Abstract
A T-box regulator or riboswitch actively monitors the levels of charged/uncharged tRNA and participates in amino acid homeostasis by regulating genes involved in their utilization or biosynthesis. It has an aptamer domain for cognate tRNA recognition and an expression platform to sense the charge state and modulate gene expression. These two conserved domains are connected by a variable linker that harbors additional secondary structural elements, such as Stem III. The structural basis for specific tRNA binding is known, but the structural basis for charge sensing and the role of other elements remains elusive. To gain new structural insights on the T-box mechanism, a molecular envelope was calculated from small angle X-ray scattering data for the Bacillus subtilis glyQS T-box riboswitch in complex with an uncharged tRNAGly. A structural model of an anti-terminated glyQS T-box in complex with its cognate tRNAGly was derived based on the molecular envelope. It shows the location and relative orientation of various secondary structural elements. The model was validated by comparing the envelopes of the wild-type complex and two variants. The structural model suggests that in addition to a possible regulatory role, Stem III could aid in preferential stabilization of the T-box anti-terminated state allowing read-through of regulated genes.
INTRODUCTION
A riboswitch is a structured RNA element found in the untranslated region of an mRNA. Upon sensing changes in levels of a particular metabolite, it regulates the level of transcription or translation of a downstream mRNA sequence (1–3). Riboswitches are widespread in gram positive bacteria and are considered good targets for inhibitor design as they regulate genes or operons involved in utilization, biosynthesis or transport of essential cellular metabolites (4). Recognition of a specific metabolite in a riboswitch is accomplished by a sensory domain, called the aptamer, which is linked to a regulatory domain, the expression platform, by a variable linker (1). The expression platform is conformationally labile and can adopt one of at least two alternate states in response to the presence or absence of a ligand bound to the aptamer (2). Different riboswitches utilize one of several distinct mechanisms for regulating either transcription or translation of downstream genes (1,2).
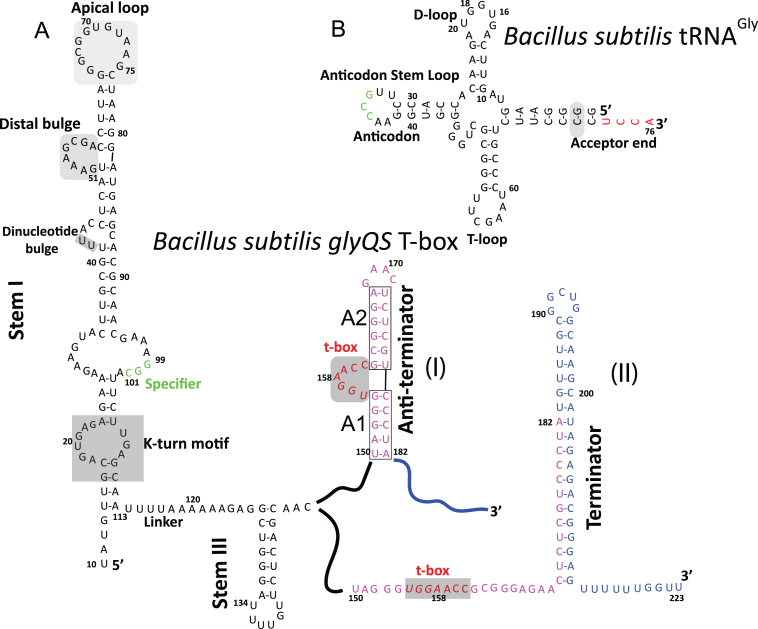
One specific type of riboswitch is the group of T-box regulators or riboswitches that participate in amino acid homeostasis by regulating genes encoding tRNA synthetases, amino acid transporters, and operons for amino acid biosynthesis (5). The availability of an amino acid in a cell is indirectly reflected by the intracellular concentration of its corresponding aminoacylated tRNA. T-box riboswitches utilize this feature by recognizing a specific tRNA as its cognate ligand and sensing its charged state to assess the availability of the amino acid corresponding to the tRNA in the cell (5). Transcriptional control is the most common mechanism for T-box-mediated regulation, although recently, T-box riboswitches that exert translational control have also been discovered (6). In general, the simplest T-box riboswitches are composed of two phylogenetically conserved domains that can fold independently into an aptamer and an expression platform. T-box riboswitches are classified into two main sub-types based on the complexity of the secondary structural elements in the variable linker region joining the aptamer and the expression platform. In the smaller and simpler T-box riboswitches, the linker contains a short single stranded region and a single stem-loop, Stem III. A prototypical T-box riboswitch of this sub-class is the Bacillus subtilis glyQS T-box riboswitch, which specifically recognizes tRNAGly as it cognate ligand and regulates the expression of glycyl-tRNA synthetase (7). The linker of the more complex T-box riboswitches harbors an additional stem-loop, Stem II, along with a highly conserved stretch of nucleotides, called Stem IIA/B, which can potentially fold into a pseudoknot (8,9). The prototypical T-box of this sub-class is the B. subtilis tyrS T-box riboswitch, the first T-box riboswitch discovered (8).
In general, a T-box aptamer is formed by an ∼100 nt long double stranded RNA helix, Stem I, which harbors several conserved structural motifs important for specific tRNA recognition (10) (Figure 1). Notable among these motifs are a K-turn motif, a specifier loop in the proximal segment, a conserved distal bulge, and an apical loop in the distal segment of Stem I. The exact role of the specifier loop in the T-box mechanism was first uncovered by biochemical studies on the tyrS T-box (8), which showed that this trinucleotide sequence motif is responsible for non-cognate tRNA discrimination and specificity for cognate tRNA. The specifier base pairs with the anti-codon loop of its cognate tRNA and uses a mechanism of recognition identical to the one used by the ribosome for decoding genetic information during translation (7). The role of the other conserved Stem I motifs was revealed more recently by crystal structures of the glyQS Stem I in complex with its cognate tRNAGly (11,12). As was anticipated (13,14), the structures showed that there is an additional crucial point of contact between Stem I and its cognate tRNA involving the two conserved distal loops, which fold into two T-loops (15) that intercalate to create a flat molecular surface of stacked nucleobases. This compact structural module is also found in other unrelated tRNA binding systems, namely the ribosome and RNase P (16,17), and in all three cases the T-loops interact with the highly conserved elbow region of the tRNA to form a stable and extended stack of nucleobases (12).
Figure 1.
Sequence and secondary structure of the Bacillus subtilis glyQS T-box riboswitch and its cognate tRNAGly. (A) Secondary structure diagram of the B. subtilis glyQS T-box in its two possible conformations, anti-terminated (I) and terminated (II). (B) Secondary structure diagram of B. subtilis tRNAGly. Typically, a T-box riboswitch interacts with its cognate tRNA using a series of conserved structural motifs. One of these involves the elbow region of the tRNA (D-loop and T-loop) and an intercalated T-loop motif formed by the apical loop and the distal bulge (shaded in gray in B) of Stem I. Specificity for a tRNA is accomplished through complimentary base-pairing between a Specifier loop (green in A) of Stem I and the tRNA anti-codon loop (green in B). The expression platform of a T-box can adopt one of two mutually exclusive conformations (I and II). Nucleotides base-paired in the anti-terminated and the terminated state are shown in purple and blue, respectively. The charged state of the bound tRNA is sensed in the anti-terminated state (I) through complimentary base-pairing between a conserved T-box sequence (red in A) and the tRNA 3΄ NCCA (red in B) sequence. The CG base-pair (shaded in gray) in the tRNA acceptor end was changed to a GC base-pair to accommodate the two guanylyl nucleotides incorporated by a T7 RNA polymerase during in vitro transcription.
The expression platform of a T-box riboswitch is formed by a conserved region that regulates downstream gene expression (9) upon sensing the charged state of the bound tRNA (Figure 1). Read-through of a T-box regulated gene is successful only when the bound tRNA is uncharged and its conserved 3΄ CCA end is unhindered, allowing it to base-pair to a conserved and complementary sequence, the T-box, which is part of a 7-nt bulged region (5,18). This key interaction allows the expression platform of a T-box to adopt a thermodynamically less stable conformation, termed the anti-terminator (5,18) (Figure 1). In contrast, when the bound tRNA is charged, this T-box:tRNA base-pairing interaction is hindered and the expression platform forms a long GC rich stem-loop, termed the terminator, which is followed by a stretch of consecutive uridines. The terminator conformation is inherently more stable than the anti-terminator conformation and it stalls a transcribing RNA polymerase causing it to dissociate from the DNA template resulting in premature transcriptional termination by a rho-independent mechanism (5,18).
The recent crystal structures of glyQS Stem I in complex with a tRNAGly (11,12), as well as the solution structure of just the proximal segment of Stem I (19), showed that the T-box aptamer uses an induced fit mechanism for tRNA binding in which both RNA binding partners undergo conformational changes around two conserved pivot points, namely the K-turn motif and the dinucleotide bulge, to achieve shape complementarity. The structures conclusively explained the rationale for strong sequence conservation at these pivot points and also implied that a full-length T-box leader would need additional conformational rearrangements in order to sense the charged state of a bound tRNA.
The mechanism of tRNA charge sensing by a T-box riboswitch has been extensively investigated by studying the B. subtilis glyQS T-box system (20–22). During read-through, the 33 nt long anti-terminator of the glyQS T-box harbors two short RNA helices (A1 and A2) that flank a highly conserved 7-nt bulge (Figure 1). Four of these seven nucleotides, the T-box sequence, detect an uncharged tRNA by base-pairing to its conserved 3΄ NCCA sequence (23). A nuclear magnetic resonance solution structure (24) of just the anti-terminator region provides valuable structural insights about tRNA charge sensing by this 7-nt bulge. The structure indicates that, similar to Stem I, charge sensing by the T-box sequence may involve a mutual-induced fit mechanism in which the more flexible T-box nucleotides may sample several different conformations to recognize and bind to the 3΄ end of the tRNA and form a rigid stacked structure that could play a role in restraining the flexibility of the T-box sequence. A recent biophysical study (20) also suggests that when an uncharged tRNA is bound to the T-box, the complementary base-paring between the 3΄ NCCA end of an uncharged tRNA and the T-box sequence could help align the A1 helix with the tRNA acceptor stem resulting in formation of a long A1/tRNA acceptor stem co-axial stack containing 29 layers of base pairs and triples that may stabilize the anti-terminator conformation and favor it over the terminator conformation.
The role of the linker region in the T-box mechanism is not known. There is no information that explains the sequence and structural variability in this region in different T-box sub-classes. For the glyQS T-box riboswitch, there is experimental evidence that shows that under certain in vitro conditions the transcription of the T-box leader would pause at position 138 (Figure 1), which resides in the loop of Stem III (25). It was therefore hypothesized that transcriptional stalling could provide additional time for Stem I to discriminate and recognize its cognate tRNA (25,26). However, there is no experimental evidence which shows that this pausing is essential in vivo and therefore the importance of transcriptional stalling in the T-box mechanism is unclear. An experimental model of Stem III depicting its location and orientation relative to other T-box domains could provide important insights about its role in the overall T-box mechanism. Particularly, a better understanding of the structure of Stem III could help us to understand whether it plays a role in tRNA discrimination, tRNA binding or charge-state sensing. In the absence of structural information on Stem III and the terminator helix, it is difficult to validate existing models and ascertain the extent of the conformational changes that occur during the recognition process. To help answer some of these questions, a molecular envelope for the full-length B. subtilis glyQS T-box leader in complex with its uncharged cognate tRNAGly was obtained using small angle X-ray scattering (SAXS). This is the first experimental model of a full-length T-box riboswitch in complex with its cognate uncharged tRNA, and it provides important clues about the relative locations of various T-box structural elements that are currently missing from the mechanistic models for T-box riboswitches. The results presented here refine the proposed models for anti-termination by this class of riboswitches and provide new insights about the mechanism of tRNA-mediated gene regulation.
MATERIALS AND METHODS
Cloning, mutagenesis and in vitro transcription
A bicistronic DNA template (Supplementary Figure S1) encoding the well-characterized B. subtilis glyQS T-box leader (+10–182) (5,7,12,22), as well as its cognate tRNAGly (5,7,12,22), was synthesized by GenScript (Piscataway, NJ, USA). In addition, another variant of this bicistronic construct was generated in which the less conserved Stem III helix was elongated by adding 10 bp just before the loop spanning nucleotides 134–139 (Figure 1) (10 bases before 134 and 10 complementary bases after 139) (S3.10), by using standard site-directed mutagenesis protocols (27). For in vitro transcription, the bicistronic DNA template was inserted into a pUC19 vector by using the EcoR1 (NEB) and Hind III (NEB) restriction sites. The resulting plasmid was transformed into DH5α cells and purified by standard protocols (28). For run-off transcription, the circular plasmid vector was linearized using two BsaI (NEB) cut sites that were placed at the end of two RNA coding regions. RNA transcription was performed by standard protocols using a purified His6-tagged T7 RNA polymerase (29). As T7 RNA polymerase incorporates two additional guanine nucleotides at the beginning of a transcript (30), in order to preserve the secondary structural features of an uncharged tRNA, a CG base-pair in the tRNA acceptor end was changed to a GC base-pair (Figure 1). For an 1 ml transcription reaction, 200 μg of linearized plasmid DNA was used and the reaction was allowed to proceed for 3 h at 37°C. The total RNA yield per 1 ml transcription reaction was ∼1.6–2 mg of which ∼400 μg was the monomeric T-box:tRNA complex.
Purification of glyQS T-box:tRNAGly complexes
All glyQS T-box complexes characterized in this study were purified under native conditions by size-exclusion chromatography (SEC). For purification of the glyQS T-box:tRNAGly complex, the crude transcription reaction was spun for 1 h at 18 000 g and directly loaded onto a gel filtration column (HiPrep 26/60, Sephacryl S-200, GE Health Sciences) that was pre-equilibrated with a pH 7.0 buffer containing 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 100 mM KCl and 10 mM MgCl2 at room temperature. In order to separate the monomeric and dimeric forms of the glyQS T-box:tRNA complex, the elution fractions from each peak were pooled, concentrated, and reinjected back onto the gel filtration column for a second round of purification. The purified T-box complex was then concentrated by centrifugation using a 10 kDa cutoff Amicon filter and stored at 4°C. For purification of the ternary complex formed by glyQS T-box, tRNAGly and the L7Ae homolog from Pyrococcus horikoshii, the monomeric T-box:tRNA binary complex was incubated with excess protein at 1:1.5 molar ratio on ice for 30 min and was purified by SEC.
Characterization of glyQS T-box:tRNAGly complexes
The purity of the glyQS T-box leader and its cognate tRNAGly in each purified complex was tested on a 7.5% polyacrylamide, 8 M urea gel. The homogeneity of each complex was ascertained by running it on a 5% native polyacrylamide gel containing 10 mM MgCl2. For characterization by SEC coupled with multi-angle light scattering (SEC-MALS), 200 μg of each purified complex was injected onto a Superdex 200 10/30 GL column (GE Health Sciences) pre-equilibrated with the purification buffer and connected to multi-angle light scattering and refractive index detectors (DAWN HELEOS and Optilab rEX, Wyatt Technology). The Wyatt detectors were kept at room temperature. Weight-averaged molar masses determined from the MALS data were calculated using the ASTRA 5.3.4 software (Wyatt Technology). Typical data are shown in Supplementary Figure S2.
SAXS data collection
All solution scattering data were collected at the DuPont Northwestern Dow Collaborative Access Team (DND CAT) beamline at the Advanced Photon Source (APS) using a SAXS, wide angle X-ray scattering (WAXS), medium angle X-ray scattering (MAXS) triple CCD detector system at room temperature (23°C) using a flow cell to minimize radiation damage (31). Data were collected at 10 keV (1.239 Å) and covered the momentum transfer range of 0.0015 < q < 2.6 Å−1 (q = 4πsinθ/λ, where 2θ is the scattering angle). The scattering data from the detectors were azimuthally integrated and merged resulting in a one-dimensional scattering profile with respect to q and normalized to the incident beam intensity. For each construct, a dilution series with at least three samples of different concentration was used for data collection. Data for an empty flow cell were collected before the start of data collection to estimate the scattering contribution made by the empty flow cell. The empty scan was also compared across samples to ensure that the flow cell was clean. Scattering data from the buffer were then collected before each sample in the dilution series and the buffer profiles were compared to ensure consistency. The buffer scattering profiles were then averaged together and subtracted from sample scattering over the entire angular range (i.e. 0.0015–2.6 Å−1) in the dilution series. The resulting sample scattering data were then normalized using the sample concentration. All data processing up to this step were carried out with local software (31). The data were truncated to a qmax of 0.3 Å−1 and all further calculations were performed using the ATSAS SAXS software package (Supplementary Table S1) (32). Normalized scattering curves for each sample were scaled together and merged to produce the final scattering curve (Supplementary Figure S3A). The program GNOM (33) was then used to obtain the radius of gyration (Supplementary Figure S3B) and the maximum particle size (Dmax) from the particle distance distribution function (PDDF), P(r) (Supplementary Figure S3C).
Ab initio envelope calculation and modeling
Bead models that fit the experimental scattering curves for the different complexes were calculated by DAMMIN (34), which is an approach that has been shown to be appropriate not only for proteins but also for nucleic acids (35). For each complex, 40 independent calculations were performed using a two-step protocol similar to the one used for solving the solution structure of the HIV-Rev response element (36). Briefly, the protocol involved an initial round of ab initio calculations using a spherical search volume whose diameter was 15 Å greater than the estimated Dmax value. The bead models were then superposed, averaged and filtered using the DAMAVER (37) package in ATSAS (32). The initial filtered model was used to estimate the dimensions of the molecule, and the new estimate was used in a second round of ab initio calculations but now using a parallelepiped as the search volume with dimensions 15 Å greater than the dimension of the initial average envelope. The averaged envelopes for each of the three glyQS T-box:tRNAGly complexes were superposed using SUPCOMB (38), and their position were manually adjusted to locally align similar features in them. A crystal structure of Stem I:tRNAGly (12) (PDB ID: 4LCK) and additional models were manually docked into the molecular envelope using PyMOL (39). Initial models of the secondary structural elements missing from the available crystal structure were generated using the web server RNAComposer (40). Alternate conformations of the models were generated by using the morphing routine implemented in CNS (41,42). All computed models were energy minimized using PHENIX (43) and were manually placed in the SAXS envelope. To evaluate the fit of various structural models to the experimental data (Supplementary Figure S4), we used CRYSOL (44) and AquaSAXS (45) that use different algorithms for modeling the primary hydration shell.
Figures
Figures for the atomic models were created using PyMOL (39).
RESULTS
Purification and characterization of glyQS T-box:tRNA complex
The full-length B. subtilis glyQS T-box leader is 223 nt long (Figure 1). Its aptamer domain (nucleotides 1–113), called Stem I, and its expression platform (nucleotides 150–223), both primarily form long double stranded RNA helices. The expression platform is conformationally labile and can switch between one of two conformations (terminated and anti-terminated) depending on the charged state of tRNAGly bound to the aptamer (Figure 1). In this study, we purified a truncated version of the glyQS T-box leader in which 41 nt (183–223) from the 3΄ end were omitted from the full-length leader (Figure 1A). This truncation allows the expression platform to adopt only the anti-terminated conformation, corresponding to the conformation during progression of transcription and read-through of the downstream regulated gene. In the truncated construct, the T-box leader (nucleotides 1–182) cannot adopt the terminated conformation, but can still interact with the conserved 3΄ CCA sequence of an uncharged tRNA through complementary base-pairing (12). Previous studies using similar constructs had shown that the B. subtilis glyQS T-box leader can refold into multiple conformations when purified under denaturing conditions (13). It is also known that refolded B. subtilis tRNAGly has a strong tendency to form non-physiological dimers (12). We therefore explored an alternate strategy to assemble and purify the B. subtilis glyQS T-box:tRNA complex using T7 RNA polymerase co-transcription of the glyQS T-box leader and its cognate tRNAGly from a linearized bicistronic DNA construct (Supplementary Figure S1) followed by purification of the natively assembled complex by SEC. This methodology takes advantage of the high affinity of the truncated B. subtilis glyQS T-box leader for its uncharged cognate tRNAGly (∼250 nM) (12), which promotes assembly of the natively folded glyQS T-box leader with tRNAGly into a homogeneous complex that is suitable for structural characterization by SAXS.
The glyQS T-box leader forms two types of complexes with its uncharged cognate tRNAGly under the experimental conditions described above. On an SEC column, these two types of complexes run as distinct peaks (Supplementary Figure S2A) that can be completely purified by a second pass of each peak fraction through the SEC column. Each peak fraction contained two RNA molecules of different size, corresponding to the truncated glyQS T-box leader and an uncharged tRNAGly (Supplementary Figure S2A, inset). Further characterization to ascertain the homogeneity and oligomeric state of the two T-box complexes was carried out by SEC-MALS, which showed that the two T-box complexes were monodisperse in solution (Supplementary Figure S2B). The calculated hydrodynamic radii of the two T-box complexes were 38 ± 1.9 and 67 ± 2.7 Å, respectively, which correspond to RNAs of molecular weight (MW) 84.2 ± 0.3 and 156 ± 1.6 kDa, respectively. This shows that the smaller complex is a monomeric (1:1) glyQS T-box:tRNA complex (theoretical MW ∼82 kDa), and during transcription it oligomerizes to form a dimeric (2:2) T-box:tRNA complex. The two T-box complexes were stable and did not repartition after purification under the experimental conditions used in this study. For the SAXS structural analysis of the T-box complex, only the monomeric (1:1) T-box:tRNA complex was used, as the exact mode for dimerization is not known. For the wild-type T-box complex, the monomeric state and monodispersity of the complex were confirmed after SAXS data collection by analyzing the same RNA sample by SEC-MALS (Supplementary Figure S2B). The sample was monomeric and monodisperse after the SAXS experiment.
In addition to the wild-type complex, we purified two other variants (L7Ae complex and S3.10) of the monomeric glyQS T-box:tRNA complex using the same strategy (Supplementary Figure S2C and D) and subsequently analyzed them by SAXS. The two variant complexes contained additional structural features that were used to locate the secondary structural elements of the glyQS T-box leader in the low resolution molecular envelope obtained through SAXS. One of the two variant complexes was a ternary complex formed by the monomeric glyQS T-box:tRNA complex and an archaeal homolog of the bacterial L7Ae protein. This 14 kDa protein specifically binds to a conserved K-turn motif found in the proximal segment of Stem I and was used to locate the base of the T-box aptamer in the molecular envelope. Interestingly, a bacterial homolog of the L7Ae protein aided in crystallization of glyQS Stem I and an uncharged tRNAGly (12), where it acted as a chaperone that facilitated the formation of crystal lattice contacts. The SEC profile of the ternary complex is shown in Supplementary Figure S2C, and it can be seen that it has a larger molecular weight and elutes before the binary T-box:tRNA complex on the SEC column. The archaeal L7Ae homolog binds with high affinity to the T-box complex, and on a native gel it runs as a single band and with lower mobility in comparison to the binary complex (Supplementary Figure S2C, inset). The second variant T-box complex (S3.10) purified was used to locate Stem III in the molecular envelope of the wild-type complex and was made by elongating Stem III by 10 bp. The SEC profile of the transcription reaction for this variant showed the same features (Supplementary Figure S2D) as the wild-type construct and the only difference was that the two main peak positions were shifted to higher molecular weight.
SAXS analysis of monomeric glyQS T-box:tRNA complexes
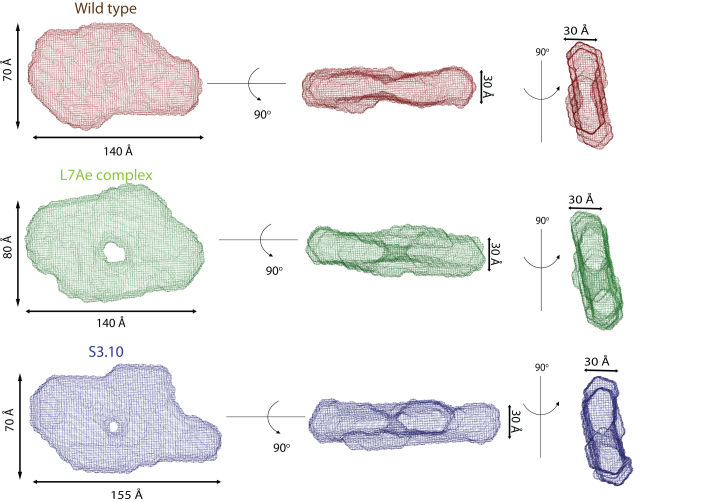
The experimental SAXS curves for the three T-box complexes are shown in Supplementary Figure S3A. The Guinier region (Supplementary Figure S3B) of the scattering curve is linear in all cases, which indicates that the three T-box complexes are homogeneous and monodisperse in solution. The Rg values of the wild-type T-box:tRNA complex, the L7Ae bound wild-type T-box complex, and the Stem III variant complex (S3.10) were 43.2 ± 1.3, 45.0 ± 1.2, and 45.5 ± 1.3 Å respectively, which indicate that the overall sizes of all three T-box complexes are similar. The PDDF evaluated from a scattering curve was used to estimate the largest intermolecular distance (Dmax) in the molecule. The plots of the PDDF (Supplementary Figure S3C) for the three T-box complexes indicate that the wild-type T-box complex and its L7Ae bound form are similar in size (Dmax ∼ 140 Å) and the Stem III variant (S3.10) of the complex with 10 bp insertion is longer with a Dmax of ∼155 Å. The overall shape of the PDDF plots for the three T-box complexes was similar and the appearance of a broad peak may indicate the presence of a flexible system with an extended molecular surface (46). This is not uncommon and SAXS-derived molecular envelopes of other non-coding RNAs, including the partial glyQS T-box:tRNA complex, also show similar structural features (13,36,47). In each plot, the distance distribution profile plateaus at ∼40 Å, which could arise due to juxtaposition of two long RNA helices, such as Stem I and the anti-codon stem of tRNAGly in the glyQS T-box complex. The differences in the distance distribution profile of each complex could be attributed to the variability in their overall flexibility, consistent with the broad shoulder. This last observation is further supported by the dimensionless Kratky plots (Supplementary Figure S3D), which suggest that the three glyQS T-box:tRNAGly complexes have open and extended structures and that the wild-type complex differs slightly from the L7Ae and S3.10 complex in its overall compactness (13,36,47). Molecular envelopes for the three glyQS T-box:tRNAGly complexes were obtained through ab initio calculations with the program DAMMIN (34) following a two-step protocol similar to the one used to study the structure of HIV-1 Rev response element (36). For each T-box complex, 40 independent calculations were carried out to obtain bead models that fit the experimental scattering curves. These bead models were then aligned and their normalized standard deviation (NSD) values were computed to identify outliers. The NSD values for the wild-type complex, L7Ae bound complex, and Stem III variant (S3.10) complex were 0.879 ± 0.055, 0.760 ± 0.041, and 0.830 ± 0.04, respectively, and they show that the computed models for each complex had excellent internal agreement. A final molecular envelope for each complex was obtained by averaging the individual bead models aligned to each other followed by filtering to remove loosely connected atoms with low occupancy. Finally, the molecular weight of a particle can be estimated from the scattering curve by evaluating its volume of correlation (32). For the three T-box complexes, the estimated molecular weight was found to be consistent with their theoretical molecular weight (Supplementary Table S1).
Molecular envelope of glyQS T-box:tRNAGly complex
The experimentally derived molecular envelopes of the three complexes of glyQS T-box leader with its cognate uncharged tRNAGly are shown in Figure 2. The most salient structural feature common to the three molecular envelopes is the flat shape of the envelopes, with a thickness of around 30 Å, which roughly corresponds to the width of an A-form RNA helix. The flat shape of the molecular envelopes suggests that all major secondary structural elements of a T-box leader along with the bound tRNA reside in the same plane and thereby create an extended surface. The overall shape of the three molecular envelopes resembles an asymmetric torus in which the two unequal halves are centered about a pinched region and enclosed different volumes. Another salient feature of the three molecular envelopes is that the leading edge of the larger asymmetric half is arched at an angle, which suggested that an RNA with a bent conformation was enclosed by this.
Figure 2.
SAXS molecular envelopes of the Bacillus subtilis glyQS T-box:tRNAGly complexes. (Top) Molecular envelope of the wild-type B. subtilis glyQS T-box:tRNAGly complex (red). (Middle) Molecular envelope of the T-box:tRNAGly:L7Ae ternary complex (green). (Bottom) Molecular envelope of the T-box (S3.10):tRNAGly complex (blue). The overall flat shape of a T-box:tRNA complexes suggests that all major secondary structural elements of the two interacting RNA molecules lie in the same plane. The diagrams show three orthogonal views of the molecular envelopes.
Location of Stem I, tRNAGly and Stem III in the structure of glyQS T-BOX:tRNAGly complex
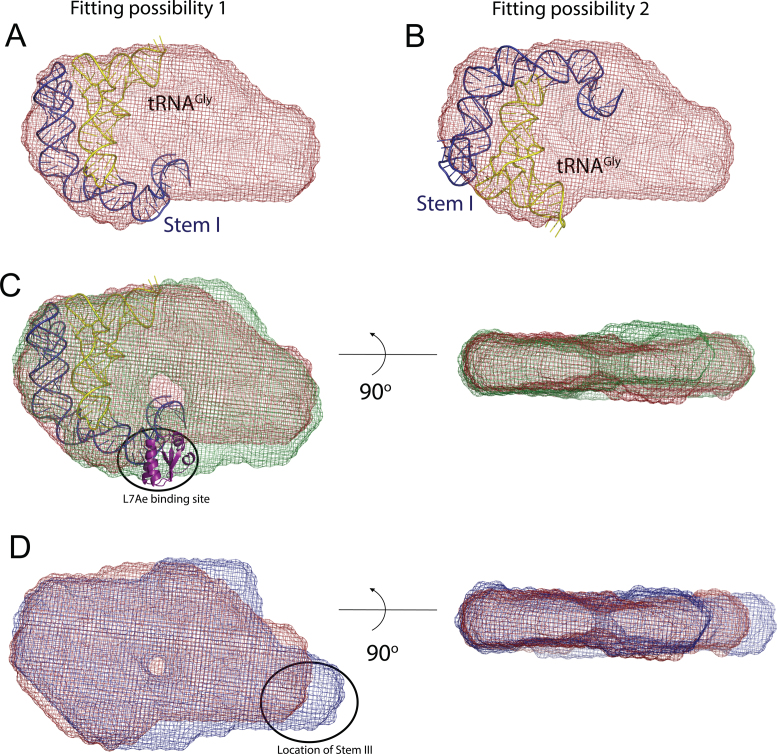
The two crystal structures (PDB IDs: 4MGN and 4LCK) (11,12) of glyQS Stem I helix in complex with its cognate tRNAGly showed that there are two pivot points around which the RNA helix changes the direction of progression and arches to follow the shape of the bound tRNA (11,12). The first major pivot point is located half way along the length of Stem I in its distal segment and is called the dinucleotide bulge. The second pivot point, which is also the binding site for the bacterial L7Ae protein, is called the K-turn and is located in the proximal segment of Stem I. These features of the crystal structures suggested that the larger half of the asymmetric envelope is the most probable location of the glyQS Stem I and tRNAGly. The crystal structure of the Stem I:tRNAGly (PDB ID: 4LCK) complex (12) fitted well in this segment of the envelope, but due to the absence of recognizable structural features in the low resolution SAXS envelope it was possible to place the partial model in two orientations (Figure 3A and B). To eliminate this ambiguity, we aligned and compared the molecular envelopes of the wild-type glyQS complex and its L7Ae bound form (Figure 3C). The 14 kDa L7Ae protein binds to the base or proximal segment of Stem I and its inclusion in the T-box complex resulted in the appearance of a protrusion, which was assigned as the location of the K-turn motif in the molecular envelope (Figure 3C).
Figure 3.
Location of Stems I and III in the molecular envelope of the Bacillus subtilis glyQS T-box:tRNAGly complex. Panels (A) and (B) show two alternate fittings of the crystal structure of Stem I:tRNAGly complex (12) (PDB ID: 4LCK) in the molecular envelope of the wild-type complex (red). (C) The orientation of Stem I was selected by aligning and comparing the molecular envelopes of the T-box:tRNAGly:L7Ae ternary (green) and wild-type (red) complexes. The 14 kDa L7Ae protein binds to the proximal segment of Stem I and represents a small protrusion in the envelope. A model of the Stem I:tRNAGly crystal structure (12), including the L7Ae structure was placed in the molecular envelope of the T-box:tRNAGly:L7Ae ternary (green), corresponding to the orientation in panel (A). (D) A similar comparison between the envelopes of the wild-type and the T-box (S3.10):tRNAGly complexes helped locate Stem III in the envelope and the model. The diagrams in (C) and (D) show two orthogonal views of the aligned molecular envelopes.
The putative position of the adjoining RNA helix, Stem III, was less ambiguous after the initial location of the 3΄ end of Stem I was identified. To confirm the location of Stem III in the molecular envelope, the molecular envelopes of tRNAGly complexed to the wild-type glyQS T-box leader and the Stem III variant (S3.10) were aligned (Figure 3D). In a linear A-form RNA helix, 10 bp roughly corresponds to a length of ∼30 Å (48). In the Stem III variant molecular envelope, a sufficiently large protrusion is evident (Figure 3D and Supplementary Figure S5A) and this area was assigned as the Stem III location. After determining the location of the anti-terminator helix (see below), a structural model of the Stem III helix with 10 additional base pairs (SIII.10) was placed in the molecular envelope of the S3.10 complex (Supplementary Figure S5B), which confirmed the location of Stem III as well as its orientation in the final model.
Location and modeling of the anti-terminator region in the glyQS T-box:tRNA complex
The anti-terminator region of glyQS T-box regulator is made up of two short RNA helices, A1 and A2, containing 5 and 6 bp, respectively (Figure 1). These two helices flank a 7-nt single stranded bulge that harbors a conserved sequence motif, called the t-box, which is directly responsible for detecting an uncharged tRNA through base-pairing with its 3΄ CCA end. Initial placement of the crystal structure of Stem I:tRNAGly (PDB ID: 4LCK) (12) and Stem III in the molecular envelope showed that there was enough available space to place the terminator helix in the anti-terminated state. Placement of the anti-terminator helix at this location would position the conserved t-box sequence in the vicinity of the 3΄ CCA end of the tRNA, which would allow it to establish this key intermolecular base-pairing interactions.
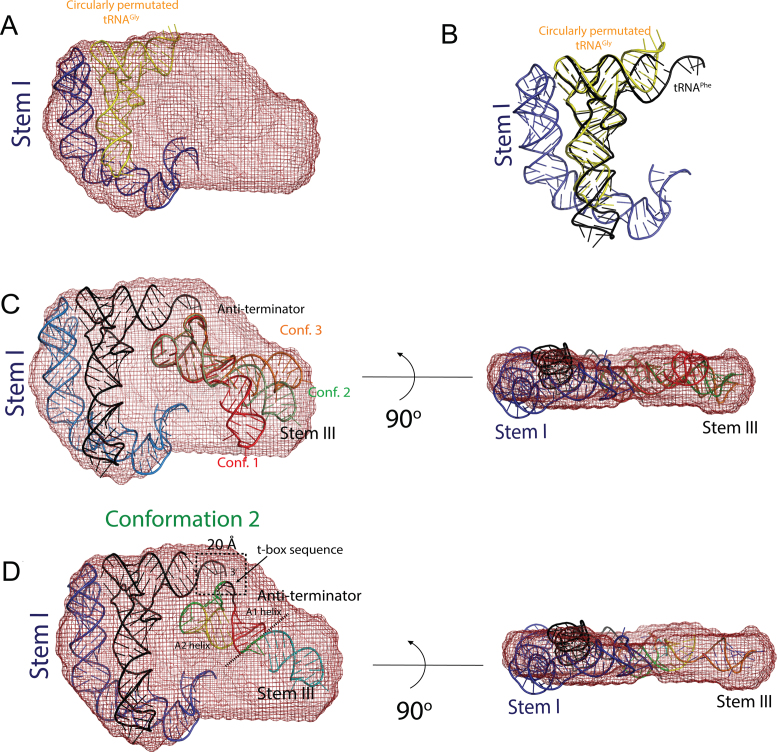
In the crystal structure of Stem I:tRNAGly complex (PDB ID: 4LCK) (12), the bound tRNA is circularly permutated and was therefore not useful for modeling the intermolecular interaction between the tRNA 3΄ CCA end and the T-box sequence (Figure 4B). Similar to a T-box riboswitch, bacterial RNase P uses the tRNA 3΄ CCA sequence to establish key intermolecular contacts with a precursor tRNA during catalysis (17). Therefore, to map the approximate location of the tRNA 3΄ CCA end in the molecular envelope, we used the coordinates of tRNAPhe from the complex with bacterial RNase P (PDB ID: 3Q1Q) and aligned it with the modified tRNAGly coordinates from the crystal structure with Stem I. The two tRNAs were superposed by aligning only their conserved T-loop motif found in the elbow region (Figure 4B).
Figure 4.
Location of the anti-terminator helix and Stem III in the molecular envelope of the wild-type Bacillus subtilis glyQS T-box:tRNAGly complex. (A) The crystal structure of Stem I in complex with a circularly permutated tRNAGly (PDB ID: 4LCK) was placed in the SAXS-derived molecular envelope. (B) The position of the tRNA 3΄ NCCA sequence was determined by aligning an uncharged tRNAPhe (17) (PDB ID: 3Q1Q) to this circularly permutated tRNAGly. (C) Two orthogonal views of the molecular envelope of the wild-type T-box complex with three representative conformer (Conf.) models with different relative orientations of Stem III and the anti-terminator helix. (D) Two orthogonal views of the molecular envelope of the wild-type T-box complex. Placement of the modeled anti-terminator helix and its orientation with respect to Stem III were guided by restraining the backbone distance between the t-box sequence of the anti-terminator helix and the tRNA CCA sequence to 20 Å. The orientation of Stem III was determined by comparing the envelopes of the wild-type complex and T-box (S3.10):tRNAGly complex and by comparing the theoretical scattering profile calculated using the final structural model (Conformer 2) with the experimental data of the wild-type complex. Despite the model being incomplete, the fit is reasonable (Supplementary Figure S4) and the scattering curve captures the main features of the experimental data.
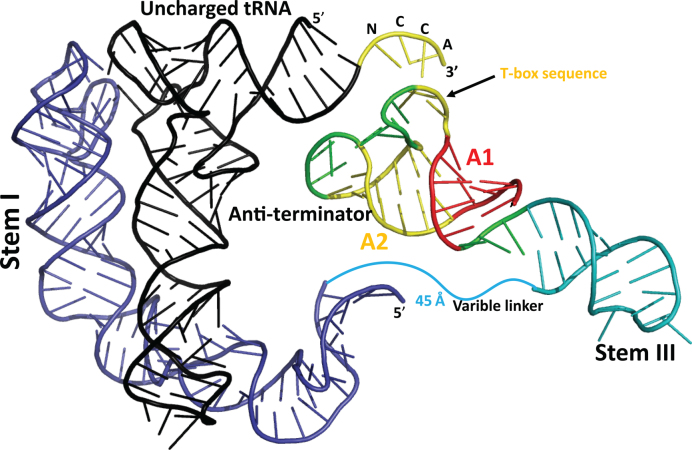
In the anti-terminated state, the A1 helix of the glyQS T-box leader is linked to Stem III by a 3 nt long single stranded region. The two major helices, Stem III and the A1/A2 anti-terminator helix, could be oriented relative to each other in many ways (Figure 4C). To determine the location and to model the position of the anti-terminator helix, we made use of two structural restrains as well as the experimental scattering data (Supplementary Figure S4) as a guide to assess the fit of our structural model. An initial model of the anti-terminator helix was generated based on its secondary structure (40) and was placed in the molecular envelope for the wild-type complex by orienting the T-box sequence bulge toward the 3΄ CCA end of the modeled tRNA. The corresponding backbone distance between the interacting nucleotides in the two RNA molecules was restrained to ∼20 Å, which corresponds to the backbone distance of base-paired nucleotides in an A-form RNA helix. Stem III was then attached to the anti-terminator helix by a 3 nt linker and its orientation was restrained based on its location in the molecular envelope (Figure 3D and Supplementary Figure S5) and the fit to the experimental data (Supplementary Figure S4). The shape of the smaller asymmetric half of the molecular envelope suggested that a long double stranded RNA helix with the same number of base pairs as Stem III and the anti-terminator helix could fit well in this region. The experimental scattering curve of the wild-type complex differed significantly from the theoretical curve computed from the crystal structure of the glyQS Stem I:tRNAGly complex (CRYSOL χ2:48; AquaSAXS χ2:4) (Supplementary Figure S4A). Inclusion of Stem III and the anti-terminator helix stacked on top of each other significantly improved the fit of the model (CRYSOL χ2:4.1; AquaSAXS χ2:1.4) to the experimental data (Supplementary Figure S4C), which supported the approximate placement of these T-box secondary structural elements. Inclusion of the 13 nt single stranded linker, which connects Stem III to Stem I, slightly improves the fit to the experimental data (CRYSOL χ2:3.8; AquaSAXS χ2:1.3), but this region was not included in the model or final analysis due to the lack of information to define its conformation. From the model, it was inferred that during read-through state the Stem III and the anti-terminator helices, could be stacked on top of each other to generate an extended co-axial stack (Figure 5). In the final model the distance between the 3΄ end of Stem I and the 5΄ end of Stem III is ∼45 Å, a gap that a 13 nt single stranded region could bridge (Figure 5). Note that these missing nucleotides account for ∼5% of the total mass, which may be part of the reason why the fit to the experimental data is not better.
Figure 5.
Molecular model of the Bacillus subtilis glyQS T-box:tRNAGly complex. The diagram depicts the position of the various secondary structural elements of the two interacting RNAs in the anti-terminated glyQS T-box riboswitch. A model for the anti-terminator conformation of the expression platform and Stem III (blue) was generated based on known secondary structure. The terminator helix in the anti-terminated state has a 7-nt loop (yellow) that harbors the T-box sequence. This conserved loop base-pairs with the 3΄ NCCA (yellow) sequence of an uncharged tRNA and is flanked by two small helices, A1 (red) and A2 (yellow). The model suggests that only in the read-through state can Stem III (blue) co-axially stack with the A1 helix, which in turn could stacks against the tRNA acceptor stem to generate an extended stack of nucleobases. The coordinates for Stem I and the uncharged tRNA were derived from PDB IDs: 4LCK (12) and 3Q1Q (17), respectively.
DISCUSSION
T-box riboswitches employs a large structured RNA molecule, the tRNA, as its effector making them unique among riboswitches. T-box riboswitches have a modular architecture where different structural elements are responsible for distinct functional roles. The two key aspects in the T-box mechanism is the recognition and binding of a cognate tRNA, and the assessment of the bound tRNA charged state. tRNA recognition is solely carried out by Stem I, which is the most conserved and longest RNA helix in a T-box, whereas the charged state sensing is executed by the expression platform, which is conformationally labile and can switch between two alternate conformations. These two major RNA helices are linked through a variable linker region that varies in complexity in different T-box riboswitches. Whereas the structural basis for tRNA recognition by Stem I is now known, the exact role of the variable linker region, the precise molecular basis for tRNA charge sensing, and the structural determinants for conformational switching of the expression platform are still poorly understood. In the absence of complete molecular models of the full-length T-box:tRNA complexes, it is not evident if the secondary structural elements in the variable region of a T-box make direct contacts with the cognate tRNA or if these elements play an indirect role in the T-box mechanism by facilitating cross-talk between the aptamer and expression platform. In this study, a molecular envelope of a glyQS T-box leader in complex with its cognate tRNAGly was calculated from SAXS data, which was used to derive a composite structural model of a T-box leader in the anti-terminated state. The most salient feature of the SAXS envelope is its relatively flat shape, which strongly suggests that all the major secondary structural elements of the two interacting RNA molecules lie in the same plane. In addition, the experimentally derived model of the T-box:tRNA complex shows a plausible location and orientation of two major stems, Stem III and the anti-terminator helix, and places them in a co-axial stack. These stems are missing from the available crystal structures, and therefore the SAXS-derived model provides new structural insights on the overall T-box architecture and their plausible role in the overall mechanism.
Stem III is a structurally conserved stem-loop in T-box riboswitches, but displays considerable variation in its sequence, length, and overall thermodynamic stability (Supplementary Table S2). The exact function of Stem III helix is not known, but it is speculated that it may act as a regulatory transcriptional pause site where an actively transcribing RNA polymerase could stall through a sequence independent backtracking mechanism (25,26). Similar transcriptional pausing is commonly found in other regulatory bacterial RNAs that operate through attenuation (26). For T-box riboswitches, it has been suggested that a transcriptional pause could provide a time window for the Stem I aptamer to fold correctly, thereby allowing it to engage with its cognate tRNA effectively (25). A specific pause event at the Stem III loop would also prevent premature transcription of the full-length terminator helix that would abolish its ability to sense an uncharged tRNA. However, the significance of transcriptional pausing in the T-box mechanism has not been completely elucidated under in vivo conditions. The structural model of glyQS T-box leader:tRNAGly complex derived from the SAXS envelope shows that Stem III does not interact directly with its cognate tRNA and could indeed fulfill the role of a transcription regulatory element. In addition to this, our model suggests a plausible role for Stem III in the T-box mechanism, where it can co-axially stack with the proximal end of the A1 helix and favor the anti-terminator conformation, thus preventing formation of the competing terminator conformation, which is thermodynamically more favorable by ∼ −16 kcal/mole (20). The current model for T-box regulators suggest that the distal end of the A1 helix in the anti-terminator stacks with the acceptor stem of an uncharged tRNA resulting in the formation of a long co-axial stack containing 29 layers of base pairs and triples (20). Based on the relative location of the A1 and A2 helices in our model, it appears that the tRNA acceptor stem could align with the distal end of A1 helix, but additional conformational changes would be required. Our model also suggests that the A2 helix is placed closer to the tRNA acceptor stem, where it can form either direct or indirect contacts with the tRNA backbone atoms. Based on these observations, we propose a model in which binding of an uncharged tRNA to Stem I triggers the formation of the anti-terminator conformation through base-pairing between the 3΄ end of an uncharged tRNA and the T-box sequence of the anti-terminator region. The anti-terminator helix is stabilized through synergistic co-axial stacking that utilizes the proximal as well as the distal end of the A1 helix. The stacking interaction involving the proximal end of the A1 helix and Stem III would further extend the proposed intermolecular stacking between the tRNA acceptor and the distal A1 helix (20) by an additional 7 bp. A charged tRNA bound to Stem I cannot stabilize the anti-terminator by similar structural rearrangements as the crucial base-pairing interactions between the 3΄ end of the tRNA and the T-box sequence are abolished due to tRNA aminoacylation. Loss of this interaction would favor the formation of a terminator helix in which the 7-nt loop harboring the T-box sequence is now part of the linker region joining Stem III to the terminator helix. In addition to this, the terminator conformation may place the terminator helix much further away from Stem III (Figure 1), as there are over 20 nt separating Stem III from the terminator helix preventing stacking of the terminator helix with Stem III.
CONCLUSION
Based on our structural model of the glyQS T-box:tRNAGly complex, we propose a new role for Stem III in the overall T-box mechanism. In the read-through or anti-terminated state, Stem III could serve a dual role and act as a transcription pause site and at the same time stabilize the thermodynamically less stable anti-terminator conformation through tertiary interactions that do not rely on stringent sequence conservation. The variability in length and sequence of Stem III could be important for its ability to fine-tune the overall stability of the anti-terminated state and different T-box riboswitches could exploit this structural feature to indirectly modulate the affinity for their cognate uncharged tRNA. Further experimental validation is still needed to corroborate these hypotheses derived from our proposed model. In addition, a higher resolution experimental model of the full-length glyQS T-box:tRNAGly is still awaited to elucidate the structural basis for tRNA charge sensing by a T-box as well as to determine the associated conformational changes needed for regulation of downstream genes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Steven Weigand of DND-CAT for his help and advice with the SAXS experiments and initial data processing. We acknowledge staff and instrumentation support from the Keck Biophysics Facility and the Structural Biology Facility at Northwestern University, and DND-CAT at the APS at Argonne National Laboratory. DND-CAT is supported by Northwestern University, E.I. DuPont de Nemours & Co., and The Dow Chemical Company. The use of the APS is supported by the Department of Energy (DOE). Support from the R.H. Lurie Comprehensive Cancer Center of Northwestern University to the Keck Biophysics and Structural Biology Facilities is acknowledged. We would like to thank Clarence Chan for providing the purified archaeal L7Ae protein used in this study and Kathryn Gunn for critical reading of the manuscript and comments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01GM058443 to A.M.] Funding for open access charge: NIH [R01GM058443].
Conflict of interest statement. None declared.
REFERENCES
- 1. Serganov A., Nudler E.. A decade of riboswitches. Cell. 2013; 152:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Breaker R.R. Riboswitches and the RNA world. Cold Spring Harb. Perspect. Biol. 2012; 4:a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sherwood A.V., Henkin T.M.. Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses. Annu. Rev. Microbiol. 2016; 70:361–374. [DOI] [PubMed] [Google Scholar]
- 4. Deigan K.E., Ferre-D’Amare A.R.. Riboswitches: discovery of drugs that target bacterial gene-regulatory RNAs. Acc. Chem. Res. 2011; 44:1329–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang J., Ferre-D’Amare A.R.. Structure and mechanism of the T-box riboswitches. Wiley Interdiscip. Rev. RNA. 2015; 6:419–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sherwood A.V., Grundy F.J., Henkin T.M.. T box riboswitches in Actinobacteria: translational regulation via novel tRNA interactions. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grundy F.J., Winkler W.C., Henkin T.M.. tRNA-mediated transcription antitermination in vitro: codon-anticodon pairing independent of the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11121–11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grundy F.J., Henkin T.M.. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993; 74:475–482. [DOI] [PubMed] [Google Scholar]
- 9. Grundy F.J., Rollins S.M., Henkin T.M.. Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: a new role for the discriminator base. J. Bacteriol. 1994; 176:4518–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rollins S.M., Grundy F.J., Henkin T.M.. Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol. Microbiol. 1997; 25:411–421. [DOI] [PubMed] [Google Scholar]
- 11. Grigg J.C., Ke A.. Structural determinants for geometry and information decoding of tRNA by T box leader RNA. Structure. 2013; 21:2025–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang J., Ferre-D’Amare A.R.. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature. 2013; 500:363–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grigg J.C., Chen Y., Grundy F.J., Henkin T.M., Pollack L., Ke A.. T box RNA decodes both the information content and geometry of tRNA to affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:7240–7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lehmann J., Jossinet F., Gautheret D.. A universal RNA structural motif docking the elbow of tRNA in the ribosome, RNAse P and T-box leaders. Nucleic Acids Res. 2013; 41:5494–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chan C.W., Chetnani B., Mondragon A.. Structure and function of the T-loop structural motif in noncoding RNAs. Wiley Interdiscip. Rev. RNA. 2013; 4:507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Korostelev A., Trakhanov S., Laurberg M., Noller H.F.. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006; 126:1065–1077. [DOI] [PubMed] [Google Scholar]
- 17. Reiter N.J., Osterman A., Torres-Larios A., Swinger K.K., Pan T., Mondragon A.. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010; 468:784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Henkin T.M. The T box riboswitch: A novel regulatory RNA that utilizes tRNA as its ligand. Biochim. Biophys. Acta. 2014; 1839:959–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang J., Nikonowicz E.P.. Solution structure of the K-turn and Specifier Loop domains from the Bacillus subtilis tyrS T-box leader RNA. J. Mol. Biol. 2011; 408:99–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang J., Ferre-D’Amare A.R.. Direct evaluation of tRNA aminoacylation status by the T-box riboswitch using tRNA-mRNA stacking and steric readout. Mol. Cell. 2014; 55:148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Caserta E., Liu L.C., Grundy F.J., Henkin T.M.. Codon-anticodon recognition in the Bacillus subtilis glyQS T box riboswitch: RNA-dependent codon selection outside the ribosome. J. Biol. Chem. 2015; 290:23336–23347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yousef M.R., Grundy F.J., Henkin T.M.. Structural transitions induced by the interaction between tRNA(Gly) and the Bacillus subtilis glyQS T box leader RNA. J. Mol. Biol. 2005; 349:273–287. [DOI] [PubMed] [Google Scholar]
- 23. Grundy F.J., Moir T.R., Haldeman M.T., Henkin T.M.. Sequence requirements for terminators and antiterminators in the T box transcription antitermination system: disparity between conservation and functional requirements. Nucleic Acids Res. 2002; 30:1646–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gerdeman M.S., Henkin T.M., Hines J.V.. Solution structure of the Bacillus subtilis T-box antiterminator RNA: seven nucleotide bulge characterized by stacking and flexibility. J. Mol. Biol. 2003; 326:189–201. [DOI] [PubMed] [Google Scholar]
- 25. Grundy F.J., Henkin T.M.. Kinetic analysis of tRNA-directed transcription antitermination of the Bacillus subtilis glyQS gene in vitro. J.Bacteriol. 2004; 186:5392–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang J., Landick R.. A two-way street: regulatory interplay between RNA polymerase and nascent RNA structure. Trends Biochem. Sci. 2016; 41:293–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kunkel T.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. U.S.A. 1985; 82:488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Birnboim H.C., Doly J.. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979; 7:1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C.. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987; 15:8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beckert B., Masquida B.. Synthesis of RNA by in vitro transcription. Methods Mol. Biol. 2011; 703:29–41. [DOI] [PubMed] [Google Scholar]
- 31. Weigand S.J., Keane D.T.. DND-CAT's new triple area detector system for simultaneous data collection at multiple length scales. Nucl. Instrum. Methods Phys. Res. A. 2011; 649:61–63. [Google Scholar]
- 32. Petoukhov M.V., Franke D., Shkumatov A.V., Tria G., Kikhney A.G., Gajda M., Gorba C., Mertens H.D., Konarev P.V., Svergun D.I.. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012; 45:342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Svergun D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992; 25:495–503. [Google Scholar]
- 34. Svergun D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999; 76:2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lipfert J., Chu V.B., Bai Y., Herschlag D., Doniach S.. Low-resolution models for nucleic acids from small-angle X-ray scattering with applications to electrostatic modeling. J. Appl. Crystallogr. 2007; 40:S229–S234. [Google Scholar]
- 36. Fang X., Wang J., O’Carroll I.P., Mitchell M., Zuo X., Wang Y., Yu P., Liu Y., Rausch J.W., Dyba M.A. et al. . An unusual topological structure of the HIV-1 Rev response element. Cell. 2013; 155:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Volkov V.V., Svergun D.I.. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003; 36:860–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kozin M.B., Svergun D.I.. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 2001; 34:33–41. [Google Scholar]
- 39. Schrodinger L.L.C. 2015; The PyMOL Molecular Graphics System, Version 1.8.
- 40. Popenda M., Szachniuk M., Antczak M., Purzycka K.J., Lukasiak P., Bartol N., Blazewicz J., Adamiak R.W.. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012; 40:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brunger A.T. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2007; 2:2728–2733. [DOI] [PubMed] [Google Scholar]
- 42. Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S. et al. . Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998; 54:905–921. [DOI] [PubMed] [Google Scholar]
- 43. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Svergun D., Barberato C., Koch M.H.J.. CRYSOL—a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995; 28:768–773. [Google Scholar]
- 45. Poitevin F., Orland H., Doniach S., Koehl P., Delarue M.. AquaSAXS: a web server for computation and fitting of SAXS profiles with non-uniformally hydrated atomic models. Nucleic Acids Res. 2011; 39:W184–W189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Burke J.E., Butcher S.E.. Nucleic acid structure characterization by small angle X-ray scattering (SAXS). Curr. Protoc. Nucleic Acid Chem. 2012; 7, doi:10.1002/0471142700.nc0718s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jones C.P., Cantara W.A., Olson E.D., Musier-Forsyth K.. Small-angle X-ray scattering-derived structure of the HIV-1 5΄ UTR reveals 3D tRNA mimicry. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:3395–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Taylor P., Rixon F., Desselberger U.. Rise per base pair in helices of double-stranded rotavirus RNA determined by electron microscopy. Virus Res. 1985; 2:175–182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.