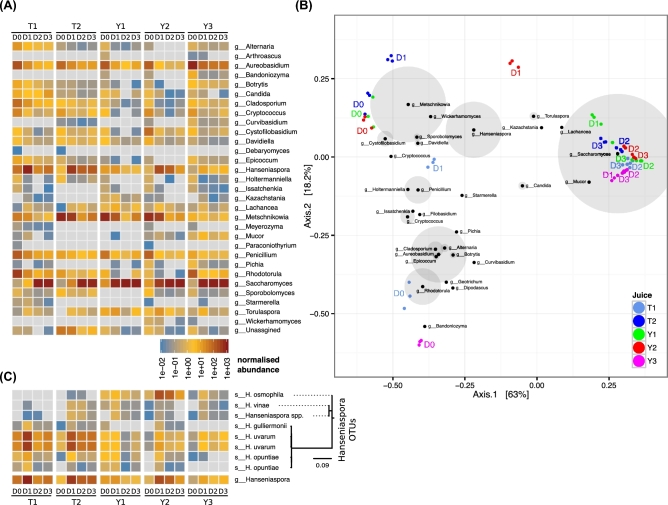
Figure 1:
ITS amplicon abundance of uninoculated ferments. (A) Laboratory-scale ferments analyzing 4 fermentation time points in 5 different musts in triplicate. ITS sequences are grouped by genus and are colored-coded by their normalized abundance (reads per thousand reads). (B) Dissimilarity analysis of ITS-amplicon abundance. Triplicate samples from each time point were subjected to Bray-Curtis dissimilarity analysis. The PCA weightings of the top 30 genera are overlaid on the plot, with the size of the gray circles around each node proportional to the total abundance of each genus across all samples (no shading for nodes >5000 counts). (C) Species-level ITS assignment for the genus Hanseniaspora. The individual abundance measurements for the 8 OTUs that comprise the g__Hanseniaspora category are shown, grouped by phylogenetic distance. Results are color-coded according to normalized abundance (reads per thousand reads).