Abstract
Approximately 15% of non-small cell lung cancer cases are associated with a mutation in the epidermal growth factor receptor (EGFR) gene, which plays a critical role in tumor progression. With the goal of treating mutated EGFR-mediated lung cancer, we demonstrate the use of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9) system to discriminate between the oncogenic mutant and wild-type EGFR alleles and eliminate the carcinogenic mutant EGFR allele with high accuracy. We targeted an EGFR oncogene harboring a single-nucleotide missense mutation (CTG > CGG) that generates a protospacer-adjacent motif sequence recognized by the CRISPR/Cas9 derived from Streptococcus pyogenes. Co-delivery of Cas9 and an EGFR mutation-specific single-guide RNA via adenovirus resulted in precise disruption at the oncogenic mutation site with high specificity. Furthermore, this CRISPR/Cas9-mediated mutant allele disruption led to significantly enhanced cancer cell killing and reduced tumor size in a xenograft mouse model of human lung cancer. Taken together, these results indicate that targeting an oncogenic mutation using CRISPR/Cas9 offers a powerful surgical strategy to disrupt oncogenic mutations to treat cancers; similar strategies could be used to treat other mutation-associated diseases.
INTRODUCTION
Lung cancer is one of the most commonly diagnosed malignant types of cancer. It also accounts for 42% of overall cancer mortality (1) (https://seer.cancer.gov/statfacts/html/lungb.html). Despite extensive research, the prognosis of lung cancer treated with conventional chemical and radiological therapies is poor. Overall, the 5-year survival rate for patients with metastatic lung cancer is <15% (2). To this end, improvement in this area requires the development of highly specific targeted therapeutics.
Several oncogenic mutations that cause lung cancer have been identified; thus, targeted cancer treatment has become a reality. Aberrant epidermal growth factor receptor (EGFR) expression has been recognized as a key driver of cellular proliferation, differentiation, migration and angiogenesis, ultimately contributing to lung cancer oncogenesis (3–9). Approximately 15% of non-small cell lung cancer (NSCLC) cases, which account for 85% of lung cancers, are associated with mutations in the EGFR gene. These mutations play critical roles in tumor progression. About 90% of EGFR activation mutations involve either deletions in exon 19 or a missense mutation in exon 21 that substitutes an arginine for a leucine (L858R) in the tyrosine kinase domain (10). This classical activating L858R mutation in exon 21 accounts for ∼40% of all EGFR mutations (11). Targeting mutant EGFR has been key in advancing lung cancer research treatment and improving patient outcomes.
EGFR inhibitors such as gefitinib and erlotinib have been developed to inhibit the tyrosine kinase activity of mutated EGFR and are currently the first-line therapies for treating lung cancer (12,13). However, this targeted therapy often fails because of secondary genetic mutations that arise after repeated drug exposure and confer resistance to the targeted drug. Indeed, most lung cancers in EGFR targeted drug treatment groups have been reported to acquire resistance within 2 years, thus requiring second- and third-generation drugs (14). To transcend the current limitations of targeted cancer treatments, an alternative strategy would be needed to eliminate cancer-causing mutations at the DNA level using targeted nucleases.
Several types of nucleases have been developed for targeted gene editing. These include RNA-guided programmable nucleases, which have been repurposed from the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9) adaptive immune system in eubacteria and archaea that protects against invading genetic elements (15–21). For gene editing, Cas9, derived from Streptococcus pyogenes (SpCas9), is complexed with target-derived CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) or with a single-guide RNA (sgRNA) composed of essential portions of crRNA and tracrRNA that are physically linked for ease of use. SpCas9 recognizes the 5΄-NGG-3΄ protospacer-adjacent motif (PAM) at a target site, whereas its associated sgRNA or crRNA, which contains 19 or 20 nucleotides (nt) of complementary target sequence, hybridizes with the target DNA sequence upstream of the PAM via Watson–Crick base pairing. As a result, Cas9 induces a DNA double-strand break (DSB) 3 nt upstream of the PAM. Repair of the DSB by error-prone non-homologous end-joining or microhomology-mediated end joining gives rise to mutations at the cleavage site (22).
Several studies have explored the use of CRISPR/Cas9 for the direct disruption of abnormally activated oncogenes or the restoration of inactivated tumor suppressor genes (23–31). To express Cas9 or sgRNA specifically in cancer cells, various strategies have been reported, such as the use of cancer-specific promoters, drug-induced sgRNA vector systems, AND gate genetic circuits regulated by a cancer-specific promoter and aptamer-liposome systems (24,29,30). However, without a system that selectively disrupts mutated oncogenes, but not the corresponding wild-type proto-oncogenes, the value of these approaches will be limited. The indiscriminate destruction of both alleles can cause serious side effects because the proto-oncogenes themselves have many biologically important roles. To overcome this limitation, an allele-specific gene editing approach has been reported to target a mutant allele (32–37). Yet there is no report in cancer treatment, demonstrating that allele specific oncogene disruption in vivo leads to tumor growth inhibition.
To address the issues of allelic heterogeneity in cancer treatment with respect to gene editing, here we demonstrate a more precise approach to selectively target the mutant allele. In this study, we targeted a single nucleotide missense mutation (CTG > CGG) in EGFR, which results in one of the major EGFR activation mutations (L858R) in NSCLC. We hypothesized that the mutant EGFR allele could be selectively destroyed by exploiting the fact that this missense mutation generates a PAM (5΄-NGG-3΄) in the genome. We found that delivery of an oncogenic mutant-specific CRISPR/Cas9 via adenovirus (Ad) results in the cleavage and disruption of the mutant EGFR allele with high accuracy, leading to a significant reduction of tumor growth in a mouse lung cancer xenograft model. To best of our knowledge, our study is first to demonstrate that CRISPR/Cas9-mediated knockout of L858R mutation in EGFR-overexpressing lung cancers can lead to tumor growth inhibition. This approach has potential for targeting cancers in which EGFR is often mutated or drug resistant and potential advantages for personalized cancer treatment relative to conventional therapeutics.
MATERIALS AND METHODS
Cell lines and cell culture
All cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Gibco BRL), L-glutamine (2 mM), penicillin (100 IU/ml), and streptomycin (50 μg/ml). HEK293 (human embryonic kidney cell line expressing the Ad E1 region) and non-small lung cancer cell lines (H1975, A549) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA).
Plasmid construction
To generate an Ad construct that expresses sgRNA targeting mutated EGFR (sgEGFR) in the E3 region of E1-deleted Ad, the U6 promoter and a GX19 sgRNA (5΄-CAAGATCACAGATTTTGGG-3΄) that hybridizes with EGFR exon 21 were synthesized and subcloned into a pSP72-E3, Ad E3 shuttle vector (38), yielding a pSP72-E3/sgEGFR shuttle vector. pSP72-E3/sgEGFR was linearized with XmnI and co-transformed into Escherichia coli BJ5183 along with SpeI-digested dE1-RGD, used as an empty vector control (39) for homologous recombination, generating a dE1-RGD/SgEGFR (Ad/sgEGFR) vector.
Adenovirus production
All viruses were propagated in HEK293 cells, and the purification, titration, and quality analysis of all Ads were performed as previously described (40,41). Viral particle (VP) numbers were calculated from measurements of optical density at 260 nm (OD260), where 1 absorbency unit is equivalent to 1012 VPs per milliliter. Infectious titers (plaque forming unit per milliliter) were determined by a limiting dilution assay on HEK293 cells. After viral generation, polymerase chain reaction (PCR) amplification and DNA sequencing were performed to verify viral genome structures. A Cas9-expressing Ad vector was purchased (Ad/Cas9; Vector Biolabs, Malvern, PA, USA).
Transfection and genomic DNA extraction
SpCas9 plasmid (500 ng) and sgRNA plasmid (1500 ng) were transfected into cells (1 × 105) with lipofectamine 2000 (Invitrogen). After 48 h of transfection, genomic DNA was isolated using a DNeasy Blood & Tissue kit (Qiagen). For DNA extraction from tumors, a tumor section was cut into eight–nine pieces and homogenized using tungsten carbide beads (3 mm; Qiagen) and a TissueLyser II (Qiagen). Then, genomic DNA was isolated using a DNeasy Blood & Tissue kit (Qiagen).
RNA extraction and qPCR
Total RNA was isolated from H1975 cells using TRIzol (Sigma) according to the manufacturer's protocol. One microgram of RNA was then reverse transcribed using a High-Capacity cDNA reverse transcription kit (Applied Biosystems). Quantitative PCR was performed using Fast Sybr Green Master mix (Applied Biosystems) with the following primers: human EGFR, 5΄-GTGACCGTTTGGGAGTTGATGA-3΄ (forward), 5΄-GGCTGAGGGAGGCGTTCTC-3΄ (reverse).
Mutation analysis
On-target loci were amplified for targeted deep sequencing. Deep sequencing libraries were generated by PCR. TruSeq HT Dual Index primers were used to label each sample. Pooled libraries were subjected to paired-end sequencing (LAS, Inc.). Wild-type and mutated sequences were discriminated based on the presence of the single nucleotide missense mutation in the EGFR allele. Indels located 3-bp upstream of the PAM were considered to be the mutations induced by SpCas9. The indel frequency was calculated as described (42). PCR was performed with the following primers; human EGFR, 5΄-CGGATGCAGAGCTTCTTCCCATG-3΄ (forward), 5΄-AAGGCAGCCT GGTCCCTGGT-3΄ (Reverse).
MTT assays
To evaluate the cytopathic effect of CRISPR/Cas9, 2-5 × 104 cells were plated onto a 24-well plate at about 70% confluence and then transduced with a combination of Ad/sgEGFR and empty Ad vector (Ad/sgEGFR), a combination of Ad/Cas9 and empty Ad vector (Ad/Cas9) or a combination of Ad/sgEGFR and Ad/Cas9 (Ad/sgEGFR + Ad/Cas9) at a multiplicity of infection (MOI) of 10-50. The killing effect of these treatments was monitored daily under a microscope. At 48 h post-treatment, a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was carried out as previously described (43). In brief, 200 μl of MTT (Sigma) in phosphate buffered saline (PBS; 2 mg/ml) was added to each well. After 4 h incubation at 37°C, the supernatant was discarded and the precipitate was dissolved in 1 mL of dimethyl sulfoxide. Plates were then read on a microplate reader at 540 nm.
Generation, treatment and analysis of tumor xenografted mice
Tumors were implanted in the abdomens of 5- to 6-week old male nude mice by subcutaneous injection of H1975 or A549 cells (1 × 107 cells in 100 μl of Hank's balanced salt solution (HBSS; Gibco BRL). When tumor volumes reached a range of 70–100 mm3 within 1–2 weeks of tumor cell injection, animals were randomly assigned to one of three groups to receive PBS, Ad/Cas9 + empty Ad vector or Ad/sgEGFR + Ad/Cas9 (seven or eight mice per group) when they were 6–8 weeks old. The first day of treatment was designated as day 1. Ad or PBS was administered intratumorally (a total of 5 × 1010 VPs in 30 μL of PBS) on days 1, 3 and 5. Tumor growth inhibition was assessed every other day by measuring the length (L) and width (w) of the tumor and determining the tumor volume, with following formula: volume = 0.523L(w)2. The percentage of surviving mice was determined by monitoring tumor growth-related events (tumor size > 800 mm3) over a period of inspection.
Histology and immunohistochemistry
Representative sections were stained with Hematoxylin and eosin (H & E) and then examined by light microscopy (Carl Zeiss Inc.). For immunohistochemistry, slides were deparaffinized in xylene and then processed as described earlier (43–46). H1975 tumor tissue sections were incubated at 4°C overnight with mouse anti-proliferating cell nuclear antigen (PCNA: DAKO, Glostrup, Denmark) or goat anti-EGFR primary antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), rinsed and then incubated for 30 min with a biotinylated anti-mouse (Cell Signaling Technology, Danvers, MA, USA) or anti-goat secondary antibody (DAKO), respectively. Sections were then rinsed in buffer and incubated with streptavidin-peroxidase. All slides were counterstained with Meyer's hematoxylin (Sigma). The results from PCNA or EGFR staining were semi-quantitatively analyzed by MetaMorph® image analysis software (Universal Image Corp., Buckinghamshire, UK).
Statistical analysis
No statistical methods were used to predetermine sample sizes for in vitro or in vivo experiments. All group results are expressed as mean ± SEM if not otherwise indicated. Comparisons between groups were made using the one-way ANOVA with Tukey's post-hoc tests for multiple groups. Statistical significance as compared to untreated controls is denoted with * (P < 0.05), ** (P < 0.01), *** (P < 0.001) in the figures and figure legends. Statistical analysis was performed in Graph Pad PRISM 5.
Data availability
The deep sequencing data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRX2866619. We have included the read depth information of the deep sequencing data in the Supplementary Tables S3–5.
RESULTS
Precise editing of an oncogenic mutant allele via CRISPR/Cas9
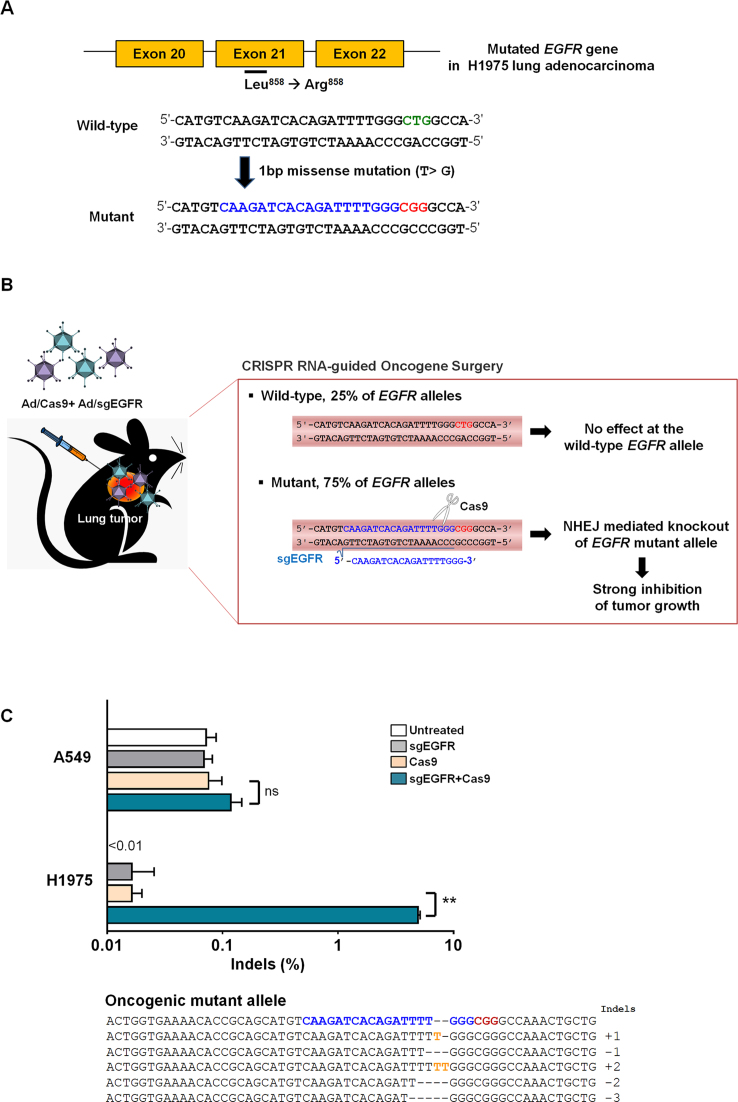
First, we designed a CRISPR/Cas9 nuclease to discriminate between the oncogenic mutant and wild-type EGFR alleles (Figure 1A). The EGFR mutant gene harbors a single nucleotide missense mutation (CTG > CGG) in EGFR exon 21, generating a PAM sequence recognized by SpCas9. Here, we generated a GX19 sgRNA that hybridizes with the EGFR mutant allele (sgEGFR). This sgRNA includes an extra guanine (G) at the 5΄-end for transcription under the control of the U6 promoter; the remaining 19 nucleotides (X19) hybridize with a 19 nt target DNA sequence upstream of the newly generated PAM (5΄-CGG-3΄) sequence in the oncogene allele.
Figure 1.
Oncogenic mutant-specific Cas9. (A) A PAM sequence (red) is generated by a single nucleotide missense mutation in the EGFR gene. The corresponding wild-type sequence is shown in green. The sgRNA target sequence is shown in blue. (B) Diagram of selective cancer cell killing by cleavage of the oncogenic mutant allele 3-bp upstream of the PAM. (C) Mutation frequencies at the EGFR target site in H1975 cells or A549 cells co-transfected with Ad/sgEGFR and Ad/Cas9 48 h post-transfection. Mismatched nucleotides are shown in yellow and PAM sequences in red. The sgRNA target sequence is shown in blue. The column on the right indicates the number of inserted or deleted bases. Bars represent the mean ± S.E.M (n = 3). **P < 0.01, ns: not significant.
Then, we examined the activity of the mutant allele-specific Cas9 nuclease in H1975 cells derived from a patient with NSCLC harboring the EGFR mutant allele (Figure 1B). We found that the H1975 cells contained the mutant and wild-type alleles at a ratio of 3:1. To confirm oncogenic mutant-specific DNA cleavage by Cas9, we transfected plasmids encoding Cas9 and sgEGFR into H1975 cells. A549 lung adenocarcinoma cells, which do not contain the mutant allele, were utilized as a negative control (Figure 1C). Cas9-induced small insertions/deletions (indels) were detected in the EGFR mutant allele in H1975 cells with a frequency of 3.6% (±0.1%) 2 days post-transfection, whereas no mutations were detectably induced in the wild-type allele in either H1975 or A549 cells.
Enhanced cancer cell killing mediated by oncogenic mutant-specific genome editing
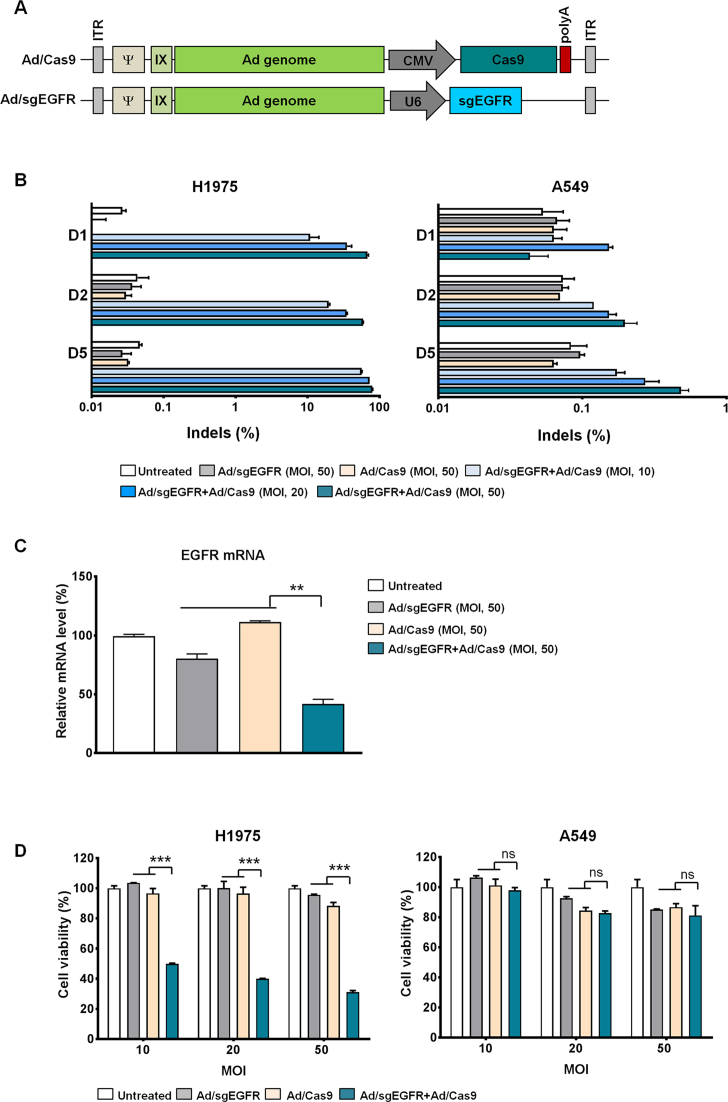
For efficient delivery of CRISPR/Cas9 to cancer cells, we utilized a replication-incompetent Ad vector. First, we constructed Cas9-expressing Ad (Ad/Cas9) and sgEGFR-expressing Ad (Ad/sgEGFR) (Figure 2A). Then, H1975 and A549 cells were transduced with a combination of Ad/sgEGFR and Ad/Cas9 at MOIs of 10, 20 and 50 (Figure 2B). Indels accumulated at the target site in a time- and dose-dependent manner with a frequency of 11% (±4%), 20% (±0.7%) and 56% (±0.7%) 1, 2 and 5 days post treatment, respectively, at an MOI of 10. At 5 days post-infection, indels were detected with a frequency of 56% (±0.7%), 73% (±0.6%) and 79% (±1.2%) in H1975 cells at MOIs of 10, 20 and 50, respectively, resulting in robust knock-down of the EGFR mRNA level (Figure 2C). Indels were minimally detected at the wild-type EGFR allele in H1975 cells, with a frequency of 2.8% (±0.1%), 4.1% (±0.1%) and 4.4% (±0.2%) at MOIs of 10, 20 and 50, respectively, at 5 days post transduction (Supplementary Figure S1). Indels were not detected in A549 cells (wild-type EGFR allele); frequencies were below 0.5% at an MOI of 50 (Figure 2B).
Figure 2.
Specific excision of the oncogenic EGFR allele induces cytopathic effect. (A) Representative diagram of E1-deleted adenovirus type 5 encoding either SpCas9 or sgRNA targeting the EGFR gene. (B) Mutation frequencies at the EGFR target site in H1975 and A549 cells were examined via targeted deep sequencing day 1, 2 and 5 after co-transduction of Ad/sgEGFR and Ad/Cas9 with MOIs of 10, 20 and 50. (C) EGFR mRNA levels, normalized with an 18S rRNA internal control, were determined day 2 after co-infection of H1975 cells with Ad/sgEGFR and Ad/Cas9. (D) CRISPR/Cas9-mediated cell killing efficacy in H1975 or A549 cells. Cells were treated with Ad/sgEGFR, Ad/Cas9, or Ad/sgEGFR + Ad/Cas9; day 2 post-treatment, an MTT assay was performed. Bars represent the mean ± S.E.M (n = 3). * (P < 0.05), ** (P < 0.01), *** (P < 0.001), ns: not significant.
To investigate whether targeted cleavage and disruption of the EGFR mutant allele could induce death of cancer cells, we conducted an MTT cell proliferation assay (Figure 2D and Supplementary Figure S2). Co-treatment with Ad/sgEGFR and Ad/Cas9 led significant cancer cell killing of H1975 cells in a dose-dependent manner, indicating that disruption of the EGFR mutant allele was essential for cancer cell killing effect. In contrast, co-treatment with Ad/sgEGFR and Ad/Cas9 did not significantly affect the viability of A549 cells carrying the wild-type EGFR allele. Collectively, these results suggest that mutant allele-specific Cas9 can efficiently distinguish the EGFR mutant allele from the wild-type allele, leading to targeted oncogene disruption and cancer cell death.
Targeted mutant allele disruption in a murine xenograft model of human lung adenocarcinoma
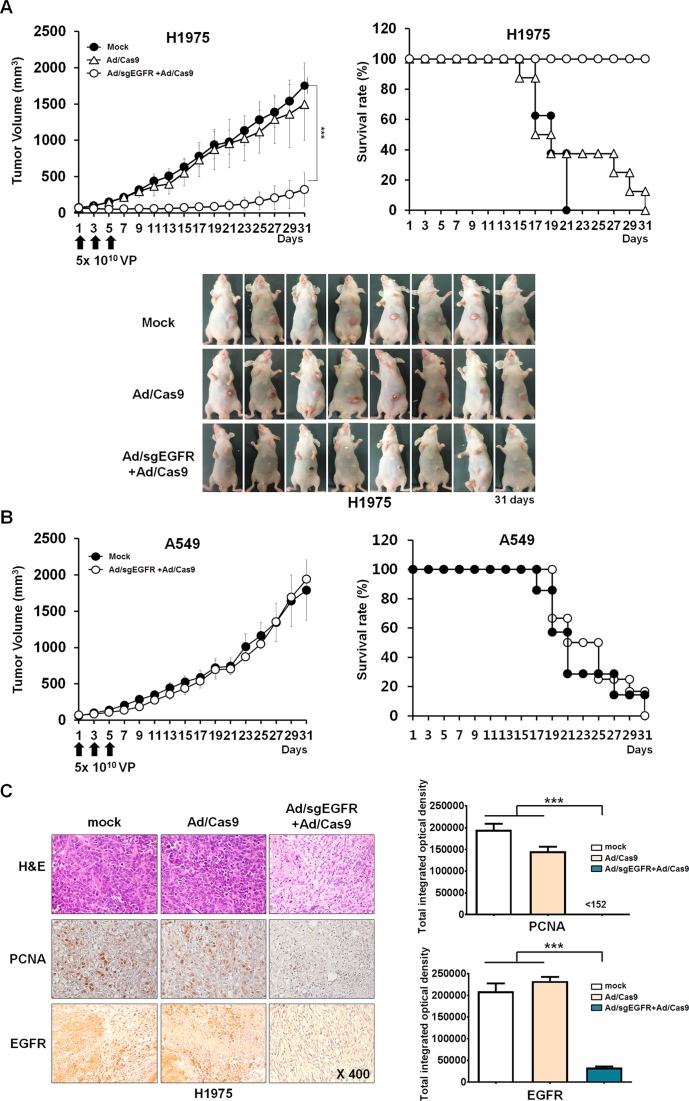
To investigate oncogenic mutant allele-specific cleavage mediated by CRISPR/Cas9, we used xenograft mice implanted with either H1975 or A549 cancer cells. When subcutaneously implanted tumors reached a volume of 80–100 mm3, we intratumorally injected a mixture of Ad/sgEGFR and Ad/Cas9 at a dose of 5 × 1010 VPs every other day for a total of three times. As negative controls, we injected either a mixture of Ad/Cas9 with Ad/empty vector or PBS alone. Co-treatment with Ad/sgEGFR and Ad/Cas9 resulted in indels at frequencies of 50% (±5.3%), 40% (±2.8%) and 35% (±4.1%) at the EGFR mutant allele in H1975-derived tumors at day 7, 9 and 11, respectively, after the first injection (Figure 3A). Interestingly, indels were not detectable in the H1975 tumor xenograft 31 days post-injection, suggesting that disruption of the mutant EGFR allele contributes to cancer cell killing. Of note, indels at the wild-type EGFR allele were minimally detected in H1975 tumors, with a frequency of 2.6% (±0.4%), 2.3% (±0.2%) and 0.9% (±0.1%) at day 7, 9 and 11 post-injection, respectively (Figure 3A). No indels were detectably induced at the wild-type EGFR mutant allele in A549-derived tumors (Figure 3B). These results suggest that Ad-mediated EGFR oncogene-specific Cas9 expression can disrupt the EGFR mutant allele in H1975 cells with high accuracy in vivo.
Figure 3.
Selective oncogene disruption using Ad/sgEGFR and Ad/Cas9 in vivo. Indels at the wild-type (gray bar) and mutant EGFR alleles (blue bar) in (A) H1975 or (B) A549 tumor xenografts co-injected with Ad/sgEGFR and Ad/Cas9 at day 7, 9, 11 and 31 after the first injection. Tumor-bearing mice were given intratumoral injections of PBS (mock) or 5 × 1010 Ad VPs (Ad/Cas9 + Ad/empty vector or Ad/sgEGFR + Ad/Cas9) on days 1, 3 and 5. Values represent the mean ± S.E.M. (n= 3 to 4). **P < 0.01. ns: not significant. (C) No off-target indels were detectably induced at eight homologous sites that differed from the on-target sites by up to 3 nt in the human genome. Mismatched nucleotides are shown in blue and PAM sequences in red, On; on-target site, OT; off-target site. Red arrow indicates cleavage position within the 19-bp target sequences. Error bar indicates S.E.M. (n = 3 to 4).
We next investigated whether the mutant allele-specific nuclease had off-target effects. Off-target nuclease activities were measured by targeted deep sequencing at 17 potential off-target sites, identified using CAS-OFFinder algorithm (http://www.rgenome.net/cas-offinder/) that differed from the on-target site by up to 3 nt in the human genome. No indels were detectably induced at these sites in Ad/sgEGFR and Ad/Cas9 co-treated H1975 tumors 7 days after the first injection (Figure 3C and Supplementary Table S1).
Rapid tumor regression and prolonged survival rate due to EGFR mutant allele disruption
We then assessed the therapeutic efficacy of Ad-CRISPR/Cas9 in tumor xenograft mice implanted with H1975 or A549 cells to investigate whether targeted disruption of the oncogenic mutant allele using Cas9 could be translated to tumor regression (Figure 4A). Co-treatment with Ad/sgEGFR and Ad/Cas9 led to significant tumor growth inhibition, resulting in an average tumor size of 324.1 (± 83.4) mm3 (P < 0.001). Tumor volumes in control groups treated with either PBS or Ad/Cas9 with Ad/empty vector increased over time and reached an average size of 1755.5 (±111.3) mm3 or 1496.5 (±177.3) mm3, respectively, 31 days after the treatment (Figure 4A). Thus, the EGFR mutation-specific Cas9 reduced tumor size by 81.5% and 78.3%, compared to the PBS and Ad/Cas9 treated controls, respectively.
Figure 4.
Antitumor effect and survival benefit of adenoviral delivery of CRISPR/Cas9 to tumor xenograft models. (A) H1975 tumor-bearing mice were given intratumoral injections of PBS or 5 × 1010 VPs (Ad/Cas9 + Ad/empty vector or Ad/sgEGFR and Ad/Cas9) on days 1, 3 and 5. Tumor growth was monitored every other day until 31 days post injection. Values represent the mean ± S.E.M. for eight animals per group. ***P < 0.001 compared with the PBS and Ad/Cas9 treated groups. The percentage of surviving mice was determined by monitoring tumor growth-related events (tumor size < 800 mm3) over a time period. (B) A549 tumor-bearing mice were given intratumoral injections of PBS or 5 × 1010 Ad VPs (Ad/sgEGFR and Ad/Cas9) on days 1, 3 and 5. Tumor growth was monitored every other day until the end of the study. Bar represent the mean ± S.E.M. for eight animals per group. NS; not significant compared with the PBS treated groups. The percentage of surviving mice was determined by monitoring tumor growth-related events (tumor size < 800 mm3) over a time period. (C) PBS, Ad/Cas9 + Ad/empty vector or Ad/sgEGFR and Ad/Cas9 was injected on days 1, 3 and 5 into established H1975 tumors in nude mice. Tumors were harvested on day 7 for histological analysis. H & E staining and immunohistochemical staining of PCNA and EGFR were performed on tumor sections from each group of mice.
The survival rate was also significantly increased in animals co-treated with Ad/sgEGFR and Ad/Cas9 compared to control groups treated with either Ad/Cas9 or PBS alone. By day 31 following treatment in H1975 tumor-bearing mice, all animals in the Ad/sgEGFR and Ad/Cas9 co-treated group were still viable, whereas all animals in control groups treated with either Ad/Cas9 or PBS were dead (P < 0.001). As expected, there was no significant difference in tumor size between CRISPR/Cas9 and PBS treated mice implanted with A549 cells harboring the wild-type EGFR allele (Figure 4B). Only 0% or 14.3% of the animals were viable in the Ad/sgEGFR and Ad/Cas9 co-treated or PBS control groups, respectively (P < 0.001). Throughout the course of the study, no signs of systemic toxicity, such as diarrhea, loss of weight or cachexia, were observed. Taken together, these results suggest that highly efficient and mutant allele-specific CRISPR/Cas9-mediated precise excision contributes to significant tumor regression, leading to prolonged survival in vivo.
Antitumor efficacy of oncogenic mutant-specific Cas9
The antitumor efficacy of oncogenic mutant-specific CRISPR/Cas9 was further investigated by histological and immunohistochemical analysis (Figure 4C). H & E staining revealed a markedly reduced number of viable tumor cells and more extensive necrotic regions in Ad/sgEGFR and Ad/Cas9 co-treated tumors than the Ad/Cas9 or PBS treated control groups. There was no noticeable immune cell infiltration in all tested tumor tissues. Moreover, Ad/sgEGFR and Ad/Cas9 co-treated tumor tissues exhibited markedly lower levels of PCNA compared with Ad/Cas9 or PBS treated tissues (P < 0.001). Importantly, tumor tissues from mice co-treated with Ad/sgEGFR and Ad/Cas9 showed a 1284- or 958-fold reduction in EGFR protein expression compared with those treated with either PBS or Ad/Cas9, respectively, implying that Ad-mediated CRISPR/Cas9 can efficiently target and attenuate EGFR mutant protein expression, leading to decreased tumor cell proliferation and enhanced cell death in H1975 tumors (P < 0.001). Taken together, these data indicate that the Ad-mediated CRISPR/Cas9 system provides strong inhibitory effects on tumor growth, resulting in an increased survival rate in EGFR-mutated tumor-bearing mice.
DISCUSSION
CRISPR/Cas9-mediated genome editing is a powerful new technique that can generate targeted genetic modifications in eukaryotic cells and organisms. This system has been utilized to correct disease-associated genetic defects and to inactivate disease-causing wild-type genes (47–50). Several studies that aim to remove oncogenes using the CRISPR/Cas9 system are underway and may have therapeutic potential (23–31). Huang et al. demonstrated that CRISPR/Cas9 system targeting EGFR exon 17 can inhibit NF-κB activation in EGFRwt/vIII glioma cells (25). However, these approaches still provide limited specificity, with indiscriminate disruption of both the wild-type proto-oncogene and mutant oncogene alleles, emphasizing the urgent need for more precise targeted therapy.
In this study, we demonstrated a novel approach, a mutated oncogene surgery, to destroy the predominant oncogenic EGFR mutation in lung cancer using CRISPR/Cas9. Delivery of EGFR mutation-specific CRISPR/Cas9 via Ad into EGFR mutant-bearing tumors resulted in precise excision at the oncogenic mutation site, demonstrating target specificity of the approach. We showed mutant allele-specific gene disruption in H1975 tumors in vivo. Disruption of the carcinogenic EGFR mutation (L858R) in H1975 tumors resulted in potent inhibition of cancer cell proliferation, followed by rapid tumor regression. Yet, we cannot rule out the possibility of disruption of neighboring cells which lack the PAM generating mutant allele. However, indels and its cytotoxicity associated with CRISPR/Cas9 were not detectably induced in A549 cells which harbor wild-type EGFR alleles only. These results are in line with in vivo data that tumor growth inhibition was not shown in Ad/sgEGFR and Ad/Cas9 co-treated A549 tumors, demonstrating the specificity of the system. This approach could also be extended to disrupt other cancer-driving mutations that generate PAM sequences recognized by SpCas9 (Supplementary Table S2) for multiple gene editing in cancers characterized by frequent mutation heterogeneity. In this study, we showed the specificity of CRISPR/Cas9 for targeting a mutant allele that generates a 5΄-NGG-3΄ PAM sequence. Cas9 variants with altered PAM recognition can be employed to target other oncogenic mutations. Cong et al. has previously reported that single-base mismatches in the PAM-proximal seed region abolished genomic cleavage by SpCas9 (17), suggesting that mutations in the seed sequence could also be targetable.
Mutated EGFR targeting CRISPR/Cas9 system induced indels with a frequency of 60–80% at the target site (L858R) in H1975 cells (Figure 2B), followed by 60% decrease in EGFR mRNA expression level (Figure 2C). We found that H1975 cells harbor 25% wild-type alleles and 75% mutant alleles of the total allele composition (Supplementary Table S5). Based on this, we speculate that indels at frequencies of 80% in mutant alleles led to 60% decrease in total EGFR expression. Furthermore, there was a negative correlation between indels and viability of H1975 cells in vitro (Figure 2B and D), suggesting that disruption of the mutant EGFR alleles leads to cell death. These results are in line with other reports demonstrating that L858R activating mutation promotes malignant pleural tumor formation and polypoid (51). Thus, the inhibition of mutated EGFR (L858R) using gefitinib leads to the death of NSCLC (52,53).
Taken together, CRISPR/Cas9-mediated death of mutated EGFR-expressing cancer cells and subsequent change in genetic composition of cancer cell population can attenuate malignancy of cancer. Similar results were obtained in vivo in which a positive correlation between indels in the mutated EGFR gene and tumor growth inhibition was observed (Figures 3 and 4). Specifically, downregulation of EGFR protein levels by CRISPR/Cas9-mediated disruption of L858R mutation resulted in attenuated tumor burden and improved survival of tumor-bearing mice. Tumors treated with mutated EGFR gene targeting CRISPR/Cas9 showed extensive necrosis and decreased proliferation (Figure 4C), indicating that decreased mutant allele composition in a H1975 tumor can efficiently inhibit tumor cell proliferation and EGFR signaling pathway. Consistent with these results, others have reported that ablation of oncogenic mutations using the CRISPR/Cas9 system can significantly improve the survival rate of tumor-bearing mice (31). Taken together, it indicates that the CRISPR/Cas9 system can be a promising method of cancer treatment to ablate or correct cancer driving mutations.
Of interest, the in vivo data of Figure 3 shows a time-dependent reduction in indel rate with the promotion of tumor regrowth observed in tumors treated with CRISPR/Cas9 from day 21 and onward (Figure 4A). Specifically, tumor growth between day 1 and 21 averaged 1.89 mm3/day, whereas average tumor growth from day 21 to 31 was 22.1 mm3/day. One possible explanation for tumor regrowth is that a time-dependent decrease in indel frequency in CRISPR/Cas9-treated tumors may lead to re-proliferation of unedited mutant EGFR expressing cancer cells, ultimately leading to tumor regrowth. Based on these results, administrating second cycle of treatment at later time period might induce tumor regrowth inhibition.
The CRISPR/Cas9 system can be delivered efficiently to target tumors via the adenoviral vector system (47,54,55). In this work, we utilized a replication-incompetent Ad as a carrier to deliver CRISPR/Cas9 to lung cancer with high accuracy. Ad has several advantages for applications in cancer gene therapy: the ability to infect both dividing and non-dividing cells, a high level of gene expression, the production of high-titer stocks and no risk of insertional mutagenesis (56–58). We demonstrate improved knock-out efficiency of CRISPR/Cas9 at the oncogenic EGFR mutation by using Ad as a vehicle compared with plasmid-based delivery. The efficacy of this approach could be further enhanced by utilizing oncolytic Ad, a cancer-specific replicating Ad (59,60), to induce preferential and highly specific expression of Ad moieties at a high level in tumor cells. Furthermore, we have utilized immune-deficient animal model to establish human xenograft tumor model, thus immune response-mediated killing of cancer cells was excluded in this study. However, oncolytic Ad, Cas9 or sgRNA may induce an anti-tumor immune response (40,61–64), ultimately instigating antitumor immune response and inhibiting tumor growth. We believe that our system in immune-competent animal model may elicit more potent antitumor efficacy and evaluation of immune response mediated by this system should be explored in future studies. No off-target effect was detectably induced in Ad/sgEGFR and Ad/Cas9 co-treated H1975 tumors. However, the potential off-target effect of this system should be further investigated for a long period of time. The use of tissue- or tumor-specific promoters to regulate Cas9 protein expression may improve target specificity for selective disruption of cancer-driving mutant alleles.
Taken together, our results demonstrate that expression of mutant allele-specific CRISPR/Cas9 in conjunction with Ad-mediated delivery can be a powerful technique for precise disruption of oncogenic mutations and inhibition of tumor growth. Our results indicate that the use of the mutant allele-specific CRISPR/Cas9 could overcome the current limitations of conventional cancer therapies with their side effects caused by unselective cell killing. Application of this platform offers promise for targeted cancer therapy and treating other mutation-associated diseases.
ACCESSION NUMBER
The deep sequencing data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRX2866619.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institute for Basic Science [IBS-R021-D1 to J.-S.K.] and National Research Foundation of Korea [2015R1A2A1A13027811, 2016M3A9B5942352 to C-O Yun]. Funding for open access charge: Institute for Basic Science [IBS-R021-D1].
Conflict of interest statement. None declared.
REFERENCES
- 1. Siegel R.L., Miller K.D., Jemal A.. Cancer Statistics, 2017. CA Cancer J. Clin. 2017; 67:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Edwards B.K., Noone A.M., Mariotto A.B., Simard E.P., Boscoe F.P., Henley S.J., Jemal A., Cho H., Anderson R.N., Kohler B.A. et al. Annual Report to the Nation on the status of cancer, 1975–2010, featuring prevalence of comorbidity and impact on survival among persons with lung, colorectal, breast, or prostate cancer. Cancer. 2014; 120:1290–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ohsaki Y., Tanno S., Fujita Y., Toyoshima E., Fujiuchi S., Nishigaki Y., Ishida S., Nagase A., Miyokawa N., Hirata S. et al. Epidermal growth factor receptor expression correlates with poor prognosis in non-small cell lung cancer patients with p53 overexpression. Oncol. Rep. 2000; 7:603–607. [DOI] [PubMed] [Google Scholar]
- 4. Inamura K., Ninomiya H., Ishikawa Y., Matsubara O.. Is the epidermal growth factor receptor status in lung cancers reflected in clinicopathologic features. Arch. Pathol. Lab. Med. 2010; 134:66–72. [DOI] [PubMed] [Google Scholar]
- 5. Hynes N.E., Lane H.A.. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer. 2005; 5:341–354. [DOI] [PubMed] [Google Scholar]
- 6. Beljanski V., Hiscott J.. The use of oncolytic viruses to overcome lung cancer drug resistance. Curr. Opin. Virol. 2012; 2:629–635. [DOI] [PubMed] [Google Scholar]
- 7. Sandler A.B., Johnson D.H., Herbst R.S.. Anti-vascular endothelial growth factor monoclonals in non-small cell lung cancer. Clin. Cancer Res. 2004; 10:4258s–4262s. [DOI] [PubMed] [Google Scholar]
- 8. Giaccone G. Epidermal growth factor receptor inhibitors in the treatment of non-small-cell lung cancer. J. Clin. Oncol. 2005; 23:3235–3242. [DOI] [PubMed] [Google Scholar]
- 9. Besse B., Adjei A., Baas P., Meldgaard P., Nicolson M., Paz-Ares L., Reck M., Smit E.F., Syrigos K., Stahel R. et al. 2nd ESMO Consensus Conference on Lung Cancer: non-small-cell lung cancer first-line/second and further lines of treatment in advanced disease. Ann. Oncol. 2014; 25:1475–1484. [DOI] [PubMed] [Google Scholar]
- 10. Fang S., Wang Z.. EGFR mutations as a prognostic and predictive marker in non-small-cell lung cancer. Drug Des. Dev. Ther. 2014; 8:1595–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fukui T., Otani S., Hataishi R., Jiang S.X., Nishii Y., Igawa S., Mitsufuji H., Kubota M., Katagiri M., Masuda N.. Successful rechallenge with erlotinib in a patient with EGFR-mutant lung adenocarcinoma who developed gefitinib-related interstitial lung disease. Cancer Chemother. Pharmacol. 2010; 65:803–806. [DOI] [PubMed] [Google Scholar]
- 12. Paez J.G., Janne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004; 304:1497–1500. [DOI] [PubMed] [Google Scholar]
- 13. Minari R., Bordi P., Tiseo M.. Third-generation epidermal growth factor receptor-tyrosine kinase inhibitors in T790M-positive non-small cell lung cancer: review on emerged mechanisms of resistance. Transl. Lung Cancer Res. 2016; 5:695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cross D.A., Ashton S.E., Ghiorghiu S., Eberlein C., Nebhan C.A., Spitzler P.J., Orme J.P., Finlay M.R., Ward R.A., Mellor M.J. et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014; 4:1046–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sung Y.H., Kim J.M., Kim H.T., Lee J., Jeon J., Jin Y., Choi J.H., Ban Y.H., Ha S.J., Kim C.H. et al. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res. 2014; 24:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R.. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013; 153:910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cho S.W., Lee J., Carroll D., Kim J.S., Lee J.. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics. 2013; 195:1177–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cho S.W., Kim S., Kim J.M., Kim J.S.. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013; 31:230–232. [DOI] [PubMed] [Google Scholar]
- 21. Kim H., Kim J.S.. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014; 15:321–334. [DOI] [PubMed] [Google Scholar]
- 22. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yi L., Li J.. CRISPR-Cas9 therapeutics in cancer: promising strategies and present challenges. Biochim. Biophys. Acta. 2016; 1866:197–207. [DOI] [PubMed] [Google Scholar]
- 24. Zhen S., Takahashi Y., Narita S., Yang Y.C., Li X.. Targeted delivery of CRISPR/Cas9 to prostate cancer by modified gRNA using a flexible aptamer-cationic liposome. Oncotarget. 2017; 8:9375–9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang K., Yang C., Wang Q.X., Li Y.S., Fang C., Tan Y.L., Wei J.W., Wang Y.F., Li X., Zhou J.H. et al. The CRISPR/Cas9 system targeting EGFR exon 17 abrogates NF-kappaB activation via epigenetic modulation of UBXN1 in EGFRwt/vIII glioma cells. Cancer Lett. 2017; 388:269–280. [DOI] [PubMed] [Google Scholar]
- 26. Liu T., Shen J.K., Li Z., Choy E., Hornicek F.J., Duan Z.. Development and potential applications of CRISPR-Cas9 genome editing technology in sarcoma. Cancer Lett. 2016; 373:109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Feng W., Li H.C., Xu K., Chen Y.F., Pan L.Y., Mei Y., Cai H., Jiang Y.M., Chen T., Feng D.X.. SHCBP1 is over-expressed in breast cancer and is important in the proliferation and apoptosis of the human malignant breast cancer cell line. Gene. 2016; 587:91–97. [DOI] [PubMed] [Google Scholar]
- 28. Feng Y., Sassi S., Shen J.K., Yang X., Gao Y., Osaka E., Zhang J., Yang S., Yang C., Mankin H.J. et al. Targeting CDK11 in osteosarcoma cells using the CRISPR-Cas9 system. J. Ortho. Res. 2015; 33:199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aubrey B.J., Kelly G.L., Kueh A.J., Brennan M.S., O’Connor L., Milla L., Wilcox S., Tai L., Strasser A., Herold M.J.. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015; 10:1422–1432. [DOI] [PubMed] [Google Scholar]
- 30. Liu Y., Zeng Y., Liu L., Zhuang C., Fu X., Huang W., Cai Z.. Synthesizing AND gate genetic circuits based on CRISPR-Cas9 for identification of bladder cancer cells. Nat. Commun. 2014; 5:5393. [DOI] [PubMed] [Google Scholar]
- 31. Valletta S., Dolatshad H., Bartenstein M., Yip B.H., Bello E., Gordon S., Yu Y., Shaw J., Roy S., Scifo L. et al. ASXL1 mutation correction by CRISPR/Cas9 restores gene function in leukemia cells and increases survival in mouse xenografts. Oncotarget. 2015; 6:44061–44071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fujita T., Yuno M., Fujii H.. Allele-specific locus binding and genome editing by CRISPR at the p16INK4a locus. Sci. Rep. 2016; 6:30485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Monteys A.M., Ebanks S.A., Keiser M.S., Davidson B.L.. CRISPR/Cas9 editing of the mutant huntingtin allele in vitro and in vivo. Mol. Ther. 2017; 25:12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shin J.W., Kim K.H., Chao M.J., Atwal R.S., Gillis T., MacDonald M.E., Gusella J.F., Lee J.M.. Permanent inactivation of Huntington's disease mutation by personalized allele-specific CRISPR/Cas9. Hum. Mol. Genet. 2016; 25:4576–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith C., Abalde-Atristain L., He C., Brodsky B.R., Braunstein E.M., Chaudhari P., Jang Y.Y., Cheng L., Ye Z.. Efficient and allele-specific genome editing of disease loci in human iPSCs. Mol. Ther. 2015; 23:570–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yoshimi K., Kaneko T., Voigt B., Mashimo T.. Allele-specific genome editing and correction of disease-associated phenotypes in rats using the CRISPR-Cas platform. Nat. Commun. 2014; 5:4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gebler C., Lohoff T., Paszkowski-Rogacz M., Mircetic J., Chakraborty D., Camgoz A., Hamann M.V., Theis M., Thiede C., Buchholz F.. Inactivation of Cancer Mutations Utilizing CRISPR/Cas9. J. Natl. Cancer Inst. 2017; 109, doi:10.1093/jnci/djw183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yun C.O., Kim E., Koo T., Kim H., Lee Y.S., Kim J.H.. ADP-overexpressing adenovirus elicits enhanced cytopathic effect by induction of apoptosis. Cancer Gene Ther. 2005; 12:61–71. [DOI] [PubMed] [Google Scholar]
- 39. Lee Y.S., Kim J.H., Choi K.J., Choi I.K., Kim H., Cho S., Cho B.C., Yun C.O.. Enhanced antitumor effect of oncolytic adenovirus expressing interleukin-12 and B7-1 in an immunocompetent murine model. Clin. Cancer Res. 2006; 12:5859–5868. [DOI] [PubMed] [Google Scholar]
- 40. Kim J., Cho J.Y., Kim J.H., Jung K.C., Yun C.O.. Evaluation of E1B gene-attenuated replicating adenoviruses for cancer gene therapy. Cancer Gene Ther. 2002; 9:725–736. [DOI] [PubMed] [Google Scholar]
- 41. Yoon A.R., Hong J., Yun C.O.. A vesicular stomatitis virus glycoprotein epitope-incorporated oncolytic adenovirus overcomes CAR-dependency and shows markedly enhanced cancer cell killing and suppression of tumor growth. Oncotarget. 2015; 6:34875–34891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park J., Lim K., Kim J.S., Bae S.. Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics. 2017; 33:286–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yoo J.Y., Kim J.H., Kim J., Huang J.H., Zhang S.N., Kang Y.A., Kim H., Yun C.O.. Short hairpin RNA-expressing oncolytic adenovirus-mediated inhibition of IL-8: effects on antiangiogenesis and tumor growth inhibition. Gene Ther. 2008; 15:635–651. [DOI] [PubMed] [Google Scholar]
- 44. Choi K.J., Kim J.H., Lee Y.S., Kim J., Suh B.S., Kim H., Cho S., Sohn J.H., Kim G.E., Yun C.O.. Concurrent delivery of GM-CSF and B7-1 using an oncolytic adenovirus elicits potent antitumor effect. Gene Ther. 2006; 13:1010–1020. [DOI] [PubMed] [Google Scholar]
- 45. Yoo J.Y., Kim J.H., Kwon Y.G., Kim E.C., Kim N.K., Choi H.J., Yun C.O.. VEGF-specific short hairpin RNA-expressing oncolytic adenovirus elicits potent inhibition of angiogenesis and tumor growth. Mol Ther. 2007; 15:295–302. [DOI] [PubMed] [Google Scholar]
- 46. Kim J.H., Lee Y.S., Kim H., Huang J.H., Yoon A.R., Yun C.O.. Relaxin expression from tumor-targeting adenoviruses and its intratumoral spread, apoptosis induction, and efficacy. J. Natl. Cancer Inst. 2006; 98:1482–1493. [DOI] [PubMed] [Google Scholar]
- 47. Koo T., Kim J.S.. Therapeutic applications of CRISPR RNA-guided genome editing. Brief. Funct. Genomics. 2017; 16:38–45. [DOI] [PubMed] [Google Scholar]
- 48. Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015; 520:186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Swiech L., Heidenreich M., Banerjee A., Habib N., Li Y., Trombetta J., Sur M., Zhang F.. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 2015; 33:102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N.. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016; 351:400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao B.X., Wang J., Song B., Wei H., Lv W.P., Tian L.M., Li M., Lv S.. Establishment and biological characteristics of acquired gefitinib resistance in cell line NCI-H1975/gefinitib-resistant with epidermal growth factor receptor T790M mutation. Mol. Med. Rep. 2015; 11:2767–2774. [DOI] [PubMed] [Google Scholar]
- 52. Tsai M.F., Chang T.H., Wu S.G., Yang H.Y., Hsu Y.C., Yang P.C., Shih J.Y.. EGFR-L858R mutant enhances lung adenocarcinoma cell invasive ability and promotes malignant pleural effusion formation through activation of the CXCL12-CXCR4 pathway. Sci. Rep. 2015; 5:13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yu J.Y., Yu S.F., Wang S.H., Bai H., Zhao J., An T.T., Duan J.C., Wang J.. Clinical outcomes of EGFR-TKI treatment and genetic heterogeneity in lung adenocarcinoma patients with EGFR mutations on exons 19 and 21. Chin. J. Cancer. 2016; 35:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cheng R., Peng J., Yan Y., Cao P., Wang J., Qiu C., Tang L., Liu D., Tang L., Jin J. et al. Efficient gene editing in adult mouse livers via adenoviral delivery of CRISPR/Cas9. FEBS Lett. 2014; 588:3954–3958. [DOI] [PubMed] [Google Scholar]
- 55. Ding Q., Strong A., Patel K.M., Ng S.L., Gosis B.S., Regan S.N., Cowan C.A., Rader D.J., Musunuru K.. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ. Res. 2014; 115:488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Russell S.J., Peng K.W., Bell J.C.. Oncolytic virotherapy. Nat. Biotechnol. 2012; 30:658–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Green N.K., Hale A., Cawood R., Illingworth S., Herbert C., Hermiston T., Subr V., Ulbrich K., van Rooijen N., Seymour L.W. et al. Tropism ablation and stealthing of oncolytic adenovirus enhances systemic delivery to tumors and improves virotherapy of cancer. Nanomedicine. 2012; 7:1683–1695. [DOI] [PubMed] [Google Scholar]
- 58. Bachtarzi H., Stevenson M., Fisher K.. Cancer gene therapy with targeted adenoviruses. Expert Opin. Drug Deliv. 2008; 5:1231–1240. [DOI] [PubMed] [Google Scholar]
- 59. Bischoff J.R., Kirn D.H., Williams A., Heise C., Horn S., Muna M., Ng L., Nye J.A., Sampson-Johannes A., Fattaey A. et al. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996; 274:373–376. [DOI] [PubMed] [Google Scholar]
- 60. Kirn D. Replication-selective oncolytic adenoviruses: virotherapy aimed at genetic targets in cancer. Oncogene. 2000; 19:6660–6669. [DOI] [PubMed] [Google Scholar]
- 61. Yoon A.R., Kasala D., Li Y., Hong J., Lee W., Jung S.J., Yun C.O.. Antitumor effect and safety profile of systemically delivered oncolytic adenovirus complexed with EGFR-targeted PAMAM-based dendrimer in orthotopic lung tumor model. J. Control. Release. 2016; 231:2–16. [DOI] [PubMed] [Google Scholar]
- 62. Chew W.L., Tabebordbar M., Cheng J.K., Mali P., Wu E.Y., Ng A.H., Zhu K., Wagers A.J., Church G.M.. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods. 2016; 13:868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kim D.H., Longo M., Han Y., Lundberg P., Cantin E., Rossi J.J.. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 2004; 22:321–325. [DOI] [PubMed] [Google Scholar]
- 64. Pichlmair A., Schulz O., Tan C.P., Naslund T.I., Liljestrom P., Weber F., Reis e Sousa C.. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5΄-phosphates. Science. 2006; 314:997–1001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The deep sequencing data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRX2866619. We have included the read depth information of the deep sequencing data in the Supplementary Tables S3–5.