Abstract
RGG/RG domains are the second most common RNA binding domain in the human genome, yet their RNA-binding properties remain poorly understood. Here, we report a detailed analysis of the RNA binding characteristics of intrinsically disordered RGG/RG domains from Fused in Sarcoma (FUS), FMRP and hnRNPU. For FUS, previous studies defined RNA binding as mediated by its well-folded domains; however, we show that RGG/RG domains are the primary mediators of binding. RGG/RG domains coupled to adjacent folded domains can achieve affinities approaching that of full-length FUS. Analysis of RGG/RG domains from FUS, FMRP and hnRNPU against a spectrum of contrasting RNAs reveals that each display degenerate binding specificity, while still displaying different degrees of preference for RNA.
INTRODUCTION
The arginine/glycine-rich (RGG/RG) domain is prevalent throughout eukaryotes and the second most common RNA-binding domain (RBD) in the human genome (1–6). The importance of decoding RGG-mediated RNA recognition is underscored by the observation that these RNPs regulate all levels of RNA metabolism including transcription, RNA processing, nucleocytoplasmic shuttling and translation (4,5,7). Furthermore, mutations in RGG/RG proteins are implicated in several neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS), fragile X syndrome and spinal muscular atrophy (4). Unlike the most common RBD, the RNA recognition motif (RRM), the RNA binding properties of RGG/RG domains are still poorly defined. A key challenge for understanding the cellular functions of the rapidly expanding set of RNAs comprising the transcriptome is to identify and characterize their interactions with RNA-binding proteins. This is particularly difficult for RNA-binding proteins that engage a large number of transcripts, suggesting ‘promiscuous’ or non-specific binding (7–12).
RGG/RG domains are intrinsically disordered (1), and thus do not adopt a single, stable structure in the absence of RNA but instead have conformational plasticity and adaptability. This feature may facilitate flexible targeting to a variety of RNAs because their own conformational flexibility provides a larger interaction surface area. While for some proteins, such as hnRNPU, the RGG/RG domain is the only identified RBD (13), this motif is most often found in proteins possessing other RBDs (4–6). This relationship between disordered RGG/RG and structured, classic RBDs, such as the RRM and KH domain, is also poorly understood.
The prevalence of RGG/RG domains among RNA-binding proteins has only recently been appreciated (14). Thus far, data suggest that RGG/RG domains may display some selectivity in RNA binding (14–19). For example, the RGG/RG domain of FMRP was found to bind tightly to an in vitro selected aptamer, Sc1, containing a G-quadruplex structure (14,15,18–19). While the G-quadruplex is the dominant feature of the Sc1 aptamer, the RGG/RG peptide sits in the interface between the duplex and quadruplex through a shape complementarity interaction, with the majority of protein-base contacts mediated by two Watson–Crick G-C pairs in the duplex (14,15).
If the preference of RGG/RG domains were actually directed to a particular conformation involving double-stranded RNA, this interaction would not be completely non-specific. Instead, the structure/sequence requirements for binding could appear frequently throughout the transcriptome (7–11). This behavior may explain reports of binding specificity for well-ordered RBDs that are inconsistent with those of a protein harboring both RBDs and RGG/RG domains. The RBDs of these RGG/RG proteins, such as the RRMs of hnRNPA1 or hnRNPA2/B1, display a robust specificity in vitro, but these target motifs are found only in a minor fraction of experimentally identified target sites within the transcriptome (20–22). These studies, however, do not shed light on how RGG/RG domains facilitate the molecular recognition of a subset of cellular RNAs (20).
RGG/RG domains are prevalent in many if not most heterogeneous ribonuclear particle (hnRNP) proteins. A particularly prominent family of RGG/RG domain proteins and subset of the hnRNP family is the FET family, comprised of FUS, EWSR1 and TAF15 (7,23,24). Each FET protein has three RGG/RG domains interspersed between structured domains. FET proteins are conserved throughout metazoans. In humans, they are ubiquitously expressed in all tissues and are among the most highly expressed proteins in the cell (7,12,25). These three nuclear proteins predominantly associate with RNA Pol II and pre-mRNA, which affects both transcription and mRNA processing (12,26,27). In addition to having multiple RGG/RG domains, the number of individual RGG or RG repeats are high, with up to nine repeats, suggesting these proteins can provide a useful model of RNA-binding by a RGG/RG domain.
Highlighting the challenges to studying RGG/RG domain activity, sequencing studies (iCLIP, CLIP-seq, HITS-CLIP and PAR-CLIP) have raised more questions than answers about the RNA-binding specificity of the Fused in Sarcoma (FUS) protein (12,23,27–31). These experimental protocols applied to FUS seek to identify all of the RNA sequences that co-immunoprecipitate with the protein, to which computational analysis is applied to detect any sequence motif and hypothetical structure that may be enriched. To date, results from these studies have failed to provide agreement regarding an RNA sequence or structure targeted by FUS (7,8). Neither have the sequence-structure motifs proposed stood up to more rigorous biochemical scrutiny (7,8). This may, in part, result from the fact that sequences identified in FUS immunoprecipitations represent the majority of transcribed RNAs in the cell. Also, FUS and other hnRNP proteins are known to oligomerize along RNA, reducing the significance of any target over that of adjacent sequences (7,20,27,29,32,33). Finally, many hnRNPs, like FUS, possess low-complexity (LC) domains, that self-assemble into large RNA-rich granules (25,32–43). Taken together, the bulk of sequences that immunoprecipitate with promiscuous RNA-binding proteins, like FUS, may not be reflective of RNA sequences or structures with higher affinity for FUS or its RBDs.
To define the RNA binding activity of RGG/RG domains, we have determined the RNA binding characteristics of each domain of the FET protein FUS using electrophoretic mobility shift assay (EMSA), isothermal titration calorimetry (ITC) and pulldown assays. For FUS, we find binding largely driven by the affinity of the RGG/RG domains for RNA and, in fact, the RGG/RG domains alone have substantial RNA binding activity as opposed to the RRM and zinc finger (ZnF) domains. Supporting this observation, we find that the RGG/RG domains drive binding affinity in vitro and in vivo. To further define the RNA binding characteristics of RGG/RG domains, those of FUS were contrasted with those of two additional proteins FMRP and hnRNPU, which showed varied degrees of preference in binding to RNAs of different sequence and structure composition. Taken together, we reveal a newly defined specificity of RGG/RG domains that is degenerate, and provide a model for the significance of intrinsic disorder to RNA binding.
MATERIALS AND METHODS
Protein expression and purification
All protein constructs were cloned into pET30b vector system with N-terminal His6-MBP tag and transformed into BL21 (DE3) Rosetta Escherichia coli cells. A total of 10 ml LB-Kanamycin bacterial culture was grown overnight, and then inoculated into 1 l LB medium. Cultures were incubated at 37°C until OD600 reached ∼0.6. Protein expression was induced by 0.5 mM Isopropyl b-D-1-thiogalactopyranoside (IPTG) and 100 μM ZnCl2 was added to the cultures if the expressed protein included the ZnF domain. The cultures were incubated at 20°C overnight. Bacterial cells were pelleted at 1500 g and resuspended in lysis buffer (1 M KCI, 1 M Urea, 50 mM Tris–HCl pH 7.4, 10 mM imidazole, with or without 100 μM ZnCl2). Cells were lysed using an Emulsiflex C3 homogenizer. The cell lysates were clarified by centrifugation at 17 000 g for 30 min and the supernatants were incubated with Ni-NTA sepharose beads on an orbital shaker for 1 h at 4°C. Beads were centrifuged at 300 g for 2 min and washed three times in lysis buffer and once in lysis buffer supplemented with 100 mM Imidazole. Proteins were eluted in lysis buffer supplemented with 250 mM Imidazole. Maltose binding protein (MBP) tag was not cleaved after the purification since it keeps proteins soluble. No RNA binding activity was observed for MBP protein (data not shown). Proteins were dialyzed into FPLC buffer (1 M KCI, 1M Urea, 50 mM Tris–HCl, 2 mM Dithiothreitol (DTT), with or without 100 μM ZnCl2) with appropriate molecular weight cutoff dialysis tubing. Size exclusion chromatography was performed using Hiload 16/600 Superdex 200 or Hiload 16/600 Superdex G-75 prep grade columns (GE Life Sciences) (Supplementary Figure S1). All proteins purified as monomers, based on comparison to size standards. Final protein concentration was calculated using molar extinction coefficient as determined using Expasy-Protparam tool and the absorbance at 280 nm. Proteins were stored at 4°C for 2 weeks.
RNA synthesis and purification
DNA template for RNA transcription of a short noncoding RNA sequence derived from the promoter of the gene DNA methyl transferase 3b (DNMT) and previously reported to bind FUS was amplified by using polymerase chain reaction (PCR) and transcribed by T7 RNA polymerase (44). Transcripts were purified using denaturing polyacrylamide gel and bands were visualized using UV shadowing. Full-length transcripts were excised from the gel and incubated in 0.5× TE buffer (10 mM Tris–HCl pH 8.0, 1 mM ethylenediaminetetraacetic acid) for 2 h at 4°C. RNA was concentrated using centrifugal concentrators with a 10 kD molecular weight cutoff (Amicon Ultra, 0.5 ml) and buffer exchange was performed by the same method. Final RNA concentration was calculated using the molar extinction coefficient as determined using an extinction coefficient calculated as the summation of the individual bases and the absorbance at 260 nm. The RNA was stored at −20°C.
RNA body labeling reaction
Consensus sequences of human and mouse RNA repeat domains (RRD; 152 and 155 nt respectively) of Firre lncRNA were generated and cloned into pUC19 vector. DNA templates for in vitro transcription reactions were amplified from RRD containing plasmids using PCR. A total of 100 μl in vitro RNA transcription reaction that included 20 μCi ATP [α-32P] was carried out with T7 RNA polymerase at 37°C for 2 h. MicroSpin G25 columns were used to remove unincorporated nucleotides from the labeling reactions. Labeled transcripts were purified using the appropriate percentage denaturing polyacrylamide gel. RNAs were visualized by phosphor imager, and excised from the gel and eluted by 0.5× TE buffer in the presence of 0.3 M sodium acetate, pH 5.3, at 4°C for 2 h. The RNAs were precipitated with ethanol and glycogen at −80°C for 30 min and centrifuged at 17 000 g for 30 min at 4°C. Precipitated RNA was resuspended in 0.5× TE buffer and quantified by liquid scintillation counting.
RNA 5΄-end labeling reaction
DNMT, Sc1, GGUG, ss (single strand), poly-A RNAs were chemically synthesized (Integrated DNA Technologies, IDT and Dharmacon). RNAs were end labeled with γ-32P using T4 polynucleotide kinase (NEB) at 37°C for 30 min. Unincorporated nucleotides were removed and labeled RNAs were purified using denaturing polyacrylamide gel. RNAs were visualized and purified from the gel as described in RNA body labeling reaction.
Electrophoretic mobility shift assay
Appropriate concentrations of each protein and trace amount of radioactive labeled RNAs (heated at 95°C for 3 min and snap cooled) were incubated with binding buffer (250 mM KCI, 250 mM Urea, 50 mM Tris–HCl pH 7.4, 2 mM DTT, 2.5 μM yeast tRNA, 0.1 mg/ml bovine serum albumin, 10% glycerol) in a 10-μl final reaction volume at room temperature for 30 min. Each reaction was loaded into a native polyacrylamide (appropriate percentage for different constructs) gel supplemented with 0.5× TB (45 mM Tris–HCI, 45 mM borate, pH 8.1) buffer. Native polyacrylamide gel electrophoresis was run at 6–7 Watt for 1 h at room temperature. Gels were dried and subsequently imaged using a Typhoon PhosphoImager (Molecular Dynamics) and the signals were quantified with ImageQuant software suite. Quantified data were fit to a standard two-state binding isotherm using Igor (Wavemetrics), allowing calculation of both dissociation constants and Hill Coefficients.
Isothermal titration calorimetry (ITC)
DNMT RNA binding affinity to RGG1-RRM-RGG2 or RRM-RGG2 proteins were measured using ITC. DNMT RNAs were synthesized and purified as described above. RNA and protein were dialyzed overnight into 2 l of ITC buffer (appropriate concentrations of KCI, 50 mM Tris–HCI, pH 7.4) at 4°C. Briefly, the desired volume of DNMT RNA or protein was mixed with an equal volume of 1× ITC buffer and dispensed into 6–8000 Dalton molecular weight cutoff dialysis tubing (Spectra/Por). The RNA and the protein (in the same beaker) were dialyzed 1 day in 2 l of ITC buffer at 4°C by gentle stirring. Buffer was changed every 8 h. Appropriate concentrations of RNA and protein were made using centrifugal concentrators. For all of the titrations C-values were between 5–10. Titrations were performed at 37°C using a MicroCal ITC200 microcalorimeter (GE Healthcare) and data were fit using the Origin software suite as previously described (45).
CD spectroscopy
RNA and ZnF domains of FUS were expressed and purified as described above. His-MBP tags of the domains were cleaved by Precission protease enzyme at 4°C. Uncleaved protein and cleaved His-MBP were removed using nickel affinity resin, and size exclusion chromatography (using a 16/600 Superdex 75 pg column) was performed to further purify isolated domains. Samples were dialyzed in sodium phosphate buffer (250 mM NaF, 50mM (NaH2PO4 and Na2HPO4), pH 7). Circular dichroism (CD) spectra of the fragments were measured on a ChirascanPlus CD instrument at 20°C. Samples were placed in a cuvette (0.5 mm path length) and four measurements were taken and averaged. Secondary structure content of the fragments was calculated using the program CDNN (46) using the CD spectra in the wavelength range of 185–260 nm.
RNase T1 probing
5΄-end-labeled RNA in 0.5× TE buffer was heated at 95°C for 3 min and snap-cooled on ice for 10 min. A total of 1 μl of RNA solution was added to 9 μl buffer containing 250 mM KCI or LiCI and Tris–HCI (pH 7.4) for 15 min at room temperature. A total of 0.1 units RNase T1 (Ambion) was added to the RNA and incubated at room temperature for 5 min. Reactions were quenched with 10 μl phenol and then phenol–chloroform extractions were performed. To generate a uniform ladder, 1 μl RNA was incubated with 9 μl alkaline hydrolysis buffer (Ambion) for 3 min at 95°C. Products were separated using appropriate percentage of denaturing 29:1 acrylamide;bisacrylamide gel by electrophoresis. Gels were dried and visualized using a Typhoon PhosphoImager (Molecular Dynamics).
In vivo analysis of FUS RNA-binding activity
HEK293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum and antibiotics. For all transfections cells were seeded at a density of 2.5 × 106 cells in T-25 flasks and allowed to grow overnight at 37°C. Cells were transfected with 10 μg of plasmid expressing FLAG-tagged FUS constructs using Lipofectamine 2000, Opti-MEM and media without antibiotics. After 24 h, each flask was washed with 1× phosphate buffered saline (PBS), trypsinized, resuspended in media with antibiotics and split into a T-75 flask to grow overnight at 37°C. Cells were harvested the next day and UV-crosslinked once at 400 mJ/cm2 to crosslink protein and RNA.
To visualize RNA–protein complexes, the UV-crosslinked cells were lysed in lysis buffer (PBS, 0.1% SDS, 0.5% deoxycholate, 0.5% NP-40) and sonicated. DNase I and RNase T1 were added to the sonicated lysate, incubated at 37°C, and centrifuged to collect supernatant. The supernatant was added to Protein A/G agarose beads (Pierce #20421) previously pre-bound to FLAG-antibody (monoclonal M2 anti-FLAG antibody, Sigma-Aldrich, #F1804) and incubated at 4°C for 2 h to specifically pull down FLAG–FUS–RNA complexes. Beads were washed to remove non-specifically bound proteins and the retained RNA was 5΄-end labeled by adding polynucleotide kinase (PNK) buffer, T4 PNK, and γ32P-ATP and incubated at 37°C. After incubation period, labeled beads were washed, 4x NuPage loading buffer added, and the beads incubated at 95°C to elute 32P-labeled FLAG–FUS–RNA complexes. Samples were electrophoresed on a 4–20% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, transferred to a PDVF membrane, exposed on the Molecular Imager FX Imaging Screen K for 5 min and imaged on a Biorad Pharos FX Plus Molecular Imager. Two technical replicates of westerns and phosphor imaging from three separate pull-downs were quantitated and results averaged together.
RESULTS
The individual RRM and ZnF domains of FUS do not bind RNA with high affinity
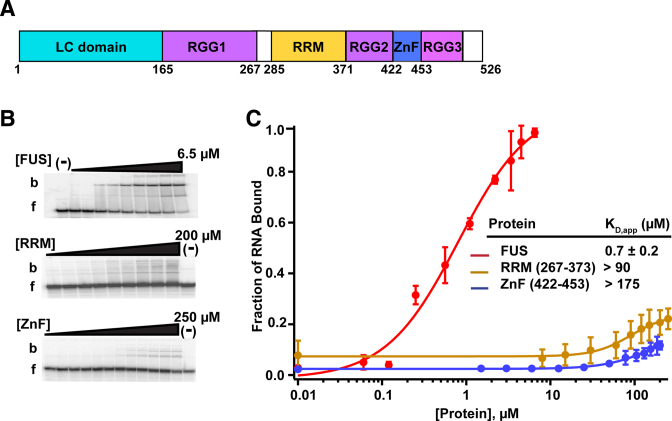
FET proteins contain the most repeats of RGG/RG and the longest RGG/RG domains. Thus, we used the FET protein FUS as a model to define the RNA binding properties these domains. FUS contains an N-terminal LC domain and five putative RBDs: an RRM, a ZnF domain and three arginine/glycine rich regions (RGG1, 2 and 3) (Figure 1A and Supplementary Figure S2). While most of the FUS mutations associated with ALS are in the nuclear localization signal, several mutations within the RGG/RG domains are directly associated with the disease (47,48).
Figure 1.
RRM and ZnF domains of FUS do not bind to the RNA with a high affinity. (A) Domain structure of FUS. (B) A trace amount of the DNMT RNA was incubated with increasing concentrations of FUS, RRM (267–373) or ZnF (422–453). Binding was analyzed by electrophoretic mobility shift assay (EMSA). b = bound DNMT RNA and f = free DNMT RNA. ‘(−)΄ shows no protein lane. (C) Binding curves of EMSA data. Error bars represent the S.D. of three independent titrations for each construct.
To quantify the contributions of the two structured RBDs, RRM and ZnF, to RNA binding, each was tested for binding activity in the absence of other domains. The purified minimal RRM (a.a. 267–373) and ZnF (a.a. 422–453) domains were determined to be folded in a manner consistent with previous CD measurements and associated structural studies (Supplementary Figure S3A) (32,49,50). From this, we concluded that our RRM and ZnF domains do not require the flanking RGG/RG domains to fold into a stable form.
Our previous studies determined FUS affinity for numerous RNA sequences and found FUS highest affinity to the 48 nt RNA, DNMT RNA (previously referred to as prD), a non-coding RNA identified to be bound to FUS in HEK293T/17 cells by CLIP-seq and ChIP-seq (8,12,32). Therefore, DNMT RNA was chosen as a model RNA to further examine the contribution of individual FUS domains on RNA binding. We observed full length FUS bound DNMT RNA with a KD,app of 0.7 ± 0.2 μM (Figure 1B and Supplementary Table S1). In contrast to previous studies, we did not adjust our KD values for the relative RNA-binding activity, which is difficult to accurately assess for FUS due to its RNA-dependent oligomerization behavior (25,32,37). Therefore, our reported KD,app's are ∼7-fold higher than that previously reported, which had been interpreted to be 15% active (8,32). Unlike full-length FUS, the RRM and ZnF domains had little observed affinity for DNMT RNA as determined by EMSA (Figure 1B and C). Isothermal titration calorimetry (ITC) was used to confirm the EMSA data, again yielding no detectable binding (Supplementary Figure S3B and C). Together, these data reveal that the minimal RRM and ZnF in the absence of other domains contribute little or no affinity for DNMT RNA.
To ensure that this result is not idiosyncratic to DNMT RNA, we tested other RNAs possessing a range of sequences and structures. These other RNAs include GGUG RNA (from an in vitro selection for FUS-binding aptamers (51)), a guanosine-deplete 36-nucleotide (nt) single stranded RNA sequence (derived from cobalamin riboswitch linker, CRL (52)), a 40-nt polyadenosine RNA, the repeating RNA domain (RRD) from human (hRRD) and mouse (mRRD) Firre lncRNA (53) and the Sc1 RNA aptamer (36 nt) (19) (Supplementary Table S1). Each of these RNAs exhibited low micromolar affinity for wild-type FUS but not the ZnF domain, while the RRM domain weakly bound Sc1 (KD,app = 48 ± 3 μM) and poly-A RNA (KD,app of 45 ± 2 μM) (Supplementary Table S2). Thus, for the RRM and ZnF domains, often annotated as nucleic acid binding domains, we found no evidence that they alone were responsible for the RNA binding activity of FUS. These results agree well with three prior structural studies of the FUS RRM (49–51).
FUS RGG/RG domains promote RNA binding
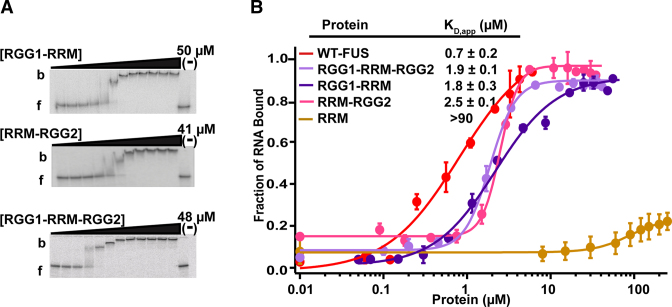
To determine whether the flanking RGG/RG domains can increase the affinity of the RRM or ZnF for RNA, we investigated the RNA-binding activity of each domain with one or both flanking RGG/RG domains. Fusion of RGG1 to the RRM (RGG1-RRM) and RRM to RGG2 (RRM-RGG2) increased the RNA binding similarly (Figure 2 and Supplementary Figure S3C). RRM with both flanking domains (RGG1-RRM-RGG2) protein showed the same affinity for RNA as RGG1-RRM and RRM-RGG2. These results reveal that modest number of RGG repeats and RRM domain are sufficient to bind enable RNA binding. The RGG/RG domains flanking the ZnF domain also restored RNA binding activity (Supplementary Figure S4A and B). However, the ZnF required both RGG2 and RGG3 for binding affinities near to that of full-length FUS, which would add a large RNA interaction surface area.
Figure 2.
Flanking RGG/RG domains impart the RNA binding activity of the RRM. (A) Representative EMSAs and (B) the corresponding binding curves showing binding of DNMT RNA to RGG1-RRM (165–373), RRM-RGG2 (267–422), RGG1-RRM-RGG2 (165–422) proteins. b = bound and f = free RNA. ‘(−)’ shows no protein lane. Error bars represent the S.D. of three independent titrations for each construct.
To establish the minimum number of RGG/RG repeats necessary to enhance the RNA binding activity of RRM, we fused to the RRM domain increasing numbers of RGG repeats from the RGG2 domain. RRM with one or two RGGs did not bind RNA, but three RGGs recovered binding close to that of the full length FUS protein (KD,app = 4 ± 0.5 μM, Table 1). Additional RGG repeats increased binding to within 3-fold of wild-type FUS.
Table 1. Binding of RRM-RGG domains to DNMT RNA.
| FUS domain | K D,app, (μM)a |
|---|---|
| RRM (267–373) | >90 |
| RRM + 1 RGG (267–380) | >60 |
| RRM + 2 RGG (267–385) | >60 |
| RRM + 3 RGG (267–390) | 4.1 ± 0.4 |
| RRM + 4 RGG (267–397) | 2.6 ± 0.8 |
| RRM + 5 RGG (267–421) | 2.5 ± 0.1 |
a K D,app values represent range of two or more independent experiments.
Electrostatic interactions are involved in RGG/RG interactions with RNA
We hypothesized that the binding for FUS is dominated by electrostatic interactions between the phosphate backbone of RNA and arginine residues. To examine this, we measured the affinity of the RGG1-RRM-RGG2 protein for DNMT RNA as a function of salt concentration using ITC (Supplementary Figure S5A). This analysis revealed two distinct linear phases. At lower salt concentrations (50–150 mM), the shallow slope (−0.84) indicated a very modest salt dependence of the RNA–protein interaction, suggesting a small electrostatic component to binding. Thus, binding at physiological salt concentrations may be driven by hydrogen bonding interactions (14,15). At higher salt concentrations (200–300 mM), the interaction was found to be more affected by salt concentration with a steeper slope (−2.5). This suggests that electrostatic interactions are a required component of RNA binding, in addition to hydrogen bonding. Alternatively, a structural change in the either the protein or the RNA at higher salt concentration may increase reliance on the electrostatic component
Individual RGG domains can bind to RNA
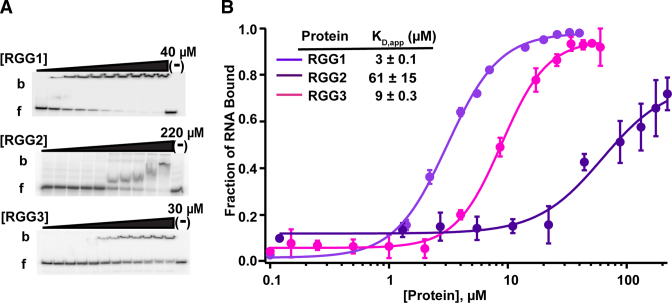
The above results suggested that the RGG/RG domains principally drive FUS affinity for the RNAs tested. We then examined each RGG/RG domain fused to the C-terminus of MBP and tested for RNA binding by EMSA. Of the three RGG/RG domains, RGG1 and RGG3 domains bound the DNMT RNA with significant affinity, 3- and 10-fold weaker than full length FUS respectively (Figure 3A and B). On the other hand, RGG2 bound RNA 100-fold weaker than full length FUS. The fact that RGG2 fused with either RRM or ZnF domains bound the RNA with markedly higher affinity (KD,app = 2.5 ± 0.1 μM for RRM-RGG2 and 13 ± 1 μM for RGG2-ZnF, Figure 2A and B; Supplementary Figure S4A and B) than either domain alone indicates a synergistic interaction between the RNA binding domains.
Figure 3.
Individual RGG/RG domains of FUS bind to RNA. (A) Representative EMSAs and (B) corresponding binding curves of individual MBP-RGG domains of FUS; RGG1 (165–267), RGG2 (372–422), RGG3 (454–501), in the presence of DNMT. b = bound and f = free RNA. ‘(−)’ shows no protein lane. Error bars represent the S.D. of three independent titrations for each construct.
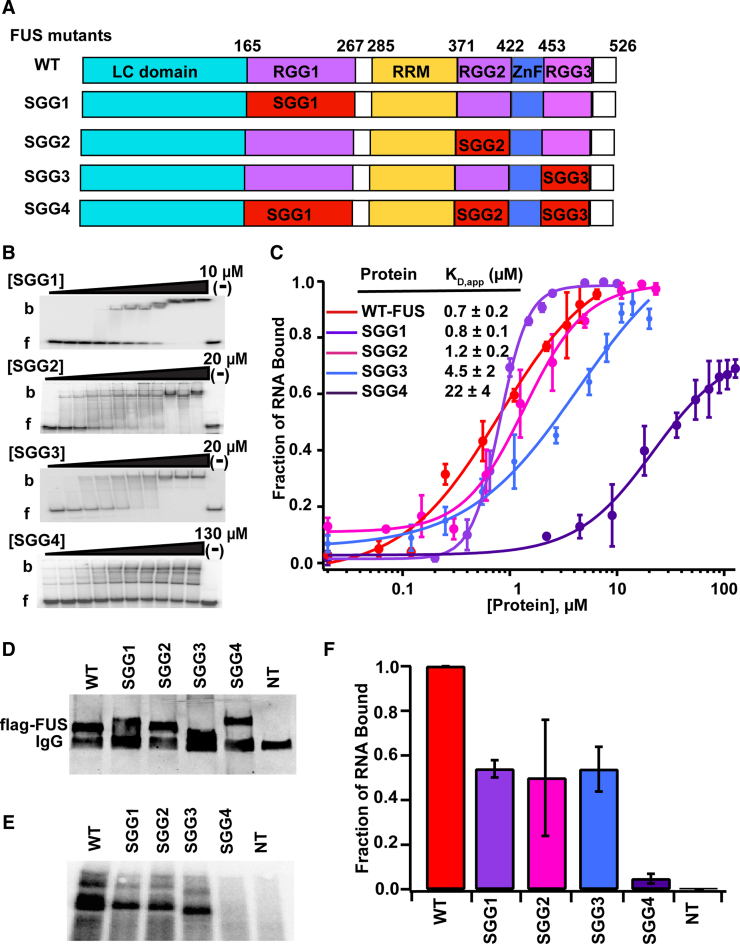
To assess the role of RGG/RG domains in the full-length protein, four mutants were created with arginine residues in either RGG1, 2 or 3 converted to serine (SGG1, 2 and 3, respectively), and a fourth with all arginine residues in these domains mutated to serine (SGG4) (Figure 4A and Supplementary Table S3). SGG1 and SGG2 bound the DNMT RNA with affinity near that of wild-type FUS (KD,app = 0.6 ± 0.1 and 1.2 ± 0.2 μM respectively, Figure 4B and C). On the other hand, SGG3 mutant bound the DNMT RNA with ∼5-fold lower affinity than wild-type FUS (KD,app = 4.3 ± 2.3 μM). This data suggested that each RGG/RG domain provides affinity for RNA and, in the case of DNMT RNA, these domains may substitute for each other. SGG4 showed a strong reduction in binding; with 30-fold lower affinity than wild-type FUS. It should be noted that the nature of the shift induced by the SGG4 mutant is different from the individual SGG mutants, suggesting a different binding mode. The relative affinity of SGG4 for RNA is suggestive that multiple weak interactions synergize to enhance the apparent binding affinity that is known as ‘avidity effect’. While the isolated RRM and ZnF domains show little individual affinity for RNA, together and in the context of this high affinity RNA partner, some binding can be observed.
Figure 4.
RGG/RG domains of FUS mediate high affinity binding to RNA. (A) Schematic illustration of RGG/RG domain mutations. Arginine amino acids of individual RGG/RG domains were converted to serine amino acids in SGG1, SGG2 and SGG3 mutants. In SGG4 mutant, arginine amino acids in all RGG/RG domains were converted to serines. (B) Representative EMSAs of mutant FUS proteins with the DNMT RNA and (C) corresponding binding curves. b = bound and f = free. ‘(−)’ shows no protein lane. Error bars represent the standard deviation of three independent titrations for each construct. (D) Western blot data of flag-tagged, wild-type and mutant FUS constructs expressed in HEK293T cells. (E) SDS-PAGE of radiolabeled RNA fragments cross-linked to flag-tagged FUS or SGG mutants of FUS. (F) Two technical replicates of three separate pull-downs were quantitated and average together. Error bars represent standard deviation.
RGG/RG domains are important for RNA binding in cells
Because DNMT RNA represents the highest affinity RNA target we have found for FUS, we hypothesized that RGG/RG domains may have varied affinities across the range of bound RNAs found in cells. To examine this, we expressed FLAG-tagged SGG mutants in HEK293T/17 cells to quantitate the amount of UV-crosslinked RNA recovered by immunoprecipitation with anti-FLAG IgG. Western blots revealed expression levels between the exogenous and endogenous FUS to be comparable. Electrophoretic mobility was affected by SGG mutations, presumably due to altered charge with fewer arginines (Figure 4D). Recovered FUS-RNA was fragmented by RNase T1 digestion and the RNA fragments protected by cross-linked protein were radiolabeled and resolved by SDS-PAGE (Figure 4E). Based on mobility, crosslinked RNA fragments were estimated to be <20 nt in length. Notably, each of the three individual RGG mutants (SGG1, 2 and 3) bound less RNA than WT FUS. Removal of all three domains in SGG4 yielded no detectable levels of bound RNA (Figure 4F). Thus, the avidity effect of RRM and ZnF was sufficient to produce measurable RNA binding in vitro, this affinity seems not to be significant in vivo.
RGG/RG domains display degenerate specificity
Whereas, mutation of any one RGG/RG in FUS did not appreciably reduce affinity for the DNMT RNA and the same mutants lost nearly half of their binding to bulk, cellular RNA, we reasoned that individual RGG/RG motifs may have different binding selectivity, resulting from their binding plasticity (Figure 4C). To characterize RNA binding properties of different RGG/RG domains, we expressed RGG/RG domains of FMRP, hnRNP-U and FUS as C-terminal MBP fusions to compare their binding to a set of RNAs representing different sequence and structural properties (Supplementary Table S1). As a benchmark for high affinity binding, we included in this panel the only structurally characterized RGG/RG domain–RNA interaction: an RNA aptamer (Sc1) containing a hybrid G-quadruplex/A-form duplex structure that was obtained in an in vitro selection for FMRP (14,15,19). As representative single stranded RNAs (ssRNAs), we employed three different RNAs: a 40 nt polyadenosine homopolymer (poly-A), a 25 nt guanosine-rich RNA that was obtained in an in vitro selection against FUS (GGUG) (51) and a heterogeneous 36 nt RNA (CRL) containing five repeats of the sequence (AUACAAC) (52). To represent dsRNA, two simple hairpins were used, one containing a hairpin exclusively composed of 12 A-U pairs (dsAU) and a second with a tract of six G-C pairs in the middle (dsGC) and each capped with a UUCG tetraloop. Finally, as a representative of an RNA with mixed secondary structure containing multiple helices and single stranded (ss-) elements, the repeating RNA domain (RRD) from human (hRRD) and mouse (mRRD) Firre lncRNA, reported targets of hnRNPU, was chosen (53).
To validate the secondary structures, RNase T1 nuclease footprinting was performed on selected RNAs (Supplementary Figure S6A–C). G-quadruplexes can be revealed by comparison of the cleavage pattern in lithium and potassium buffers, as lithium does not support quadruplex formation (54,55). These experiments confirmed the structure of most model RNAs used in this study. Unexpectedly, these experiments revealed that the DNMT RNA, which was proposed to have no stable secondary structural features, forms a G-quadruplex at its 3΄-end, likely through the association of multiple RNAs. Thus, DNMT RNA was considered by our analysis as a second example of G-quadruplex RNA, with some duplex character as well.
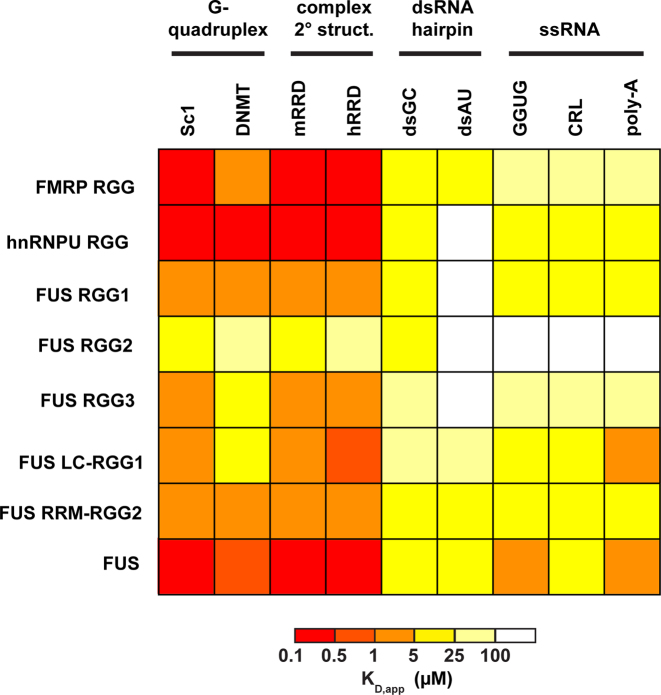
We measured the KD,app by EMSA analysis, revealing that each RGG/RG domain possessed different RNA-binding characteristics but had an overall similar pattern of behavior (Figure 5 and Supplementary Table S4). The highest affinity interaction we observed was between FMRP-RGG and Sc1 RNA (0.09 ± 0.02 μM). FMRP-RGG bound all of the RNA sequences with KD,app values spanning a 500-fold range (Figure 5 and Supplementary Table S4), showed a strong preference for G-quadruplex containing and complex RNAs, with the lowest affinities for ssRNA. This is consistent with the solved structures that show direct interactions with several G-C pairs adjacent to the G-quadruplexes (14,15). Similarly, the hnRNPU-RGG, FUS-RGG1 and FUS-RGG3 displayed a stronger preference for G-quadruplex and complex RNAs than simple hairpins or ssRNAs. Finally, the FUS-RGG2 domain had the lowest affinity for any of the RNAs tested, despite having as many RGG repeats as FMRP or hnRNP-U (Supplementary Figure S2). We concluded that these domains have more complex functional requirements than just a critical amount of RGG repeats to impart RNA binding.
Figure 5.
RGG/RG domains represent moderate preference for structured RNAs. Heat-map for affinity of RGG/RG domains and FUS constructs for each RNA. EMSA experiments were performed with DNMT and Sc1 RNAs containing G-quadruplexes, hRRD and mRRD RNAs with complex secondary structures, dsAU and dsGC simple double stranded RNAs, and GGUG, CRL and poly-A single stranded RNAs. KD,app (μM) was represented with a color code for all combinations of RNA–protein with a data range from two or more independent experiments.
G-quadruplex is not requisite for RGG/RG binding
G-quadruplexes have been proposed to be a major biological target of both FMRP and FUS (19,56,57). This perspective is supported by our observation that Sc1 RNA bound strongly to all RGG/RG domains tested. To evaluate RGG/RG domains affinity for G-quadruplex structures, we compared binding affinities of Sc1 RNA to RGG/RG peptides in buffers with either KCl or G-quadruplex disrupting LiCl. In LiCl, FMRP-RGG bound to Sc1 with ∼30-fold lower affinity, consistent with previous studies (Supplementary Figure S7) (14,15,19). Similarly, hnRNP-U RGG binding decreased ∼8-fold in the presence of LiCl. The RGG1 and RGG2 domains of FUS did not exhibit any difference in the binding affinity, but the RGG3 domain exhibited a ∼3-fold lower affinity. The binding affinity of full length FUS to Sc1 did not change in LiCl containing buffer, indicating that the G-quadruplex of Sc1 is not required for binding by the full-length protein.
To determine whether other RGG domains have the same base-specific contacts with Sc1 RNA as observed in the bound structures with FMRP-RGG, we created individual mutants of the nucleotides (G7A-C30U and C5U-G31A) that disrupt hydrogen bonding with FMRP-RGG without disrupting RNA structure (14,15). Results of binding to mutant Sc1 RNAs were diverse for different RGG domains (Supplementary Table S5). Consistent with previous work, FMRP-RGG binding affinity decreased around 25-fold for each mutant compared to the wild-type (14,15). Other RGG domains showed a lesser response from a 10-fold lower affinity to almost no change. Wild-type FUS displayed a moderate sensitivity to the G-C to A-U pair mutations, with an ∼5.5-fold decrease in binding affinity.
DISCUSSION
The RGG/RG domain is the second most common recurrent RNA binding domain (3,6). This investigation of the putative RNA binding domains of FUS has revealed these domains to be the principal drivers of RNA binding in vitro and critical for RNA binding in cells. This study has illustrated that these domains present an unexpected degree of flexibility in their recognition of RNA sequences and structures. However, these domains are not wholly indiscriminate, as they prefer GC-rich sequences and complex RNA structures featuring double-stranded helices. Similarly, the behavior of these RGG/RG proteins in cells has been shown to not be wholly non-specific (7,8,12,23,29–31,58). However, the observation that RGG/RG proteins, like FUS, bind a majority of RNAs in the transcriptome of cells has been described as promiscuous binding (7–9,11,12,32). This study's observations suggest that the RNA-binding behavior of RGG/RG domains is more accurately described as ‘degenerate specificity’, as we define features important to binding but those can be exhibited by a large number of RNA sequences and structural contexts throughout the transcriptome.
Our results provide new details regarding degenerate RNA-binding that contrasts from or compares favorably to conclusions drawn by recent comprehensive studies. A proteome-wide map of RNA-binding sites, RBDmap, recovered several novel examples of RGG/RG peptides bound to cellular RNAs and RGG3 of FUS was chosen for validation as representative of these disordered RBDs (59). Published iCLIP and CLIP-seq studies for FUS report enriched GU-rich motifs (27,29), which was also a consensus found by a SELEX study (51), but a third HITS-CLIP study found an AU-rich consensus (23), an in vitro competitive binding assay, RNAcompete, found a GC-rich motif bound to FUS RRM (58) and remaining studies report no consensus (12,30,31). While reported sequence motifs have no agreed sequence identity between them, our results support that FUS does show preference for G-rich RNAs, which may partly result from arginine hydrogen bonding to guanine.
In sharp contrast with our findings, structure-predictive analysis of iCLIP data concluded that sites bound by FUS were predominantly single-stranded (29). Two other studies conclude that FUS targets tend to lie in structured regions of RNA (23,30). Recently improved in vivo SHAPE techniques have been able to correlate evidence of complex secondary structures to specific FUS binding sites revealed through CLIP approaches (60,61). Indeed, the agreement of our work with the conclusions inferred by SHAPE-MaP would suggest that this approach currently holds the lead in resolving demonstrable binding specificity for this protein.
Taken together, there are no clear winners among comprehensive approaches to resolving the complex, degenerate binding specificity of a disordered RBD, such as the RGG/RG domain described in this work. Caution remains the key when interpreting transcriptome-wide binding profiles. Our results clearly show that validation of structure-function relationships within and without cells remains essential to provide context for interpreting global RNA-binding data. Future work should likely focus on revising overly simplified assumptions employed to interpret and extrapolate meaning from such sequencing-based studies, which do not necessary hold true for novel or underappreciated modes of specificity employed by many RNA-binding proteins.
Degenerate specificity was first used to describe flexible (but not non-specific) binding behavior of T-cell receptors, since they recognize a large number of different peptides with no amino acid homology (62). There are many examples for protein–protein interactions including kinases, phosphatases and even glutathione peroxidases that have also been defined as having degenerate specificity (63,64). We propose that the current view of ‘promiscuous binding’ of many RNA binding proteins may be better described by the term ‘degenerate specificity’. This speaks to the differences between individual domains with individual binding characteristics and the complex interactions between domains. These properties can only be fully revealed when the isolated activities of each domain are compared to that of the full-length protein.
A broadly important question concerning these data is why the second most common RBD should be an intrinsically disordered domain. These domains lack sufficient numbers of hydrophobic residues to allow a compacted core to form; thus, the peptide chain is largely solvent exposed and allowed freedom to sample many conformational states over short time scales (65–67). We find that RGG/RG preferences in binding are strongly biased against single-stranded sequences, simple A-form RNA helices or dsDNA (8,68,69). Instead, the preference of RGG/RG domains to bind to the complex FIRRE elements with higher affinity than simple hairpins and ssRNAs suggests that these domains interact with a limited and relatively common core element, likely tandem G-C pairs, in the context of more complicated or heterogeneous secondary structure. Taken together, these data suggest that the intrinsically disordered property of the RGG/RG domains imparts a great degree of plasticity providing numerous complex conformations with which to associate with helical RNA targets in a variety of structural contexts (14,15,70,71). This property provides a mechanism by which the FUS protein can bind a wide variety of RNAs as demonstrated by a number of CLIP-seq experiments (7,8,12,23,27–32,72). Taken together, the RGG/RG interactions with RNA described here are examples of intrinsically disordered domains that preferentially bind well-structured partners over less structure (1,15,63,73–75).
Comparison of analogous interactions between intrinsically disordered domains and well folded protein targets show parallels to these data. As one example, the disordered proline-rich domain (PRD) of NEF weakly binds to the Hck SH3 (Kd = 91 μM) but full length NEF binds the Hck SH3 domain with >300-fold greater affinity due to a reduction in the disorder of the PRD, thereby paying part of the entropic penalty of binding (76,77). Analogous to this, we observe that RGG2 bound RNA only weakly (KD,app = 61 μM), but with the added structural stability of the folded RRM the affinity was greatly enhanced (KD,app = 2.5 μM) beyond that of the RGG1 domain alone. It should be noted that the FUS protein is driven into phase-separation upon RNA binding and these data suggests that a conformation change upon binding may provide an underlying mechanism to this model (7,25,32,37,43,78,79).
Our establishment of a degenerate mechanism of binding may be more broadly representative of RGG/RG domain interactions with RNA. Structures of FRMP-RGG bound to Sc1 RNA reveal a β-hairpin inserted into the major grove, as well as hydrogen bonding with guanine bases (14,15). A similar type of recognition was also observed in an arginine rich peptide derived from bovine immunodeficiency virus tat protein bound to the transactivation domain of the genomic RNA (80). In our findings, the combination of hydrogen and ionic bonds as shown by FMRP-RGG presents an attractive explanation for the salt titration data. In light of a recent study indicating that the G-quadruplex forming regions in eukaryotic cells are overwhelmingly unfolded in vivo (81), the role of the G-quadruplex in Sc1 is likely to provide an unusual structure that opens the major groove for recognition. Thus, it is in fact the perturbation of the A-form helix that is particularly accommodating for RGG/RG insertion into the deep major grove. Consistent with this paradigm of selectivity for helices perturbed by local features, a recent study correlating protein–RNA binding to secondary structure within cells has concluded that FUS prefers dsRNA features adjacent to non-canonical regions (60,61). Considering the study that suggests the mRNAs were less structured or more dynamic in vivo (82), RGG/RG domains interaction with structured RNAs might be regulated by dynamic changes in structures of the RNA molecules during RNA processing.
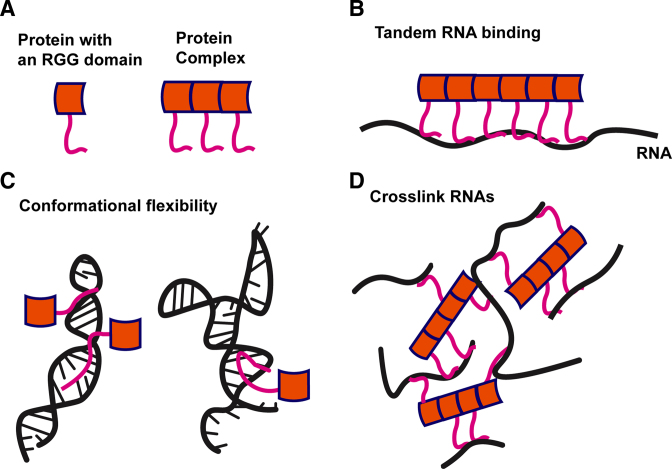
Taken together along with previously published models, we propose that conformational flexibility combined with degenerate specificity of RGG/RG domains can confer new RNA-binding activity to RBPs in at least three ways. First, degeneracy in RNA sequence recognition, particularly in the context of higher order homogeneous or heterogeneous complexes of RBPs, may allow repeated binding of RGG/RG domains in tandem along an RNA molecule (Figure 6A and B). CLIP-seq studies identifying cellular targets have suggested oligomerization of RGG/RG proteins along pre-mRNAs, particularly for FUS as well as by RGG/RG containing hnRNPs, including hnRNPA1, A2/B1 and hnRNP-U (7–8,12,20,27,32). Second, the lack of robust folding for RGG/RG domains can afford the protein flexibility to bind a variety of structural conformations of RNA (Figure 6C). It is particularly well supported because all RGG/RG domains tested here show their highest affinity for perturbed dsRNA elements as well as highly structured RNAs comprised of multiple dsRNA and ssRNA features. Third and finally, while not demonstrated in this study, the possibility has not been ruled out that long, flexible RGG/RG domains, particularly those interspersed through the RBP, can allow multivalent interactions with more than one RNA (Figure 6D). Such interactions might form a crosslinked RNP matrix such as those suggested to comprise non-membrane bound organelles, including p-bodies, stress granules and nucleoli (5,17,36,83,84).
Figure 6.
Model for RNA recognition by RGG/RG domains. (A) RGG/RG proteins may bind RNA in a 1:1 interaction or as a member of a higher order complex. (B) Multiple RGG/RG domains as part of a larger protein complex may recognize long RNA sequences. (C) RGG/RG proteins have the flexibility to accommodate and bind tightly to a variety of complex RNA structures. (D) Higher order RGG/RG proteins may allow binding to multiple RNAs at the same time.
The exceptionally broad functions of many proteins containing RGG/RG domains can be considered to be consistent with a degenerate specificity in their RNA recognition. Many hnRNP proteins possess RGG/RG domains and their function as members of ribonuclear particles is to broadly coat pre-mRNAs while they are processed within the nucleus (85,86). FUS protein regulates transcription for thousands of genes in the cell while also requiring RNA binding to trigger binding to the C-terminal domain of RNA Pol II (7,12,32). Other examples of RGG/RG domain proteins include Scd1, Sbp1, Npl3 and Ded1, which broadly affect translation (5,17). In each case, if the protein were considerably more selective in binding to RNA, its ability to act on a large number of RNAs throughout the transcriptome would become limited. RGG/RG domains might, in conjunction with other domains, broaden RNA-binding specificity, like the RGG/RG domain in FMRP broadens RNA-binding specificity beyond that typical of KH domains (19). Future research should reveal how subtle differences in the degenerate specificity of RGG/RG domains might confer broad shifts in populations of RNAs targeted, a property that might be inferred from the high conservation of each RGG/RG domain throughout vertebrates.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Prof. Deborah Wuttke for her useful discussions and Dr Annette Erbse for her help performing the CD experiments.
Author contributions. B.A.O. conducted and analyzed all of the in vitro experiments. V.F.T. and N.S.A. contributed equally to the in vivo analysis of FUS-RNA binding. C.I.W. designed and produced RS mutant constructs. R.T.B. and J.C.S. have designed experiments and analyzed data. B.A.O., R.T.B. and J.C.S. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Education of Turkey Fellowship (to B.A.O.) (in part); National Institute of General Medical Sciences [T32-GM008659 to N.S.A.]; National Institutes of Health [R01 GM12008118 to R.T.B., R00 NS082376 to J.C.S.]. Funding for open access charge: National Institute of Neurological Disorders and Stroke [R00 NS082376].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jarvelin A.I., Noerenberg M., Davis I., Castello A.. The new (dis)order in RNA regulation. Cell Commun. Signal. 2016; 14:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beckmann B.M., Castello A., Medenbach J.. The expanding universe of ribonucleoproteins: of novel RNA-binding proteins and unconventional interactions. Pflugers Arch. 2016; 468:1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gerstberger S., Hafner M., Tuschl T.. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014; 15:829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thandapani P., O’Connor T.R., Bailey T.L., Richard S.. Defining the RGG/RG motif. Mol. Cell. 2013; 50:613–623. [DOI] [PubMed] [Google Scholar]
- 5. Rajyaguru P., Parker R.. RGG motif proteins: modulators of mRNA functional states. Cell Cycle. 2012; 11:2594–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fornerod M. RS and RGG repeats as primitive proteins at the transition between the RNA and RNP worlds. Nucleus. 2012; 3:4–5. [DOI] [PubMed] [Google Scholar]
- 7. Schwartz J.C., Cech T.R., Parker R.R.. Biochemical properties and biological functions of FET proteins. Annu. Rev. Biochem. 2015; 84:355–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang X., Schwartz J.C., Cech T.R.. Nucleic acid-binding specificity of human FUS protein. Nucleic Acids Res. 2015; 43:7535–7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davidovich C., Wang X., Cifuentes-Rojas C., Goodrich K.J., Gooding A.R., Lee J.T., Cech T.R.. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol. Cell. 2015; 57:552–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davidovich C., Cech T.R.. The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA. 2015; 21:2007–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davidovich C., Zheng L., Goodrich K.J., Cech T.R.. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013; 20:1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schwartz J.C., Ebmeier C.C., Podell E.R., Heimiller J., Taatjes D.J., Cech T.R.. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 2012; 26:2690–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kiledjian M., Dreyfuss G.. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992; 11:2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Phan A.T., Kuryavyi V., Darnell J.C., Serganov A., Majumdar A., Ilin S., Raslin T., Polonskaia A., Chen C., Clain D. et al. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat. Struct. Mol. Biol. 2011; 18:796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vasilyev N., Polonskaia A., Darnell J.C., Darnell R.B., Patel D.J., Serganov A.. Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E5391–E5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fellows A., Deng B., Mierke D.F., Robey R.B., Nichols R.C.. Peptides modeled on the RGG domain of AUF1/hnRNP-D regulate 3΄ UTR-dependent gene expression. Int. Immunopharmacol. 2013; 17:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rajyaguru P., She M., Parker R.. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol. Cell. 2012; 45:244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ramos A., Hollingworth D., Pastore A.. G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA. 2003; 9:1198–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B.. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001; 107:489–499. [DOI] [PubMed] [Google Scholar]
- 20. Huelga S.C., Vu A.Q., Arnold J.D., Liang T.Y., Liu P.P., Yan B.Y., Donohue J.P., Shiue L., Hoon S., Brenner S. et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012; 1:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burd C.G., Dreyfuss G.. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994; 265:615–621. [DOI] [PubMed] [Google Scholar]
- 22. Swanson M.S., Dreyfuss G.. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol. Cell. Biol. 1988; 8:2237–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoell J.I., Larsson E., Runge S., Nusbaum J.D., Duggimpudi S., Farazi T.A., Hafner M., Borkhardt A., Sander C., Tuschl T.. RNA targets of wild-type and mutant FET family proteins. Nat. Struct. Mol. Biol. 2011; 18:1428–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tan A.Y., Manley J.L.. The TET family of proteins: functions and roles in disease. J. Mol. Cell Biol. 2009; 1:82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M. et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015; 162:1066–1077. [DOI] [PubMed] [Google Scholar]
- 26. Tan A.Y., Riley T.R., Coady T., Bussemaker H.J., Manley J.L.. TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:6030–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lagier-Tourenne C., Polymenidou M., Hutt K.R., Vu A.Q., Baughn M., Huelga S.C., Clutario K.M., Ling S.C., Liang T.Y., Mazur C. et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 2012; 15:1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kapeli K., Pratt G.A., Vu A.Q., Hutt K.R., Martinez F.J., Sundararaman B., Batra R., Freese P., Lambert N.J., Huelga S.C. et al. Distinct and shared functions of ALS-associated proteins TDP-43, FUS and TAF15 revealed by multisystem analyses. Nat. Commun. 2016; 7:12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rogelj B., Easton L.E., Bogu G.K., Stanton L.W., Rot G., Curk T., Zupan B., Sugimoto Y., Modic M., Haberman N. et al. Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci. Rep. 2012; 2:603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ishigaki S., Masuda A., Fujioka Y., Iguchi Y., Katsuno M., Shibata A., Urano F., Sobue G., Ohno K.. Position-dependent FUS-RNA interactions regulate alternative splicing events and transcriptions. Sci. Rep. 2012; 2:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Colombrita C., Onesto E., Megiorni F., Pizzuti A., Baralle F.E., Buratti E., Silani V., Ratti A.. TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J. Biol. Chem. 2012; 281:15635–15647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schwartz J.C., Wang X., Podell E.R., Cech T.R.. RNA seeds higher-order assembly of FUS protein. Cell Rep. 2013; 5:918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kwon I., Kato M., Xiang S., Wu L., Theodoropoulos P., Mirzaei H., Han T., Xie S., Corden J.L., McKnight S.L.. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013; 155:1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xiang S., Kato M., Wu L.C., Lin Y., Ding M., Zhang Y., Yu Y., McKnight S.L.. The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid-like droplets, and nuclei. Cell. 2015; 163:829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., Taylor J.P.. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015; 163:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin Y., Protter D.S., Rosen M.K., Parker R.. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell. 2015; 60:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burke K.A., Janke A.M., Rhine C.L., Fawzi N.L.. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell. 2015; 60:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim H.J., Kim N.C., Wang Y.D., Scarborough E.A., Moore J., Diaz Z., MacLea K.S., Freibaum B., Li S., Molliex A. et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013; 495:467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J. et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012; 149:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Han T.W., Kato M., Xie S., Wu L.C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G. et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012; 149:768–779. [DOI] [PubMed] [Google Scholar]
- 41. Wu H., Fuxreiter M.. The structure and dynamics of higher-order assemblies: amyloids, signalosomes, and granules. Cell. 2016; 165:1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bergeron-Sandoval L.P., Safaee N., Michnick S.W.. Mechanisms and consequences of macromolecular phase separation. Cell. 2016; 165:1067–1079. [DOI] [PubMed] [Google Scholar]
- 43. Banani S.F., Rice A.M., Peeples W.B., Lin Y., Jain S., Parker R., Rosen M.K.. Compositional control of phase-separated cellular bodies. Cell. 2016; 166:651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Edwards A.L., Garst A.D., Batey R.T.. Determining structures of RNA aptamers and riboswitches by X-ray crystallography. Methods Mol. Biol. 2009; 535:135–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gilbert S.D., Batey R.T.. Monitoring RNA-ligand interactions using isothermal titration calorimetry. Methods Mol. Biol. 2009; 540:97–114. [DOI] [PubMed] [Google Scholar]
- 46. Bohm G., Muhr R., Jaenicke R.. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 1992; 5:191–195. [DOI] [PubMed] [Google Scholar]
- 47. Deng H., Gao K., Jankovic J.. The role of FUS gene variants in neurodegenerative diseases. Nat. Rev. Neurol. 2014; 10:337–348. [DOI] [PubMed] [Google Scholar]
- 48. Renton A.E., Chio A., Traynor B.J.. State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 2014; 17:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Iko Y., Kodama T.S., Kasai N., Oyama T., Morita E.H., Muto T., Okumura M., Fujii R., Takumi T., Tate S. et al. Domain architectures and characterization of an RNA-binding protein, TLS. J. Biol. Chem. 2004; 279:44834–44840. [DOI] [PubMed] [Google Scholar]
- 50. Liu X., Niu C., Ren J., Zhang J., Xie X., Zhu H., Feng W., Gong W.. The RRM domain of human fused in sarcoma protein reveals a non-canonical nucleic acid binding site. Biochim. Biophys. Acta. 2013; 1832:375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lerga A., Hallier M., Delva L., Orvain C., Gallais I., Marie J., Moreau-Gachelin F.. Identification of an RNA binding specificity for the potential splicing factor TLS. J. Biol. Chem. 2001; 276:6807–6816. [DOI] [PubMed] [Google Scholar]
- 52. Polaski J.T., Holmstrom E.D., Nesbitt D.J., Batey R.T.. Mechanistic insights into cofactor-dependent coupling of RNA folding and mRNA transcription/translation by a cobalamin riboswitch. Cell Rep. 2016; 15:1100–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hacisuleyman E., Goff L.A., Trapnell C., Williams A., Henao-Mejia J., Sun L., McClanahan P., Hendrickson D.G., Sauvageau M., Kelley D.R. et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014; 21:198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Campbell N.H., Neidle S.. G-quadruplexes and metal ions. Met. Ions Life Sci. 2012; 10:119–134. [DOI] [PubMed] [Google Scholar]
- 55. Bhattacharyya D., Mirihana Arachchilage G., Basu S.. Metal cations in G-quadruplex folding and stability. Front. Chem. 2016; 4:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Takahama K., Miyawaki A., Shitara T., Mitsuya K., Morikawa M., Hagihara M., Kino K., Yamamoto A., Oyoshi T.. G-quadruplex DNA- and RNA-specific-binding proteins engineered from the RGG domain of TLS/FUS. ACS Chem. Biol. 2015; 10:2564–2569. [DOI] [PubMed] [Google Scholar]
- 57. Chen E., Sharma M.R., Shi X., Agrawal R.K., Joseph S.. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol. Cell. 2014; 54:407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ray D., Kazan H., Cook K.B., Weirauch M.T., Najafabadi H.S., Li X., Gueroussov S., Albu M., Zheng H., Yang A. et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013; 499:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Castello A., Fischer B., Frese C.K., Horos R., Alleaume A.M., Foehr S., Curk T., Krijgsveld J., Hentze M.W.. Comprehensive identification of RNA-binding domains in human cells. Mol. Cell. 2016; 63:696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Smola M.J., Christy T.W., Inoue K., Nicholson C.O., Friedersdorf M., Keene J.D., Lee D.M., Calabrese J.M., Weeks K.M.. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:10322–10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Smola M.J., Calabrese J.M., Weeks K.M.. Detection of RNA-protein interactions in living cells with SHAPE. Biochemistry. 2015; 54:6867–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bhardwaj V., Kumar V., Geysen H.M., Sercarz E.E.. Degenerate recognition of a dissimilar antigenic peptide by myelin basic protein-reactive T cells. Implications for thymic education and autoimmunity. J. Immunol. 1993; 151:5000–5010. [PubMed] [Google Scholar]
- 63. Roy J., Cyert M.S.. Cracking the phosphatase code: docking interactions determine substrate specificity. Sci. Signal. 2009; 2:re9. [DOI] [PubMed] [Google Scholar]
- 64. Toppo S., Flohe L., Ursini F., Vanin S., Maiorino M.. Catalytic mechanisms and specificities of glutathione peroxidases: variations of a basic scheme. Biochim. Biophys. Acta. 2009; 1790:1486–1500. [DOI] [PubMed] [Google Scholar]
- 65. Choy M.S., Page R., Peti W.. Regulation of protein phosphatase 1 by intrinsically disordered proteins. Biochem. Soc. Trans. 2012; 40:969–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dyson H.J. Making sense of intrinsically disordered proteins. Biophys. J. 2016; 110:1013–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Espinoza-Fonseca L.M. Reconciling binding mechanisms of intrinsically disordered proteins. Biochem. Biophys. Res. Commun. 2009; 382:479–482. [DOI] [PubMed] [Google Scholar]
- 68. Kipp M., Gohring F., Ostendorp T., van Drunen C.M., van Driel R., Przybylski M., Fackelmayer F.O.. SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol. Cell. Biol. 2000; 20:7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ashley C.T. Jr, Wilkinson K.D., Reines D., Warren S.T.. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993; 262:563–566. [DOI] [PubMed] [Google Scholar]
- 70. Tompa P., Csermely P.. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004; 18:1169–1175. [DOI] [PubMed] [Google Scholar]
- 71. Leulliot N., Varani G.. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001; 40:7947–7956. [DOI] [PubMed] [Google Scholar]
- 72. Masuda A., Takeda J., Okuno T., Okamoto T., Ohkawara B., Ito M., Ishigaki S., Sobue G., Ohno K.. Position-specific binding of FUS to nascent RNA regulates mRNA length. Genes Dev. 2015; 29:1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Varadi M., Zsolyomi F., Guharoy M., Tompa P.. Functional advantages of conserved intrinsic disorder in RNA-binding proteins. PLoS One. 2015; 10:e0139731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Corbin-Lickfett K.A., Souki S.K., Cocco M.J., Sandri-Goldin R.M.. Three arginine residues within the RGG box are crucial for ICP27 binding to herpes simplex virus 1 GC-rich sequences and for efficient viral RNA export. J. Virol. 2010; 84:6367–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lu C.C., Wu C.W., Chang S.C., Chen T.Y., Hu C.R., Yeh M.Y., Chen J.Y., Chen M.R.. Epstein-Barr virus nuclear antigen 1 is a DNA-binding protein with strong RNA-binding activity. J. Gen. Virol. 2004; 85:2755–2765. [DOI] [PubMed] [Google Scholar]
- 76. Lee C.H., Saksela K., Mirza U.A., Chait B.T., Kuriyan J.. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell. 1996; 85:931–942. [DOI] [PubMed] [Google Scholar]
- 77. Lee C.H., Leung B., Lemmon M.A., Zheng J., Cowburn D., Kuriyan J., Saksela K.. A single amino acid in the SH3 domain of Hck determines its high affinity and specificity in binding to HIV-1 Nef protein. EMBO J. 1995; 14:5006–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., Parker R.. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016; 164:487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Berry J., Weber S.C., Vaidya N., Haataja M., Brangwynne C.P.. RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E5237–E5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Puglisi J.D., Chen L., Blanchard S., Frankel A.D.. Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex. Science. 1995; 270:1200–1203. [DOI] [PubMed] [Google Scholar]
- 81. Guo J.U., Bartel D.P.. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 2016; 353:1382–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rouskin S., Zubradt M., Washietl S., Kellis M., Weissman J.S.. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014; 505:701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mitchell S.F., Parker R.. Principles and properties of eukaryotic mRNPs. Mol. Cell. 2014; 54:547–558. [DOI] [PubMed] [Google Scholar]
- 84. Ramaswami M., Taylor J.P., Parker R.. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013; 154:727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Dreyfuss G., Kim V.N., Kataoka N.. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002; 3:195–205. [DOI] [PubMed] [Google Scholar]
- 86. Pinol-Roma S., Dreyfuss G.. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992; 355:730–732. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.