Abstract
The CRISPR–Cas9 system is a powerful genome-editing tool useful in a variety of biotechnology and biomedical applications. Here we developed a synthetic RNA-based, microRNA (miRNA)-responsive CRISPR–Cas9 system (miR-Cas9 switch) in which the genome editing activity of Cas9 can be modulated through endogenous miRNA signatures in mammalian cells. We created miR-Cas9 switches by using a miRNA-complementary sequence in the 5΄-UTR of mRNA encoding Streptococcus pyogenes Cas9. The miR-21-Cas9 or miR-302-Cas9 switches selectively and efficiently responded to miR-21-5p in HeLa cells or miR-302a-5p in human induced pluripotent stem cells, and post-transcriptionally attenuated the Cas9 activity only in the target cells. Moreover, the miR-Cas9 switches could differentially control the genome editing by sensing endogenous miRNA activities within a heterogeneous cell population. Our miR-Cas9 switch system provides a promising framework for cell-type selective genome editing and cell engineering based on intracellular miRNA information.
INTRODUCTION
The bacterial and archaeal clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) system provided a powerful genome editing tool for a variety of biotechnology and biomedical applications (1,2). The engineered CRISPR–Cas9 system derived from Streptococcus pyogenes contains two components: the Cas9 endonuclease and a single guide RNA (sgRNA), which itself is a fusion of a designable CRISPR RNA (crRNA) and an universal trans-activating CRISPR RNA (tracrRNA). The Cas9 complex is recruited to a target DNA sequence by the sgRNA, forming an RNA–DNA hybrid. Subsequently, the endonuclease activity of Cas9 creates a DNA double strand break (DSB) at the target site and triggers a host DNA repair pathways to induce genomic alterations. To introduce the system into mammalian cells, several delivery approaches for Cas9 and the sgRNA have been tested, including viral vectors, plasmid DNAs, synthetic RNAs, and ribonucleoproteins (RNPs) (3–5). DNA-based delivery systems may induce unwanted side effects. For example, gene therapy using virus vectors may integrate the transgene into host genomic regions randomly, and induce oncogenesis in some cases (6). It has also been reported that plasmid delivery of the CRISPR–Cas9 system may cause genomic integration of the DNA fragment derived from the plasmid at off-target sites (7). Detection of the inserted DNA fragment at off-target sites is difficult, and the insertion might cause problems to host cells. In contrast, RNA-based delivery approaches are proposed to be safer than DNA-based delivery, since the limited expression window for RNA could reduce the risk of off-target mutagenesis while also avoiding the possibility of random genomic integration (3,4,8). Additionally, by using synthetic genetic circuits delivered by modified mRNAs (8), one may be able to control Cas9 protein expression post-transcriptionally by sensing intracellular signals. However, the post-transcriptional regulation of Cas9 activity by employing synthetic mRNA has remained a challenge.
Multicellular organisms consist of various cell types, hence cell type-specific genome editing will be an important tool for restricting genetic modifications to target cells and regulating the cell-fate within sub-populations in vitro and in vivo. The ideal system is one that modifies the target cells without affecting other cell types. In order to regulate the endonuclease activity of Cas9, several methods have been reported, including optogenetic or small molecule/protein-inducible systems (9–11), a tissue-specific promoter system (12), and transcriptional regulatory circuits using split-Cas9 (13). However, these reports have predominantly used DNA delivery approaches. And cell type-selective genome editing in a heterogeneous cell population has not been implemented previously.
MicroRNAs (miRNA) are small non-coding RNA that regulates gene expression through translational repression and/or mRNA degradation. The miRNA activities differ in each cell type and can therefore be used as a signature for a given target cell (14). We have previously reported a miRNA-responsive, synthetic mRNA (miRNA switch) that enables the detection and purification of target cells based on endogenous miRNA activities. By inserting a copy of the anti-sense sequence for the target miRNA (anti-miRNA) into the 5΄-UTR of the mRNAs, the switch allows for sensitive detection of the target miRNA upon hybridization and subsequent post-transcriptional repression of a downstream protein (15). Additionally, these miRNA switches function with RNA delivery to avoid random genome integration and purify target cell populations with high efficiency and selectivity (15,16).
We hypothesized that a combination of miRNA switch technology with the CRISPR–Cas9 system (i.e. miRNA-responsive Cas9 system: miR-Cas9 switch) would enable the regulation of genome engineering in a miRNA-dependent manner (Figure 1). In the present study, we newly designed a miR-Cas9 switch that controlled Cas9 activity based on an endogenous miRNA expressed in target human cells. Cas9 protein was translated from a miR-Cas9 switch in the absence of the target miRNA (ON-state), but was repressed in its presence (OFF-state). We used this system to attenuate Cas9 activity in HeLa cells, human induced pluripotent stem cells (hiPSCs), iPSC-derived differentiated neuronal cells. In turn, we demonstrate that cell-type selective genome editing can be achieved in heterogeneous cell populations.
Figure 1.
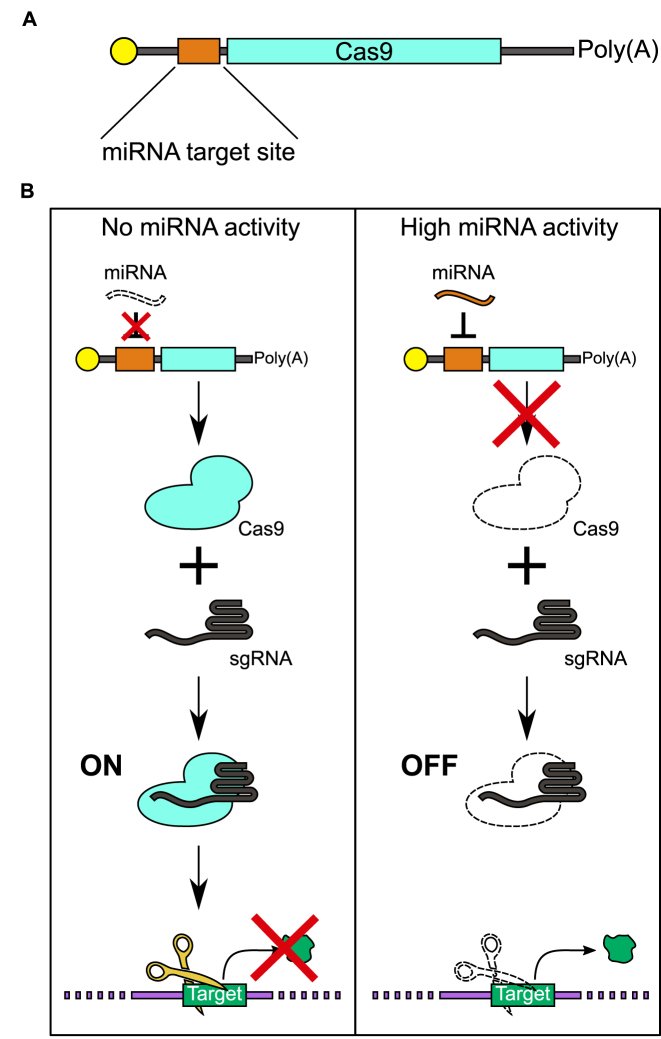
Schematic illustrations of the miR-Cas9 switch. (A) Design of miRNA-responsive Cas9 mRNA (miR-Cas9 switch). The synthetic mRNA contains an anti-reverse cap analog (yellow), miRNA target site (orange), Cas9 encoding sequence (cyan), and 120 nucleotide poly(A) tail. The miRNA target site is completely complementary to the miRNA of interest. (B) Overview of the miR-Cas9 switch system. The miR-Cas9 switch and sgRNA are introduced to cells by RNA transfection. Cas9 protein expressed from the mRNA forms a Cas9–sgRNA complex and digests the DNA in the case of no miRNA activity (ON, left). In contrast, in the case of high miRNA activity, interaction between the miRNA and Cas9 mRNA reduces Cas9 expression (OFF, right).
MATERIALS AND METHODS
Preparation of template DNA for IVT (in vitro transcription)
pAM-L7Ae was prepared according to the same method as pAM-tagBFP described in a previous report (15). The PCR product of tagBFP was inserted into modified pUC19 vector at a multi-cloning site to obtain pA9-tagBFP. The original sources of the genes and plasmid sequences are described in Supplementary Table S6. For the preparation of Cas9 mRNA, L7Ae mRNA and BFP mRNA templates, a 5΄-UTR without miRNA target sequences (control 5΄-UTR) and a 3΄-UTR were synthesized by hybridizing oligo-DNAs (oligo-DNAs lists are shown in Supplementary Table S1) followed by elongation; (94°C for 2 min, 13 cycles of 98°C for 10 s and 68°C for 10 s, and hold at 4°C). Cas9, L7Ae and BFP protein-coding regions were amplified by PCR with the appropriate primers (Supplementary Table S4) from pHL-EF1a-SphcCas9-iC-A (Addgene, Plasmid #60599), pAM-L7Ae and pA9-tagBFP, respectively. The plasmid DNA was removed following PCR by Dpn I treatment. All PCR products were purified by MinElute PCR purification kit (QIAGEN). The PCR products were then fused to construct a full DNA template for IVT via an additional PCR reaction. We conducted gel extraction when non-specific bands appeared.
For the sgRNA template, a modified protocol (17) was used. Briefly, a forward primer containing the T7 promoter sequence immediately followed by the gene-targeting sequence and a reverse primer encoding the remainder of the sgRNA sequence were used.
The complete list of combined primers and template for the PCR reactions is shown in Supplementary Table S4.
Cas9-, L7Ae- and BFP-mRNAs and sgRNA synthesis and purification
Cas9 mRNAs (with or without miRNA target sequences and with kink-turn motif), L7Ae mRNAs (with or without miRNA target sequences) and BFP mRNA (without miRNA target sequences) were prepared by using a MEGAscript kit (Ambion). In order to reduce the interferon response caused by long RNA, pseudouridine-5΄-triphosphate and 5-methylcytidine-5΄-triphosphate (TriLink Bio Technologies) were used instead of natural rUTP and rCTP, respectively (18). Guanosine-5΄-triphosphate was 5-fold diluted with an Anti Reverse Cap Analog (TriLink Bio Technologies) before the IVT reaction. The sgRNA was constructed using a MEGAshortscript kit (Ambion) according to the instruction manual. Because sgRNA with modified bases may cause downregulation of Cas9 activity, natural rNTPs were used for preparing sgRNA. The template DNA was degraded by TURBO DNase (Ambion), and the mRNAs and sgRNA were purified using a FavorPrep Blood/Cultured Cells total RNA extraction column (Favorgen Biotech) incubated with Antarctic Phosphatase (New England Biolabs) at 37°C for 30 min and then purified again using an RNeasy MinElute Cleanup Kit (QIAGEN). For further purification, sgRNA was electrophoresed, extracted from gel (10% polyacrylamide gel, 8.3 M urea), and ethanol-precipitated.
Cell culture
The heterozygously targeted AAVS1-CAG::GFP human iPSCs 317-12 line (hiPSC-EGFP) (19) was maintained in StemFit (Ajinomoto) on laminin-511 E8 (iMatrix-511, nippi). HeLa-EGFP cells were cultured in DMEM High Glucose (nacalai tesque) supplemented with 10% FBS (JBS) and Hygromycin B (50 μg/ml). Normal HeLa cells were cultured in DMEM High Glucose (nacalai tesque) supplemented with 10% FBS (JBS). All cell lines were cultured at 37°C with 5% CO2.
RNA transfection
For all assays, HeLa-EGFP and normal HeLa cells were seeded in uncoated 24-well plates, while iPS-EGFP cells were seeded in laminin-coated 24-well plates (all cells were seeded at a density of 5 × 104 cells/well). Appropriate synthetic RNAs were transfected into the cells using a Stemfect RNA transfection kit according to the manufacturer's protocol (Stemgent). The medium was changed 4 h after transfection. Cells were cultured for 24 h (Evaluation of transfection efficiency, Evaluation of gene expression level and Evaluation of Cas9 protein expression level), 48 h (Cell killing assay by targeting Alu1 element) or 72 h (T7E1 assay, Cas9 activity assay, Co-culture assay for cell-type-selective genome editing and Construction of miR-Cas9-ON switch) after transfection. Before analysis, cultured cells were captured by an IX81 microscope with output to a CCD camera (Olympus).
Cas9 activity assay
100 ng Cas9 mRNA, 300 ng sgRNA and 5 pmol miRNA inhibitor/negative control (ThermoFisher, optional) were used. MiRNA negative control (referred to as negative miRNA inhibitor henceforth) was provided by ThermoFisher as mirVana™ miRNA Mimic Negative Control and is a random sequence Anti-miR molecule that has been extensively tested in human cell lines and tissues and validated to not produce identifiable effects on known miRNA function. At 72 h after RNA transfection of miR-Cas9 switch, cells were washed with PBS. HeLa-EGFP cells were treated with 100 μl 0.25% trypsin–EDTA, incubated at 37°C with 5% CO2 for 5 min, and then 100 μl cultured medium was added. iPS-EGFP cells were treated with 200 μl Accumax (Funakoshi) and incubated at 37°C with 5% CO2 for 10 min. These cells were collected into 1.5 ml tubes, stained with SYTOX Red dead-cell stain (ThermoFisher), and incubated at room temperature for 15 min with protection from ambient light. Samples were analyzed on an Aria-II (BD Biosciences), Accuri C6 (BD Biosciences) or a LSRFortessa (BD Biosciences) flow cytometer.
From histogram or dot plots (SSC-A versus EGFP), we gated for EGFP negative and positive populations, which correspond to successful or non-successful knock-out of EGFP gene by Cas9, respectively.
Co-culture assay for cell-type-selective genome editing
Before collection, cells were washed with PBS. Cells were treated with 200 μl Accumax (Funakoshi) and incubated at 37°C with 5% CO2 for 10 min. These cells were collected into 1.5 ml tubes and centrifuged at 1000 rpm and room temperature for 5 min. After supernatant was discarded, 50 μl PBS + PI (BD Biosciences) mixture and 2.5 μl Alexa Fluor® 647 mouse anti-human TRA-1-60 antigen (BD Biosciences) were added and incubated at room temperature for 20 min with protection from ambient light. After incubation, 1 ml PBS was added and centrifuged at 1000 rpm and room temperature for 5 min (2 times). After adding 400 μl PBS, the samples were analyzed on an LSR (BD Biosciences) flow cytometer.
For immunostaining, transfected cells were treated with the following protocol. Cells were washed with PBS (2 times) and treated with 200 μl BD Cytoxfix™ fixation buffer (BD Biosciences) for 10 min at room temperature. After 10 min, buffer was removed, and cells were washed (2 times). Cells were stained with 200 μl antibody (Alexa Fluor® 647 mouse anti-human TRA-1-60 antigen (BD Biosciences)) diluted in PBS (5 μl antibody/200 μl PBS) for 1 h at room temperature with protection from ambient light. After staining, cells were washed with PBS (2 times). Upon adding 500 μl PBS, cells were captured by the IX81 microscope with output to the CCD camera (Olympus).
Construction of miR-Cas9-ON switch
Harvesting and analysis were done as in Cas9 activity assay above. Briefly, sample cells were collected 72 h after RNA transfection and stained with SYTOX Red dead-cell stain. These samples were analyzed on an Accuri flow cytometer.
Flow cytometry
An Accuri flow cytometer with FL1 (533/30 nm) and FL4 (675/25 nm) filters was used. EGFP and annexin V, Alexa Fluor 488 conjugate were detected with FL1, and SYTOX Red dead-cell stain was detected with FL4. EGFP, SYTOX Red dead-cell stain, PI and Alexa Fluor® 647 mouse anti-human TRA-1-60 antigen were detected with a blue laser (488 nm) and FITC filter (530/30 nm), a red laser (633 nm) and APC filter (660/20 nm), a blue laser and PE filter (575/25 nm), and a red laser and APC filter, respectively, using Aria-II or LSR flow cytometers.
Statistical analysis
Student's t test or Welch's t test (Supplementary Figure S3 only) were used for the statistical analysis. The levels of significance are denoted as *P < 0.05, **P < 0.01, ***P < 0.001, n.s., not significant. Student's t test and Welch's t test were performed using Excel (Microsoft).
RESULTS
Construction of miR-Cas9 switch
We first constructed Cas9 mRNA that contains a miRNA-responsive anti-sense sequence (anti-miR) positioned within the 5΄-UTR (Figure 1A). When the miRNA is inactive in cells, Cas9 protein will be produced from Cas9 mRNA and result in endonuclease activity that cleaves a target DNA sequence (Figure 1B, left). In contrast, when the miRNA is active in the cell, the expression from the Cas9 mRNA will be repressed through the interaction between the anti-miR region of the Cas9 mRNA and the endogenous miRNA. This interaction will cause post-transcriptional repression of Cas9 expression, reducing the level of DNA cleavage (Figure 1B, right). To evaluate the activity of Cas9, we employed sgRNAGFP, which cleaves an enhanced green fluorescent protein (EGFP) reporter gene integrated into the host cell's genome, and quantified Cas9 activity by assessing the loss of the EGFP fluorescence intensity (20).
We investigated whether the designed miR-Cas9 switch could control Cas9 endonuclease activity in HeLa-EGFP cells that constitutively express EGFP (Figure 2). In this study, we focused on miR-21-5p (miR-21) and miR-302a-5p (miR-302). MiR-21 is highly expressed in many cancer cells including HeLa cells and regulates tumor suppressor genes associated with proliferation, apoptosis and invasion (e.g. PTEN, PDCD4, BCL-2) (21,22). MiR-302 is predominantly expressed in human pluripotent stem cells including hiPSCs (15,23) and regulates pluripotency, proliferation, and cell cycle-related genes (e.g. MBD2, AKT1, CDK4). Indeed, we confirmed that miR-21 and miR-302 are highly expressed in HeLa cells and hiPSCs, respectively (Supplementary Figure S1). Thus, we chose miR-21 and miR-302 as target miRNAs to conduct HeLa cell- or hiPSC-specific genome editing.
Figure 2.
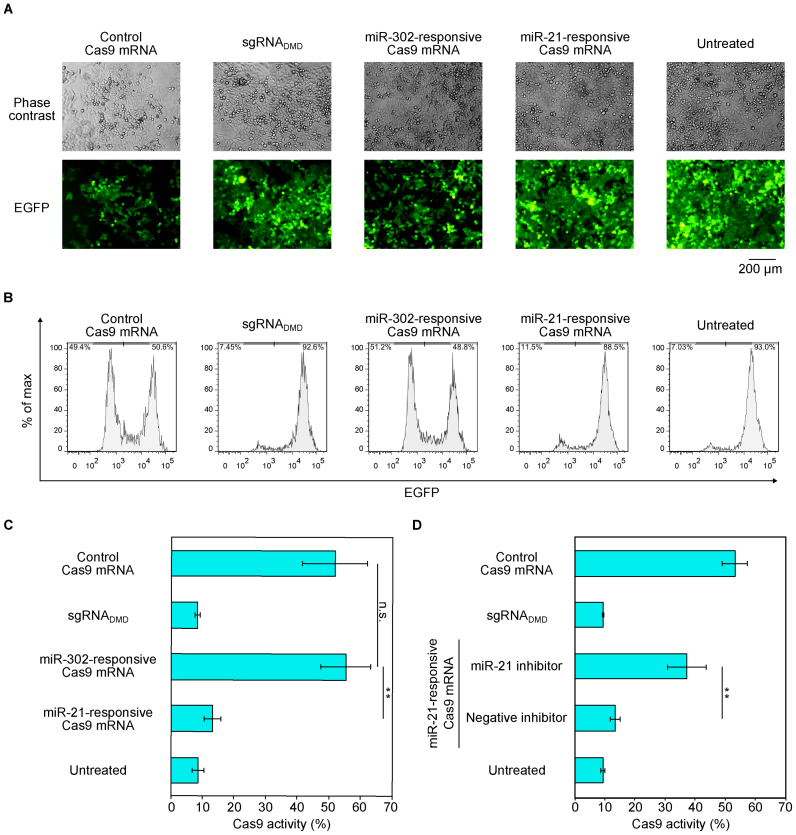
Regulation of Cas9 activity in HeLa cells by miR-21-Cas9 switch. (A) Representative phase contrast and fluorescent microscopic images at 72 h after RNA transfection. Scale bar represents 200 μm. (B) Representative flow cytometry histograms of EGFP expression. x- and y-axes indicate EGFP intensity and % of max, respectively. (C) Quantification of Cas9 activity from (B). The percentage of EGFP negative cells was defined as Cas9 activity. Cas9 activity was reduced in miR-21-responsive Cas9 mRNA. (D) Verification of endogenous miR-21-dependent Cas9 silencing by using miR-21 inhibitor. The miR-21 inhibitor rescued Cas9 activity in miR-21-responsive Cas9 mRNA, whereas a negative miRNA inhibitor did not. For the negative control, cells were transfected with Cas9 mRNA and a non-targeting sgRNA (sgRNADMD). Error bars indicate the mean ± SD (n = 3).
We prepared three Cas9 mRNAs, control Cas9 mRNA without anti-miR sequence, miR-21-responsive Cas9 mRNA (miR-21-Cas9 switch), and miR-302-responsive Cas9 mRNA (miR-302-Cas9 switch). We expected that Cas9 expression from the miR-21-Cas9 switch would be repressed in the presence of endogenous miR-21, resulting in a loss of Cas9 activity in HeLa cells. We co-transfected either control Cas9 mRNA or miR-21-Cas9 switch and sgRNAGFP into HeLa-EGFP cells. Three days after transfection, we analyzed the EGFP intensity by fluorescence microscopy and flow cytometry. These analyses showed a decrease in EGFP expression in control Cas9 mRNA-transfected cells compared with untreated cells, indicating successful knock-out of the EGFP gene by Cas9 mRNA and sgRNAGFP (Figure 2A and B). In contrast, there was no significant loss in EGFP expression in miR-21-Cas9 switch-transfected cells. The fluorescence intensity was similar to that of untreated cells or cells transfected with Cas9 mRNA and negative control sgRNADMD, which targets an unrelated genomic region (human Dystrophin gene, DMD) (24) (Figure 2A and B). To investigate the selectivity of our miR-Cas9 switch, we also transfected the miR-302-Cas9 switch, which was designed to respond to miR-302, a miRNA not expressed in HeLa cells (Supplementary Figure S1B). We observed a decrease in EGFP intensity to a level similar to positive control Cas9 mRNA, suggesting that the insertion of unrelated miRNA-responsive sequences in the 5΄UTR region of Cas9 mRNA does not affect its translation efficiency (Figure 2A and B). We further quantified Cas9 activities based on the fluorescence intensity obtained by flow cytometry analysis (Figure 2C). We observed a decrease in EGFP intensity in control Cas9 mRNA and miR-302-Cas9 switch transfected cells (∼50% of the cell population was EGFP-low), but EGFP intensity from miR-21-Cas9 switch transfected-cells was again similar to that of untreated cells or cells transfected with Cas9 mRNA and negative control sgRNADMD (∼90% of cells were EGFP-positive). In addition, we confirmed Cas9 protein expression levels from each mRNA by capillary-based immunodetection. We detected Cas9 protein produced from Cas9 mRNA and miR-302-Cas9 switch but not from miR-21-Cas9 switch (Supplementary Figure S2). These results indicate that Cas9 expression from miR-21-Cas9 switch was regulated by endogenous miR-21 in HeLa cells.
To confirm whether endogenous miR-21 is responsible for controlling Cas9 activity in HeLa-EGFP cells, the miR-21-Cas9 switch was co-transfected with either miR-21 inhibitor or miRNA inhibitor unrelated to miR-21 (negative inhibitor). We observed a decrease in EGFP intensity (i.e. increase of Cas9 activity) in the case of co-transfection with miR-21-Cas9 switch and miR-21 inhibitor, but not with miR-21-Cas9 switch and negative inhibitor (Figure 2D), indicating the rescue of Cas9 activity by inhibiting endogenous miR-21 activity. These results indicated that endogenous miR-21 in HeLa-EGFP cells is indispensable in the repression of Cas9 protein expression from the miR-21-Cas9 switch. Thus, we succeeded in constructing a synthetic RNA-delivered, miRNA-responsive Cas9 system in mammalian cells.
miR-Cas9 switch that targets endogenous Alu1 element
Next, we investigated the versatility of our switch by targeting a different region of the endogenous genome. We focused on the Alu1 element, the most common short interspaced element (SINE) in humans, distributed as thousands of copies throughout the human genome (25). We hypothesized that the cleavage of Alu1 regions by Cas9 would induce genotoxic stress resulting in cell death (Supplementary Figure S3A). We designed sgRNA targeting the Alu1 region (sgRNAAlu1), which exists in >200 000 sites in the human genome (219 114 sites in hg19), and transfected sgRNAAlu1 and Cas9 mRNA in HeLa cells. As expected, the number of dead cells increased in the case of treatment with control Cas9 mRNA and sgRNAAlu1, whereas cells untreated or treated with control Cas9 mRNA and sgRNADMD maintained a normal cell phenotype. Importantly, cells treated with miR-21-Cas9 switch and sgRNAAlu1 did not increase the number of dead cells, indicating that our miR-Cas9 switch can target endogenous genomic sequences and selectively control cell death signals (Supplementary Figure S3B–D).
miR-302-Cas9 switch modulates genome editing in human iPS cells
To further test our miR-Cas9 switch in different cell types, we next chose a human iPS cell line (201B7) that contains a single copy of the EGFP gene at the AAVS1 locus, which is often considered a safe-harbor for transgene targeting (hiPSC-EGFP) (19). We used the miR-302-Cas9 switch to selectively repress Cas9 activity in hiPSCs. Although the miR-302-Cas9 switch did not affect the Cas9 activity in HeLa cells (Figure 2A–C), we observed a repression of Cas9 activity (recovery of EGFP intensity) in hiPSCs transfected with miR-302-Cas9 switch and negative miRNA inhibitor, and de-repression of Cas9 activity in hiPSCs transfected with miR-302-Cas9 switch and miR-302 inhibitor. These results suggest that the suppression is dependent on endogenous miR-302 in hiPSCs (Supplementary Figure S4A).
We also performed T7E1 assays to investigate whether the change in EGFP intensity observed in the flow cytometry analysis was caused by mutations in the target EGFP gene due to the Cas9 activity. Treating the DNA products from genomic PCR with T7 Endonuclease I, we observed the target cleaved bands in cells transfected with Cas9 mRNA, or with miR-302-Cas9 switch and miR-302 inhibitor (Supplementary Figure S5). However, we did not observe these bands in cells treated with miR-302-Cas9 switch and negative miRNA inhibitor. The percentage of insertions and deletions (indels) at the EGFP gene as assessed by T7EI analysis showed similar trends as the flow cytometry analysis (Supplementary Figure S4A versus Supplementary Figure S5B), indicating that the loss of EGFP fluorescence intensity correlated with Cas9 nuclease activity. Moreover, we performed DNA sequencing and observed mutations at the target EGFP region in cells transfected with control Cas9 mRNA and miR-302-Cas9 switch with miR-302 inhibitor, but did not observe them with miR-302-Cas9 switch and negative miRNA inhibitor (Supplementary Figure S6). These results confirm that miR-302-Cas9 switch repressed Cas9-mediated genome engineering by detecting endogenous miR-302 in hiPSCs. Thus, we selectively controlled the activity of Cas9, in two different cell-types (HeLa and hiPS cells) by using miR-21- and miR-302-Cas9 switches, respectively. In addition, these two switches had negligible effect on endogenous mRNA expressions regulated by miR-21 or miR-302 (Supplementary Figure S7).
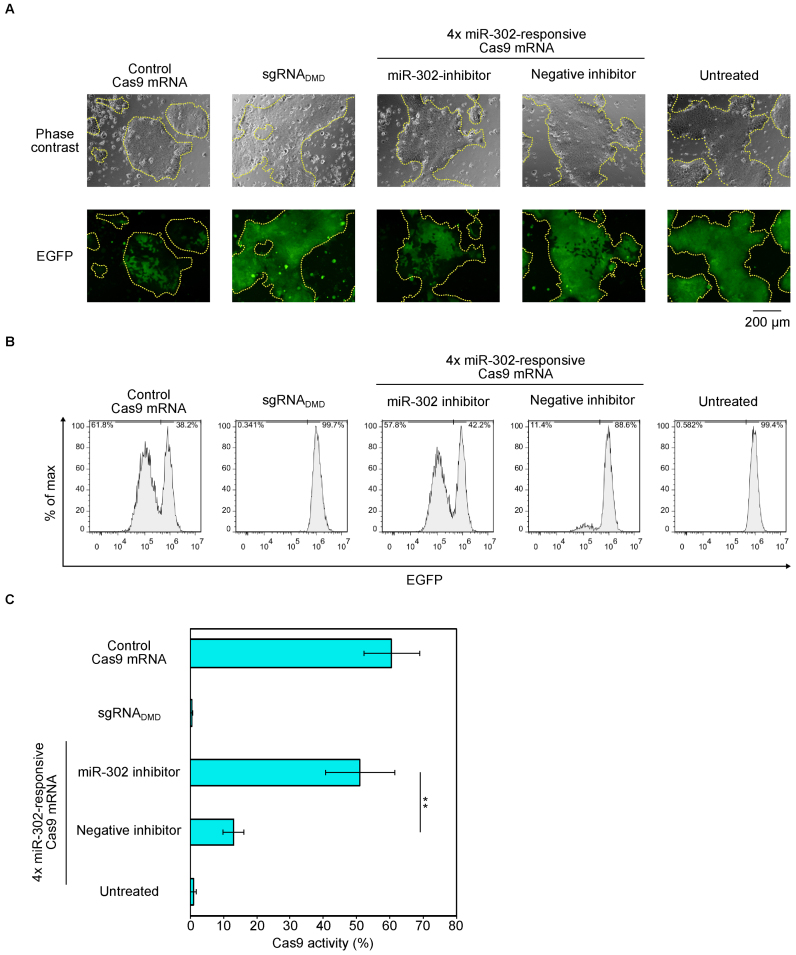
The Cas9 activity in hiPSCs was slightly lower (∼30%) than that in HeLa cells (∼50%), even though the sgRNA and mRNA transfection efficiency was high in both cells (∼90% efficiency for both cell types, Supplementary Figure S8) (16). To further increase the Cas9 activity and miR-302 sensitivity in hiPSCs, we engineered sgRNA and miRNA-responsive elements embedded in the 5΄UTR of Cas9 mRNA (Supplementary Text). We redesigned sgRNAGFP by removing the first tandem GG sequences (these do not hybridize with the target EGFP sequence), in order to increase the binding efficiency between the sgRNA and the target sequence (26). To enhance the miRNA binding, we inserted four tandem copies of anti-miR-302 sequences (instead of one copy) in the 5΄-UTR region of the switch (4× miR-302-Cas9 switch). The resulting switch increased Cas9 activity (∼60%) and responded to endogenous miR-302 to modulate Cas9 production, because miR-302 inhibitor recovered Cas9 activity (Figure 3). Thus, our miRNA-responsive Cas9 switch could be fine-tuned the balance between Cas9 activity and miRNA sensitivity.
Figure 3.
Regulation of Cas9 activity in iPS cells by 4x miR-302-Cas9 switch and improved sgRNA. (A) Representative phase contrast and fluorescent microscopic images at 72 h after RNA transfection. iPS-EGFP cells formed colonies (yellow dashed lines). Scale bar represents 200 μm. (B) Representative flow cytometry histograms of EGFP expression. x- and y-axes indicate EGFP intensity and % of max, respectively. (C) Quantification of Cas9 activity from (B). A miR-302 inhibitor rescued Cas9 activity expressed from miR-302-responsive Cas9 mRNA, whereas a negative miRNA inhibitor did not. To improve this system, 4x miR-302-responsive Cas9 mRNA (4 copies of miR-302 target site were tandemly inserted into 5΄-UTR) and sgRNA2GFP (removal of GG from 5΄ terminal) were used. Negative control is Cas9–sgRNADMD that targets DMD gene. Error bars indicate the mean ± SD (n = 3).
miR-302-Cas9 switch distinguishes hiPSCs and differentiated neurons
The activities of miRNAs dynamically change during differentiation. For example, miR-302 is highly expressed in undifferentiated hiPSCs but repressed to basal level in midbrain dopaminergic (mDA) neuronal cells differentiated from hiPSCs (16) (Supplementary Figure S1B, iPS versus mDA). We expected that miR-302-Cas9 switch could modulate Cas9 activity before and after differentiation and cleave a target gene in differentiated neurons but not in hiPSCs. Therefore, we differentiated hiPSCs into mDA neurons (16) and transfected the miR-302-Cas9 switch and sgRNA2GFP into these cells. As expected, in contrast to the results from hiPSCs, we observed Cas9 activity that was similar between control Cas9 mRNA and miR-302-responsive Cas9 mRNA (Supplementary Figure S9).
Cell-type selective genome editing in mixed cell populations
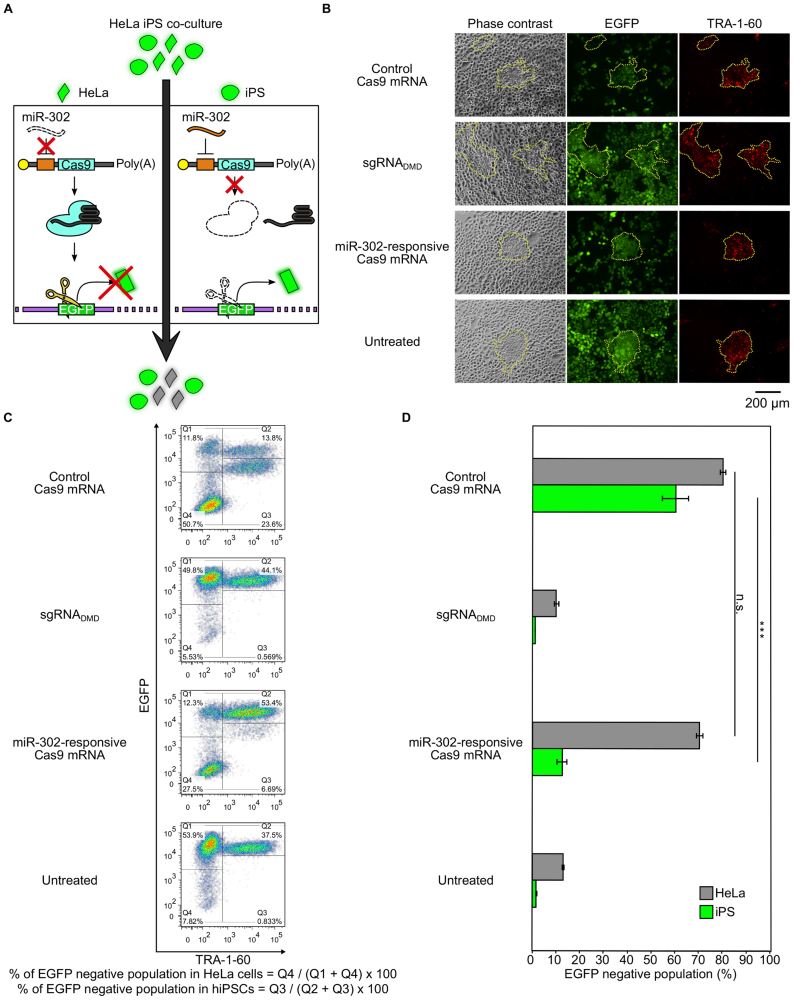
We next investigated whether miR-Cas9 switch could distinguish cells in mixed heterogeneous cell populations. Because the translation from the miR-21- and miR-302-responsive Cas9 mRNAs was selectively repressed in HeLa cells and hiPSCs, respectively, we applied our system to distinguish these two cell types. We mixed HeLa-EGFP and hiPS-EGFP cells in a dish and aimed to knockout HeLa-EGFP cells but not hiPS-EGFP cells, by employing the 4× miR-302-Cas9 switch (Figure 4A). To distinguish the two cell types, we stained the mixed cells with the TRA-1-60 antibody (a cell surface marker specific to undifferentiated pluripotent stem cells) for flow cytometry analysis. When control (non-regulated) Cas9 mRNA was transfected into the cell mixture, EGFP negative (knocked-out) cells were observed in both HeLa cells and hiPSCs. In contrast, when the 4× miR-302-Cas9 switch was transfected into the mixed cell population, the EGFP negative population was mainly observed in HeLa cells but not in hiPSCs (Figure 4B–D). This result showed that the miR-302-responsive Cas9 mRNA was preferentially translated to produce Cas9 in HeLa cells compared with hiPSCs, facilitating the genome editing activity in HeLa but preventing it in hiPSCs. Thus, our miRNA-responsive Cas9 system distinguishes different cell types within a heterogeneous cell population.
Figure 4.
Cell-type-selective genome editing in a mixed cell population. (A) Schematic illustration of cell-type-selective gene knockout in a mixed cell population. HeLa-EGFP and iPS-EGFP cells are mixed (HeLa/iPS co-culture), and 4× miR-302-responsive Cas9 mRNA is used. Because miR-302 activity is high in iPSCs, but not in HeLa cells, Cas9 protein is selectively produced in HeLa cells (low EGFP intensity). (B) Representative fluorescent microscopic images at 72 h after RNA transfection. Phase contrast, EGFP and TRA-1-60 indicate phase contrast, EGFP fluorescent images and Alexa Fluor 647 signal, respectively. Areas in the yellow dashed lines indicate TRA-1-60 positive hiPSCs. Scale bar represents 200 μm. (C) Representative dot plots. x- and y-axes indicate TRA-1-60 and EGFP, respectively. Q1 and Q4 indicate HeLa cells (TRA-1-60 negative). Q2 and Q3 indicate hiPSCs (TRA-1-60 positive). (D) EGFP knockout efficiencies of hiPSCs and HeLa cells in a co-culture condition. Bar graphs show EGFP negative populations. Gray and light green boxes show HeLa and hiPSCs, respectively. 4× miR-302-responsive Cas9 mRNA was used for the experiments. Negative control is Cas9–sgRNADMD that targets DMD gene. Error bars indicate the mean ± SD (n = 3).
miR-Cas9-ON switch that enhances Cas9 production by sensing target miRNA
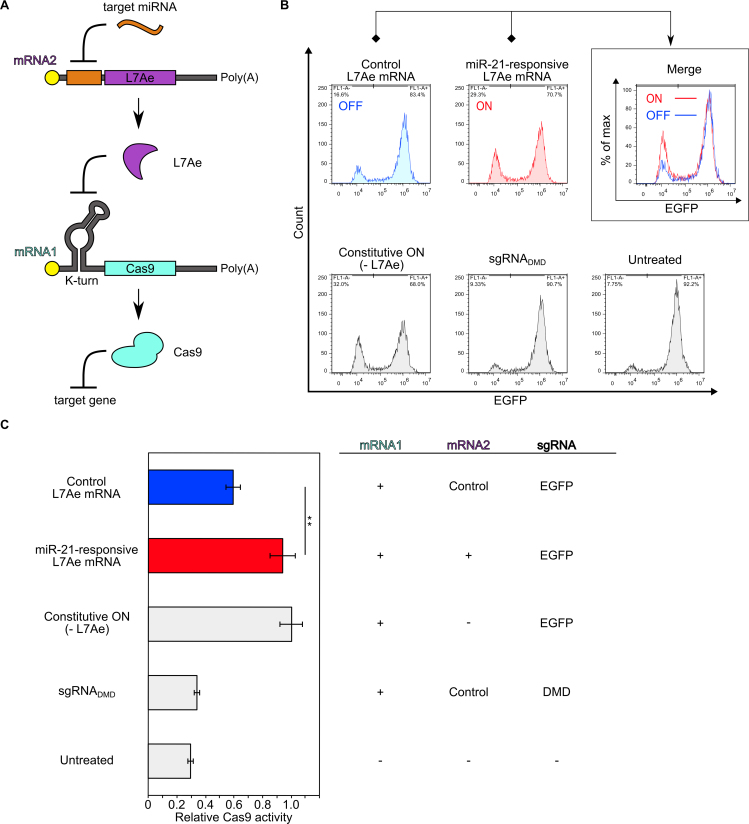
Thus far, our miR-Cas9 switch system was able to achieve potent Cas9 repression in target cells. In order to address Cas9 activation in a cell-type specific manner, we constructed a miR-Cas9-ON switch, which up-regulates the translation of Cas9 mRNA by responding to a target miRNA (Figure 5). We mimicked the topology of a cell-classifier circuit that is composed of two mRNA species (Cas9-coding mRNA1 and RNA-binding protein (L7Ae)-coding mRNA2) (8). We designed the 5΄-UTR sequence of each mRNA to contain either the L7Ae-binding kink-turn RNA motif (mRNA1) or the miR-21-responsive antisense sequence (mRNA2). In the absence of the target miRNA activity, L7Ae will be produced from mRNA2 and repress the translation of Cas9 from mRNA1 through the L7Ae-kink-turn RNA interaction. In contrast, in the presence of the target miRNA, L7Ae translation is repressed, which in turn de-represses Cas9 translation (Figure 5A). As designed, the co-transfection of both mRNAs and the sgRNA increased the level of Cas9 activity by responding to miR-21 in HeLa cells compared with untreated cells or cells transfected with mRNA1 and control L7Ae mRNA without miR-21-responsive element (Figure 5B, C). Thus, this system can selectively increase the level of Cas9 activity in target cells, by sensing the miRNA. Thus, both miR-Cas9-OFF and miR-Cas9-ON switches can be constructed to detect endogenous miRNA signatures in mammalian cells, expanding the possibilities for selective genome editing.
Figure 5.
Construction of miR-Cas9-ON switch. (A) Mechanism of the ON system. The expressions of L7Ae from mRNA2 and Cas9 protein from mRNA1 are suppressed by target miRNA (here, miR-21) and L7Ae, respectively. L7Ae suppresses the translation of Cas9 protein by interacting with its binding motif (K-turn), and miR-21 suppresses the translation of L7Ae by interacting with its anti-sense region, resulting in Cas9 translation. (B) Flow cytometry histograms of EGFP expression. The x- and y-axes indicate EGFP intensity and cell number, respectively. An increase of the EGFP negative population was observed in miR-21-responsive L7Ae mRNA (ON) compared with control L7Ae mRNA (OFF). This experiment was repeated three times and representative histograms are shown. Control L7Ae mRNA (L7Ae mRNA without miRNA-responsive element). Constitutive ON (without L7Ae mRNA). (C) Quantification of relative Cas9 activity from (B). An increase of the EGFP negative population was observed in miR-21-responsive L7Ae mRNA (ON) compared with control L7Ae mRNA (OFF). Each Cas9 activity was normalized by Constitutive ON (- L7Ae). Control L7Ae mRNA (L7Ae mRNA without miRNA-responsive element). Constitutive ON (without L7Ae mRNA). Negative control is Cas9–sgRNADMD that targets DMD gene. Error bars indicate the mean ± SD (n = 3).
DISCUSSION
In this study, we developed a miRNA-responsive Cas9 switch system (miR-Cas9 switch) that is capable of performing cell-type selective genome editing by detecting an endogenous miRNA signature, via a synthetic RNA delivery approach. We can repress or activate Cas9 expression in the presence or absence of the target miRNA, respectively, by employing our miR-Cas9-OFF switch. Our method can selectively control Cas9 activity that cleaved either an exogenous reporter gene (EGFP) integrated into the genome or an endogenous repetitive genomic sequence (Alu1) by detecting endogenous miRNA signatures: miR-21-5p and miR-302a-5p in HeLa and hiPSCs, respectively (Figures 2 and 3). We also demonstrated the application of our miR-Cas9 switch for the selective induction of genome editing in HeLa cells within a mixed (hiPSCs and HeLa) cell population (Figure 4). Additionally, by employing a cell classifier circuit with an RNA-binding protein (L7Ae), we constructed a miR-Cas9-ON switch, which up-regulates Cas9 activity when the target miRNA is present in the cell (Figure 5).
We believe that our miR-Cas9 switch could provide a promising framework to engineer genomes of target cells for the following reasons: (1) RNA delivery approaches are valuable for CRIPSR-Cas9 systems, as synthetic RNA may avoid potentially harmful genomic integration (8). (2) Our system can be used as a cell-type selective genome-editing tool that responds to endogenous miRNA signatures in target cells. Unlike non-cell autonomous regulation systems (e.g. small chemicals or light), Cas9 activity is controllable dependent on cell-state (miRNA activity). In fact, we demonstrated cell-type-selective genome editing capability in a heterogeneous cell population (Figure 4), although further improvement of the performance of the OFF and ON switches (e.g. avoiding translation of Cas9 protein from kink-turn-Cas9 mRNA in the presence of L7Ae) is important to reduce leaky expression of Cas9 in non-target cells. This improvement could be achieved by employing switches that detect multiple miRNAs (15). (3) MiRNA activities dynamically change with development and cell reprograming. Thus, by choosing an appropriate miRNA signature that represents the target cell type or the cell state, we could selectively control genome editing activity in a variety of cell types. To reach this potential, it will be important to expand our knowledge about active and distinctive miRNA signature(s) in different cell types (15). (4) Finally, our technology should be applicable to other Cas9 variants (e.g. orthologs, engineered Cas9/dCas9 proteins (27,28)) and other miRNAs in a plug-and-play manner. Additionally, we believe that our miR-Cas9 switch could have important medical applications. For example, we could maintain or purify target cells (e.g. iPS cells) but selectively eliminate non-target cells by targeting repetitive elements (e.g. Alu1) using miR-Cas9 switch. Also, in order to develop a safe and selective genome editing system in vivo, we may be able to avoid unexpected off-target effects in non-target cells (e.g. germ cells), by using germ cell-specific miRNA-responsive switch. The transient and conditional expression of our system using synthetic RNA could provide genome editing as a potentially ‘one-shot therapy’ to treat numerous genetic diseases. In conclusion, our miR-Cas9 switch combined with a RNA-delivery approach can selectively engineer genomes of target cells by detecting an endogenous miRNA signature, with potential benefits for synthetic biology, genome editing field and future therapeutic applications.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Hongmei Lisa Li (Kyoto University), Dr Kanae Mitsunaga (Kyoto University) and Saito laboratory members for kind advice about the experimental conditions, data analysis and discussion. We also thank Dr Peter Karagiannis (Kyoto University) and Ms Yukiko Nakagawa & Miho Nishimura for critical reading of the manuscript and administrative support, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
JSPS KAKENHI [15H05722, 24104002, 15H02376 to H.S. and 15H05581 to A.H.]; Naito Foundation (to H.S.); Canon Foundation (to H.S.); Nakatani Foundation (to H.S.); JSPS Fellows [16J10715 to M.H.]. Funding for open access charge: JSPS KAKENHI [15H05722 to H.S.].
Conflict of interest statement. Kyoto University has filed a patent application broadly relevant to this work. M. Hirosawa and H. Saito are the investigators of record listed on the patent application.
REFERENCES
- 1. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liang X., Potter J., Kumar S., Zou Y., Quintanilla R., Sridharan M., Carte J., Chen W., Roark N., Ranganathan S. et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015; 208:44–53. [DOI] [PubMed] [Google Scholar]
- 4. Yin H., Song C.-Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A. et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016; 34:328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Rivera R.M.C., Madhavan S., Pan X., Ran F.A., Yan W.X. et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016; 351:403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thomas C.E., Ehrhardt A., Kay M.A.. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003; 4:346–358. [DOI] [PubMed] [Google Scholar]
- 7. Kim S., Kim D., Cho S.W., Kim J., Kim J.-S.. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014; 24:1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wroblewska L., Kitada T., Endo K., Siciliano V., Stillo B., Saito H., Weiss R.. Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat. Biotechnol. 2015; 33:839–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hemphill J., Borchardt E.K., Brown K., Asokan A., Deiters A.. Optical Control of CRISPR/Cas9 Gene Editing. J. Am. Chem. Soc. 2015; 137:5642–5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zetsche B., Volz S.E., Zhang F.. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 2015; 33:139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y., Zhan Y., Chen Z., He A., Li J., Wu H., Liu L., Zhuang C., Lin J., Guo X. et al. Directing cellular information flow via CRISPR signal conductors. Nat. Methods. 2016; 13:938–944. [DOI] [PubMed] [Google Scholar]
- 12. Liu Y., Zeng Y., Liu L., Zhuang C., Fu X., Huang W., Cai Z.. Synthesizing AND gate genetic circuits based on CRISPR–Cas9 for identification of bladder cancer cells. Nat. Commun. 2014; 5:5393. [DOI] [PubMed] [Google Scholar]
- 13. Ma D., Peng S., Xie Z.. Integration and exchange of split dCas9 domains for transcriptional controls in mammalian cells. Nat. Commun. 2016; 7:13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mullokandov G., Baccarini A., Ruzo A., Jayaprakash A.D., Tung N., Israelow B., Evans M.J., Sachidanandam R., Brown B.D.. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods. 2012; 9:840–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miki K., Endo K., Takahashi S., Funakoshi S., Takei I., Katayama S., Toyoda T., Kotaka M., Takaki T., Umeda M. et al. Efficient detection and purification of cell populations using synthetic MicroRNA switches. Cell Stem Cell. 2015; 16:699–711. [DOI] [PubMed] [Google Scholar]
- 16. Parr C.J.C., Katayama S., Miki K., Kuang Y., Yoshida Y., Morizane A., Takahashi J., Yamanaka S., Saito H.. MicroRNA-302 switch to identify and eliminate undifferentiated human pluripotent stem cells. Sci. Rep. 2016; 6:32532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bassett A.R., Tibbit C., Ponting C.P., Liu J.-L.. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013; 4:220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Warren L., Manos P.D., Ahfeldt T., Loh Y., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A. et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010; 7:618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oceguera-Yanez F., Kim S.-I., Matsumoto T., Tan G.W., Xiang L., Hatani T., Kondo T., Ikeya M., Yoshida Y., Inoue H. et al. Engineering the AAVS1 locus for consistent and scalable transgene expression in human iPSCs and their differentiated derivatives. Methods. 2016; 101:43–55. [DOI] [PubMed] [Google Scholar]
- 20. Fu Y., Foden J.a, Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D.. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013; 31:822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feng Y., Tsao C.. Emerging role of microRNA-21 in cancer (review). Biochem. Rep. 2016; 5:395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xie Z., Wroblewska L., Prochazka L., Weiss R., Benenson Y.. Multi-Input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011; 333:1307–1311. [DOI] [PubMed] [Google Scholar]
- 23. Subramanyam D., Lamouille S., Judson R.L., Liu J.Y., Bucay N., Derynck R., Blelloch R.. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 2011; 29:443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H.L., Fujimoto N., Sasakawa N., Shirai S., Ohkame T., Sakuma T., Tanaka M., Amano N., Watanabe A., Sakurai H. et al. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR–Cas9. Stem Cell Rep. 2015; 4:143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cordaux R., Batzer M.A.. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009; 10:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S., Kim J.-S.. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014; 24:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V. V, Nguyen N.T., Zheng Z., Gonzales A.P.W., Li Z., Peterson R.T., Yeh J.-R.J. et al. Engineered CRISPR–Cas9 nucleases with altered PAM specificities. Nature. 2015; 523:481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F.. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016; 351:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.