gain-of-function scenario, however, they were capable of rescuing the toxicity induced by TBPH overexpression in the fly eye.
Second, they are less characterized hnRNPs with respect to hnRNP C and F/H, and might thus represent a novel research area worthy of exploration. In particular, in the pre-mRNA processing field no general study has been performed with regards to DAZAP1 targets to this date and very little on hnRNP Q and R (64).
Most importantly, however, it is interesting to note that previous studies have identified these proteins as being important for neuronal development. For example, hnRNP R has been reported to localize in presynaptic compartments at neuromuscolar endplates and can interact with SMN protein both in vitro and in vivo (65), where TDP-43 is known to play important roles (66–68). Similarly, Syncrip/hnRNP Q has been demonstrated to regulate synaptic transmission signaling at the Drosophila synapse and regulates the expression of mRNAs for key synaptic proteins (69,70). On the other hand, although initially identified as a binding partner of the germ-specific factor DAZ (71), DAZAP1 has been recently shown to play a very important role in numerous cell types by regulating mRNA translation and pre-mRNA splicing process (72–74).
The conclusion of our experiments on these proteins has confirmed the central role of DAZAP1 as a central modifier of TDP-43 toxicity that has been previously reported by Ritson et al. (35). The reason for this activity probably relies on the fact that both proteins TDP-43 and DAZAP1, can target a very similar set of transcripts.
The ID mapping of transcripts regulated by these two nuclear factors (Supplementary Figure S4) shows a high level of overlap in the targeted functional categories and suggest that one of the reasons DAZAP1 can modify TDP-43 effects on RNA metabolism and its depletion is lethal in flies could be because both proteins target the same potentially critical pathways for brain metabolism.
Intriguingly, our RNA sequencing analysis has further uncovered that most of these transcripts can be implicated with brain functions and potentially with neurodegeneration (Figure 6D and Supplementary Tables S1–4). Of course, there are still several open questions that will need to be addressed. First and foremost, the observation that a majority of genes relating to neurodegeneration were regulated into the same direction by silencing of TDP-43 and DAZAP1. Nonetheless, abnormalities in splicing that are induced by the silencing of TDP-43 in human cells could be rescued by the silencing of DAZAP1. The most likely possibility, of course, could be that the transcripts which allow the rescue of the TDP-43 splicing phenotype by DAZAP1 are within the minority of genes which are differentially regulated by the two proteins. However, further work will be required to clarify this issue.
A second issue that also remains to be investigated regards the possible influence of non-coding RNAs (lncRNAs) in modulating these hnRNP interactions. It has been described, in fact, that both TDP-43 and FUS can interact with several lncRNAs and can regulated both their functions and stability (75). In particular, TDP-43 has been shown to interact and regulate important lncRNAs such as gadd7 (76), MALAT1 (77), NEAT1-2 (78), transposable elements general (79) and two SPA lncRNA involved in Prader-Willi syndrome (80). How these hnRNPs may help or hinder TDP-43 in regulating these molecules is something that will need to be determined.
These future connections imply that connections between TDP-43 and hnRNP proteins may well go well beyond the splicing process that has represented the focus of this work.
In a more general context, and in addition to the considerations that relate specifically to TDP-43 functions, the question that our work has also partially answered regards the extent at which results obtained in fly overexpression or knock-down models can be translated to human neuronal cell lines. This is obviously a very important issue considering the massive use of fly Drosophila models in neurodegeneration and the need to eventually ‘translate’ these results in the human context. In our opinion, our results paint a rather optimistic view. Of course, differences will always exist between the two systems, but it is encouraging that, in our splicing assays, proteins such as DAZAP1 and hnRNP Q can rescue the disregulation caused by TDP-43 absence in a manner that is reminiscent of the TDP-43 OE rescue in the fly eye by Hrp27c and Syncrip. In particular, based on our RNAseq data, the reason for these similar properties probably relies in the degree of functional conservation by these proteins as described in this work.
In the future, the molecular characterization of the co-regulated transcripts potentially playing a major role in neurodegeneration and how they might be regulated in different brain regions by differing hnRNP expression levels will hopefully provide us with a better ability to follow disease onset/progression and might help to pinpoint novel key therapeutic targets that are still currently lacking.
ACCESSION NUMBER
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (45) and are accessible through GEO Series accession number GSE97262.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Francisco Baralle for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Thierry Latran Fondation (REHNPALS); EU Joint Programme-Neurodegenerative Diseases JPND (RiMod-FTD, Italy, Ministero della Sanita’/MIUR) (to E.B.); International Scientific Co-operation Agreement between Italy and Israel (SCREENCELLS4ALS, Ministero Affari Esteri, MAE, Italy); ARISLA (CHRONOS) (to F.F.). Funding for open access charge: JPND (RIMOD-FTD).
Conflict of interest statement. None declared.
REFERENCES
- 1. Hanson K.A., Kim S.H., Tibbetts R.S.. RNA-binding proteins in neurodegenerative disease: TDP-43 and beyond. Wiley Interdiscip. Rev. RNA. 2011; 3:265–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. King O.D., Gitler A.D., Shorter J.. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012; 1462:61–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Campos-Melo D., Droppelmann C.A., Volkening K., Strong M.J.. RNA-binding proteins as molecular links between cancer and neurodegeneration. Biogerontology. 2014; 15:587–610. [DOI] [PubMed] [Google Scholar]
- 4. Ling S.C., Polymenidou M., Cleveland D.W.. Converging Mechanisms in ALS and FTD: Disrupted RNA and Protein Homeostasis. Neuron. 2013; 79:416–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nussbacher J.K., Batra R., Lagier-Tourenne C., Yeo G.W.. RNA-binding proteins in neurodegeneration: Seq and you shall receive. Trends Neurosci. 2015; 38:226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sephton C.F., Yu G.. The function of RNA-binding proteins at the synapse: implications for neurodegeneration. Cell Mol. Life Sci. 2015; 72:3621–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dreyfuss G., Kim V.N., Kataoka N.. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002; 3:195–205. [DOI] [PubMed] [Google Scholar]
- 8. Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M. et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006; 314:130–133. [DOI] [PubMed] [Google Scholar]
- 9. Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y. et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006; 351:602–611. [DOI] [PubMed] [Google Scholar]
- 10. Polymenidou M., Lagier-Tourenne C., Hutt K.R., Huelga S.C., Moran J., Liang T.Y., Ling S.C., Sun E., Wancewicz E., Mazur C. et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011; 14:459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sephton C.F., Good S.K., Atkin S., Dewey C.M., Mayer P. 3rd, Herz J., Yu G.. TDP-43 Is a developmentally regulated protein essential for early embryonic development. J. Biol. Chem. 2010; 285:6826–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tollervey J.R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., Konig J., Hortobagyi T., Nishimura A.L., Zupunski V. et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011; 14:452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xiao S., Sanelli T., Dib S., Sheps D., Findlater J., Bilbao J., Keith J., Zinman L., Rogaeva E., Robertson J.. RNA targets of TDP-43 identified by UV-CLIP are deregulated in ALS. Mol. Cell Neurosci. 2011; 47:167–180. [DOI] [PubMed] [Google Scholar]
- 14. Colombrita C., Onesto E., Buratti E., de la Grange P., Gumina V., Baralle F.E., Silani V., Ratti A.. From transcriptomic to protein level changes in TDP-43 and FUS loss-of-function cell models. Biochim. Biophys. Acta. 2015; 1849:1398–1410. [DOI] [PubMed] [Google Scholar]
- 15. Stalekar M., Yin X., Rebolj K., Darovic S., Troakes C., Mayr M., Shaw C.E., Rogelj B.. Proteomic analyses reveal that loss of TDP-43 affects RNA processing and intracellular transport. Neuroscience. 2015; 293:157–170. [DOI] [PubMed] [Google Scholar]
- 16. Amlie-Wolf A., Ryvkin P., Tong R., Dragomir I., Suh E., Xu Y., Van Deerlin V.M., Gregory B.D., Kwong L.K., Trojanowski J.Q. et al. Transcriptomic changes due to cytoplasmic TDP-43 expression reveal dysregulation of histone transcripts and nuclear chromatin. PLoS One. 2015; 10:e0141836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Freibaum B.D., Chitta R.K., High A.A., Taylor J.P.. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 2010; 9:1104–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ling S.C., Albuquerque C.P., Han J.S., Lagier-Tourenne C., Tokunaga S., Zhou H., Cleveland D.W.. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:13318–13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ewing R.M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M.D., O’Connor L., Li M. et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 2007; 3:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blokhuis A.M., Koppers M., Groen E.J., van den Heuvel D.M., Dini Modigliani S., Anink J.J., Fumoto K., van Diggelen F., Snelting A., Sodaar P. et al. Comparative interactomics analysis of different ALS-associated proteins identifies converging molecular pathways. Acta Neuropathol. 2016; 132:175–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Budini M., Baralle F.E., Buratti E.. Targeting TDP-43 in neurodegenerative diseases. Expert Opin. Ther. Targets. 2014; 18:617–632. [DOI] [PubMed] [Google Scholar]
- 22. Buratti E., Brindisi A., Giombi M., Tisminetzky S., Ayala Y.M., Baralle F.E.. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through Its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 2005; 280:37572–37584. [DOI] [PubMed] [Google Scholar]
- 23. Kim H.J., Kim N.C., Wang Y.D., Scarborough E.A., Moore J., Diaz Z., Maclea K.S., Freibaum B., Li S., Molliex A. et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013; 495:467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gilpin K.M., Chang L., Monteiro M.J.. ALS-linked mutations in ubiquilin-2 or hnRNPA1 reduce interaction between ubiquilin-2 and hnRNPA1. Hum. Mol. Genet. 2015; 24:2565–2577. [DOI] [PubMed] [Google Scholar]
- 25. Berson A., Barbash S., Shaltiel G., Goll Y., Hanin G., Greenberg D.S., Ketzef M., Becker A.J., Friedman A., Soreq H.. Cholinergic-associated loss of hnRNP-A/B in Alzheimer's disease impairs cortical splicing and cognitive function in mice. EMBO Mol. Med. 2012; 4:730–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Honda H., Hamasaki H., Wakamiya T., Koyama S., Suzuki S.O., Fujii N., Iwaki T.. Loss of hnRNPA1 in ALS spinal cord motor neurons with TDP-43-positive inclusions. Neuropathology. 2015; 35:37–43. [DOI] [PubMed] [Google Scholar]
- 27. Mori K., Lammich S., Mackenzie I.R., Forne I., Zilow S., Kretzschmar H., Edbauer D., Janssens J., Kleinberger G., Cruts M. et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013; 125:413–423. [DOI] [PubMed] [Google Scholar]
- 28. Lee Y.B., Chen H.J., Peres J.N., Gomez-Deza J., Attig J., Stalekar M., Troakes C., Nishimura A.L., Scotter E.L., Vance C. et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013; 5:1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Prudencio M., Belzil V.V., Batra R., Ross C.A., Gendron T.F., Pregent L.J., Murray M.E., Overstreet K.K., Piazza-Johnston A.E., Desaro P. et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat. Neurosci. 2015; 18:1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cooper-Knock J., Higginbottom A., Stopford M.J., Highley J.R., Ince P.G., Wharton S.B., Pickering-Brown S., Kirby J., Hautbergue G.M., Shaw P.J.. Antisense RNA foci in the motor neurons of C9ORF72-ALS patients are associated with TDP-43 proteinopathy. Acta Neuropathol. 2015; 130:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Conlon E.G., Lu L., Sharma A., Yamazaki T., Tang T., Shneider N.A., Manley J.L.. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. Elife. 2016; 5:e17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Romano M., Buratti E., Romano G., Klima R., Del Bel Belluz L., Stuani C., Baralle F., Feiguin F.. Evolutionarily conserved heterogeneous nuclear ribonucleoprotein (hnRNP) A/B proteins functionally interact with human and Drosophila TAR DNA-binding protein 43 (TDP-43). J. Biol. Chem. 2014; 289:7121–7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suzuki H., Shibagaki Y., Hattori S., Matsuoka M.. Nuclear TDP-43 causes neuronal toxicity by escaping from the inhibitory regulation by hnRNPs. Hum. Mol. Genet. 2015; 24:1513–1527. [DOI] [PubMed] [Google Scholar]
- 34. He F., Krans A., Freibaum B.D., Taylor J.P., Todd P.K.. TDP-43 suppresses CGG repeat-induced neurotoxicity through interactions with HnRNP A2/B1. Hum. Mol. Genet. 2014; 23:5036–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ritson G.P., Custer S.K., Freibaum B.D., Guinto J.B., Geffel D., Moore J., Tang W., Winton M.J., Neumann M., Trojanowski J.Q. et al. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J. Neurosci. 2010; 30:7729–7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mohagheghi F., Prudencio M., Stuani C., Cook C., Jansen-West K., Dickson D.W., Petrucelli L., Buratti E.. TDP-43 functions within a network of hnRNP proteins to inhibit the production of a truncated human SORT1 receptor. Hum. Mol. Genet. 2015; 25:534–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Romano M., Feiguin F., Buratti E.. Drosophila answers to TDP-43 proteinopathies. J. Amino Acids. 2012; 2012:356081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Casci I., Pandey U.B.. A fruitful endeavor: modeling ALS in the fruit fly. Brain Res. 2015; 1607:47–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Goina E., Skoko N., Pagani F.. Binding of DAZAP1 and hnRNPA1/A2 to an exonic splicing silencer in a natural BRCA1 exon 18 mutant. Mol. Cell. Biol. 2008; 28:3850–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marcucci R., Baralle F.E., Romano M.. Complex splicing control of the human Thrombopoietin gene by intronic G runs. Nucleic Acids Res. 2007; 35:132–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mi H., Poudel S., Muruganujan A., Casagrande J.T., Thomas P.D.. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016; 44:D336–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 43. The UniProt C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roy P.J., Stuart J.M., Lund J., Kim S.K.. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature. 2002; 418:975–979. [DOI] [PubMed] [Google Scholar]
- 45. Edgar R., Domrachev M., Lash A.E.. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., Perrimon N., Mohr S.E.. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011; 12:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cragnaz L., Klima R., Skoko N., Budini M., Feiguin F., Baralle F.E.. Aggregate formation prevents dTDP-43 neurotoxicity in the Drosophila melanogaster eye. Neurobiol. Dis. 2014; 71:74–80. [DOI] [PubMed] [Google Scholar]
- 48. Fiesel F.C., Weber S.S., Supper J., Zell A., Kahle P.J.. TDP-43 regulates global translational yield by splicing of exon junction complex component SKAR. Nucleic Acids Res. 2012; 40:2668–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. De Conti L., Akinyi M.V., Mendoza-Maldonado R., Romano M., Baralle M., Buratti E.. TDP-43 affects splicing profiles and isoform production of genes involved in the apoptotic and mitotic cellular pathways. Nucleic Acids Res. 2015; 43:8990–9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dennis G. Jr, Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A.. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003; 4:P3. [PubMed] [Google Scholar]
- 51. Rodriguez M.J., Mahy N.. Neuron-microglia interactions in motor neuron degeneration. the inflammatory hypothesis in amyotrophic lateral sclerosis revisited. Curr. Med. Chem. 2016; 23:4753–4772. [DOI] [PubMed] [Google Scholar]
- 52. Budini M., Romano V., Quadri Z., Buratti E., Baralle F.E.. TDP-43 loss of cellular function through aggregation requires additional structural determinants beyond its C-terminal Q/N prion-like domain. Hum. Mol. Genet. 2015; 24:9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Romano V., Quadri Z., Baralle F.E., Buratti E.. The structural integrity of TDP-43 N-terminus is required for efficient aggregate entrapment and consequent loss of protein function. Prion. 2015; 9:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Prpar Mihevc S., Baralle M., Buratti E., Rogelj B.. TDP-43 aggregation mirrors TDP-43 knockdown, affecting the expression levels of a common set of proteins. Sci. Rep. 2016; 6:33996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bakay M., Wang Z., Melcon G., Schiltz L., Xuan J., Zhao P., Sartorelli V., Seo J., Pegoraro E., Angelini C. et al. Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb-MyoD pathways in muscle regeneration. Brain. 2006; 129:996–1013. [DOI] [PubMed] [Google Scholar]
- 56. Dadgar S., Wang Z., Johnston H., Kesari A., Nagaraju K., Chen Y.W., Hill D.A., Partridge T.A., Giri M., Freishtat R.J. et al. Asynchronous remodeling is a driver of failed regeneration in Duchenne muscular dystrophy. J. Cell Biol. 2014; 207:139–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Anko M.L., Neugebauer K.M.. RNA-protein interactions in vivo: global gets specific. Trends Biochem. Sci. 2012; 37:255–262. [DOI] [PubMed] [Google Scholar]
- 58. Jankowsky E., Harris M.E.. Specificity and nonspecificity in RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 2015; 16:533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dormann D., Madl T., Valori C.F., Bentmann E., Tahirovic S., Abou-Ajram C., Kremmer E., Ansorge O., Mackenzie I.R., Neumann M. et al. Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J. 2012; 31:4258–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cohen T.J., Hwang A.W., Restrepo C.R., Yuan C.X., Trojanowski J.Q., Lee V.M.. An acetylation switch controls TDP-43 function and aggregation propensity. Nat. Commun. 2015; 6:5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liachko N.F., Guthrie C.R., Kraemer B.C.. Phosphorylation promotes neurotoxicity in a Caenorhabditis elegans model of TDP-43 proteinopathy. J. Neurosci. 2010; 30:16208–16219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ratti A., Buratti E.. Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J. Neurochem. 2016; 1:95–111. [DOI] [PubMed] [Google Scholar]
- 63. Kalsotra A., Cooper T.A.. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011; 12:715–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Venables J.P., Koh C.S., Froehlich U., Lapointe E., Couture S., Inkel L., Bramard A., Paquet E.R., Watier V., Durand M. et al. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol Cell Biol. 2008; 28:6033–6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dombert B., Sivadasan R., Simon C.M., Jablonka S., Sendtner M.. Presynaptic localization of Smn and hnRNP R in axon terminals of embryonic and postnatal mouse motoneurons. PLoS One. 2014; 9:e110846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Alami N.H., Smith R.B., Carrasco M.A., Williams L.A., Winborn C.S., Han S.S., Kiskinis E., Winborn B., Freibaum B.D., Kanagaraj A. et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014; 81:536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Narayanan R.K., Mangelsdorf M., Panwar A., Butler T.J., Noakes P.G., Wallace R.H.. Identification of RNA bound to the TDP-43 ribonucleoprotein complex in the adult mouse brain. Amyotroph. Lateral Scler. Frontotemporal Degener. 2013; 14:252–260. [DOI] [PubMed] [Google Scholar]
- 68. Moisse K., Volkening K., Leystra-Lantz C., Welch I., Hill T., Strong M.J.. Divergent patterns of cytosolic TDP-43 and neuronal progranulin expression following axotomy: implications for TDP-43 in the physiological response to neuronal injury. Brain Res. 2009; 1249:202–211. [DOI] [PubMed] [Google Scholar]
- 69. Halstead J.M., Lin Y.Q., Durraine L., Hamilton R.S., Ball G., Neely G.G., Bellen H.J., Davis I.. Syncrip/hnRNP Q influences synaptic transmission and regulates BMP signaling at the Drosophila neuromuscular synapse. Biol. Open. 2014; 3:839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McDermott S.M., Yang L., Halstead J.M., Hamilton R.S., Meignin C., Davis I.. Drosophila Syncrip modulates the expression of mRNAs encoding key synaptic proteins required for morphology at the neuromuscular junction. RNA. 2014; 20:1593–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tsui S., Dai T., Roettger S., Schempp W., Salido E.C., Yen P.H.. Identification of two novel proteins that interact with germ-cell-specific RNA-binding proteins DAZ and DAZL1. Genomics. 2000; 65:266–273. [DOI] [PubMed] [Google Scholar]
- 72. Choudhury R., Roy S.G., Tsai Y.S., Tripathy A., Graves L.M., Wang Z.. The splicing activator DAZAP1 integrates splicing control into MEK/Erk-regulated cell proliferation and migration. Nat. Commun. 2014; 5:3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Smith R.W., Anderson R.C., Smith J.W., Brook M., Richardson W.A., Gray N.K.. DAZAP1, an RNA-binding protein required for development and spermatogenesis, can regulate mRNA translation. RNA. 2011; 17:1282–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang Y., Ma M., Xiao X., Wang Z.. Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules. Nat. Struct. Mol. Biol. 2012; 19:1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lou H., Neugebauer K.M., Gagel R.F., Berget S.M.. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol. Cell. Biol. 1998; 18:4977–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Liu X., Li D., Zhang W., Guo M., Zhan Q.. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J. 2012; 31:4415–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Guo F., Jiao F., Song Z., Li S., Liu B., Yang H., Zhou Q., Li Z.. Regulation of MALAT1 expression by TDP43 controls the migration and invasion of non-small cell lung cancer cells in vitro. Biochem. Biophys. Res. Commun. 2015; 465:293–298. [DOI] [PubMed] [Google Scholar]
- 78. Nishimoto Y., Nakagawa S., Hirose T., Okano H.J., Takao M., Shibata S., Suyama S., Kuwako K., Imai T., Murayama S. et al. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain. 2013; 6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Li W., Jin Y., Prazak L., Hammell M., Dubnau J.. Transposable elements in TDP-43-mediated neurodegenerative disorders. PLoS One. 2012; 7:e44099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wu H., Yin Q.F., Luo Z., Yao R.W., Zheng C.C., Zhang J., Xiang J.F., Yang L., Chen L.L.. Unusual processing generates SPA LncRNAs that sequester multiple RNA binding proteins. Mol. Cell. 2016; 64:534–548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.