Abstract
Piwi in a complex with Piwi-interacting RNAs (piRNAs) triggers transcriptional silencing of transposable elements (TEs) in Drosophila ovaries, thus ensuring genome stability. To do this, Piwi must scan the nascent transcripts of genes and TEs for complementarity to piRNAs. The mechanism of this scanning is currently unknown. Here we report the DamID-seq mapping of multiple Piwi-interacting chromosomal domains in somatic cells of Drosophila ovaries. These domains significantly overlap with genomic regions tethered to Nuclear Pore Complexes (NPCs). Accordingly, Piwi was coimmunoprecipitated with the component of NPCs Elys and with the Xmas-2 subunit of RNA transcription and export complex, known to interact with NPCs. However, only a small Piwi fraction has transient access to DNA at nuclear pores. Importantly, although 36% of the protein-coding genes overlap with Piwi-interacting domains and RNA-immunoprecipitation results demonstrate promiscuous Piwi binding to numerous genic and TE nuclear transcripts, according to available data Piwi does not silence these genes, likely due to the absence of perfect base-pairing between piRNAs and their transcripts.
INTRODUCTION
The harmful transpositions of transposable elements (TEs) in Drosophila gonads are strictly controlled by the Piwi-interacting RNA (piRNA) pathway, of which the PIWI family proteins are the key components (reviewed in (1)). The nuclear localized protein Piwi executes transcriptional silencing of TEs in both somatic and germline cells being the sole piRNA-binding protein in somatic cells of Drosophila ovaries (2–5). In the current model, Piwi induces the transcriptional silencing of TEs by recognizing their nascent transcripts via complementarity with the loaded piRNAs (6–10). The RNA-binding protein Asterix/dmGtsf1 has been proposed to assist Piwi at this stage (11,12). The recognition of multiple complementary sites (13) in nascent TE transcripts by Asterix/Piwi/piRNA complexes leads to the involvement of the adaptor protein Panoramix/Silencio with the further recruitment of the general cell silencing machinery, repressing TE transcription (14,15). Importantly, at least in model transgenic systems, Piwi is not required for the latter stages of silencing. To achieve target recognition, Piwi must scan all nascent transcripts. The mechanism of this scanning is currently unknown; however, it is evident that Piwi should be closely localized with the chromatin of both genes and TEs to efficiently access their nascent transcripts.
Previous attempts to identify Piwi-bound genomic regions were performed using ChIP (16–18). Huang et al. (17) demonstrated that Piwi, guided by piRNAs, can bind to numerous genomic loci containing TEs. However, later studies demonstrated that these Piwi binding sites were artefacts of incorrect bioinformatics analysis (19). More recently, the same group identified approximately one hundred Piwi binding sites in the Drosophila genome through a newly performed and analyzed ChIP-seq experiment (18). These regions corresponded to the transcription start sites (TSSs) of protein-coding genes, but not to the piRNA-targeted TEs. Importantly, the rarity of these sites did not allow for explanation of Piwi's scanning mechanism. Additionally, there is some evidence indicating that Piwi interacts with chromatin regions via RNA, but not DNA (7,9), that could hamper their detection by ChIP. Therefore, there are likely to be many more Piwi binding regions in the genome that have yet to be identified.
In this study, we have identified multiple Piwi-interacting chromosomal domains in the somatic cells of Drosophila ovaries using the DamID technique, which allows for the detection of not only constant, but also transient protein-DNA interactions. These domains significantly overlap with the genomic regions bound by nuclear pore complexes (NPCs), including those containing promoters with highly paused RNA polymerase II (Pol II). More than a third of protein-coding genes reside in the Piwi-interacting domains. Moreover, our Piwi RNA-immunoprecipitation (RIP) experiments revealed promiscuous Piwi binding to many nuclear transcripts. However, the presence of Piwi at these genes and transcripts does not result in their repression. Our findings underscore the necessity of the perfect complementarity between piRNAs and their targets for transcriptional silencing of TEs and uncover the functioning of Piwi at nuclear pores.
MATERIALS AND METHODS
Maintenance of fly stocks and generation of transgenic lines
Fly stocks were maintained under standard conditions at 25°C. Transgenic strains carrying pUAST-attB-Dam, pUAST-attB-Dam-Piwi and pUAST-attB-nos-Cre constructs were generated by φC31-mediated site-specific integration at the attP40 site on the chromosome 2 in the y, w; P{y[+t7.7] = CaryP}attP40; M{3xP3-RFP.attP}ZH-86Fb; M{vas-int.B}ZH-102D line (20) as previously described (21).
Cell cultures
Ovarian somatic cells (OSCs) (22) kindly provided by M. Siomi were grown at 25°C in Shields and Sang M3 insect medium (Sigma-Aldrich) supplemented with 10% heat inactivated FBS (Gibco), 10% fly extract (http://biology.st-andrews.ac.uk/sites/flycell/flyextract.html), 10 μg/ml insulin (Sigma-Aldrich), 0.6 mg/ml glutathione (Sigma-Aldrich), 50 units/ml penicillin and 50 μg/ml streptomycin. Kc167 cell culture obtained from Drosophila Genomics Resource Center was grown in Schneider's Drosophila Medium (Gibco) supplemented with 10% heat-inactivated FBS (Gibco), 50 units/ml penicillin and 50 μg/ml streptomycin.
Plasmid construction
The pUAST-attB-Dam and pUAST-attB-nos-Cre (nos-Cre) plasmids have been previously described (GenBank accession numbers JN993988 and KC845567, respectively) (23,24). To obtain pUAST-attB-Dam-Piwi construct, the ∼0.3-kb 5΄-fragment of Piwi open reading frame (ORF) was amplified by polymerase chain reaction (PCR) from Piwi cDNA GM05853 (Drosophila Genomics Resource Center) using primers 5΄-TAATGCGGCCGCTGGCTGATGATCAGGGACGTGGACG-3΄ and 5΄-TATTGGTCACCCCACGGCTTTCGCT-3΄ and its ∼170-bp fragment was cloned into NotI and XhoI sites of pBluescript II vector (Stratagene). In parallel, the ∼2.5-kb XhoI-NsiI 3΄-fragment of Piwi cDNA GM05853 was cloned into the XhoI and PstI sites of pBluescript II. Further, the ∼0.17-kb NotI-XhoI 5΄-fragment and the ∼2.5-kb XhoI-XbaI 3΄-fragment of Piwi ORF were re-cloned from these two plasmids into the pUAST-attB-Dam vector by NotI and XbaI sites giving the resultant pUAST-attB-Dam-Piwi construct (GenBank accession number KX908210), with the Piwi whole ORF being in frame with Dam ORF. The construct was verified by sequencing.
Piwi DamID-seq
A total of 160–200 ovaries from 0–4 days old females were manually dissected using dissection needles and collected in phosphate buffered saline (PBS) at 4°C. Isolation of genomic DNA was performed by DNeasy Blood and Tissue kit (Qiagen), following the protocol for cultured cells. DNA was concentrated by centrifugation in 0.5 ml Amicon-Ultra columns (Millipore) for 10 min at 18000 × g and its concentration was measured using a nanodrop spectrophotometer. For the selective amplification of methylated GATC-GATC fragments, genomic DNA was first digested by DpnI, followed by ligation of adaptors, DpnII digestion and PCR-amplification as described previously (25) with a minor modification: ∼0.5 μg instead of 2.5 μg of genomic DNA was used as an input. We applied 14 cycles of PCR (1 min at 94°C, 1 min at 65°C and 2 min at 68°C) for Dam-only DNA samples and 19 cycles of PCR for Dam-Piwi DNA samples. Adaptors were removed by DpnII digestion for 2 h at 37°C. The amplified methylated DNA fragments were purified using QIAquick PCR Purification kit (Qiagen), concentrated by centrifugation in 0.5 ml Amicon-Ultra columns (Millipore) for 10 min at 18000 × g and quantified on a nanodrop spectrophotometer. A total of 0.5 μg of Dam-only and Dam-Piwi DNA samples (each in two biological replicates) were subjected to next generation sequencing (NGS) on Illumina HiSeq 2500 by Evrogen (http://evrogen.com/) resulting in ∼30 million 101-nt single-end reads per sample.
Piwi RNA-immunoprecipitation
Piwi RNA-immunoprecipitation with further NGS (RIP-seq; reviewed in (26)) was performed with OSCs treated for 1 h with actinomycin D (1 μg/ml), or with untreated control cells, as described previously (27). The efficiency of transcriptional repression after actinomycin D treatment was confirmed by Click-iT® RNA Alexa Fluor® 594 Imaging Kit (Thermo Fisher Scientific) that enables the detection of global nascent RNA transcription (Supplementary Figure S1A). At the same time, the steady state RNA level was not significantly altered (Supplementary Figure S1B).
Briefly, approximately 1 × 108 cells per sample were incubated for 15 min at room temperature in lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 2 mM MgOAc, 1 mM dithiothreitol (DTT), 1% Triton-X100, 0.1% NP-40, 1 mM ethylenediaminetetraacetic acid (EDTA), 2 × Complete Protease Inhibitor Cocktail (Roche), 100 U/ml RNaseOUT™ Ribonuclease Inhibitor (Thermo Fisher Scientific)). Samples were cleared by centrifugation at 16000 × g for 5 min at 4°C and extracts were incubated with rabbit polyclonal α-Piwi antibodies (4 μg per sample; kindly provided by R. Pillai), immobilized on the Protein G Dynabeads (Thermo Fisher Scientific) for 3 h at 4°C with rotation. In parallel, RNA-immunoprecipitation was carried out in the absence of the antibody (‘mock’ sample). After incubation, the beads were washed five times with the lysis buffer. Precipitated RNA was isolated by treatment of the beads with 0.7 mg/ml proteinase K for 2 h at room temperature, followed by phenol/chloroform extraction and ethanol precipitation. For protein elution, approximately one fifth of the beads per sample after RNA-immunoprecipitation were incubated for 10 min at 70°C in sodium dodecylsulphate (SDS)-sample buffer, and then loaded onto SDS-polyacrylamide gel electrophoresis for further western-blot analysis. Equal amounts of Piwi were detected in the actinomycin D-treated and control RIP extracts and no Piwi signal was observed in the ‘mock’ sample (Supplementary Figure S2, right panel). The precipitated RNA was chemically fragmented, converted into cDNA using random hexamer priming, prepared for NGS by TruSeq RNA library preparation kit (Illumina) and sequenced on Illumina HiSeq 2000 in the Laboratory of Evolutionary Genomics, Faculty of Bioengineering and Bioinformatics, Lomonosov Moscow State University, resulting in ∼18–19 million 51-nt single-end reads obtained per sample.
For RIP-seq validation we carried out Piwi RIP and PiwiactD RIP in three biological replicates with another α-Piwi antibody (2 μg per sample; Santa Cruz Biotechnology, sc-98264, Supplementary Figure S2 left panel). cDNAs were prepared and several genes and TEs were analyzed by RT-qPCR using specific primer pairs (Supplementary Table S5). Normalization was done on the 28S rRNA because its amount is only barely changed after actinomycin D treatment (Supplementary Figure S1B).
Generation of α-Elys antibodies
A 633-bp fragment of Drosophila Elys (CG14215) cDNA clone LD14710 (Drosophila Genomics Resource Center), corresponding to the C-terminal 210 aa of Elys protein, was cloned into pET21d vector and expressed as a hexahistidine fusion protein in Escherichia coli strain BL21(DE3), purified using standard protocols and used for antibody production in rabbits. Affinity purified antibodies were used for further immunostaining and western-blotting.
Piwi coimmunoprecipitation and western-blotting
Approximately 1 × 107 grown to confluence OSCs per sample were washed in PBS and homogenized in lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% NP-40 and Complete Protease Inhibitor Cocktail (Roche)). The lysate was incubated for 20 min on ice prior to clearing by centrifugation at 16000 × g for 10 min at 4°C. Inputs were taken from the supernatant. For RNase-treated samples, RNase A was added to a final concentration of 100 μg/ml. A total of 1.5 μg of mouse monoclonal α-Piwi (27) or rabbit polyclonal α-Xmas-2 (28), α-Thoc5 (29), α-Elys (this work) antibodies per sample were immobilized on Protein G Dynabeads (Thermo Fisher Scientific), incubated with lysates for 90 min at 25°C, after that beads were washed three times with washing buffer (PBS, 0.1% Tween-20 and 0.3% Triton X-100). The bound proteins were eluted by adding SDS and urea (4 M) loading buffer and heating at 95°C for 10 min. Control immunoprecipitation experiments using mouse or rabbit non-immune serum were performed in parallel. For western-blot analysis the following antibodies were used: rabbit polyclonal α-Piwi (1:1000; Santa Cruz Biotechnology, sc-98264), α-Pol II (1:1000; ab5408 (4H8), Abcam), α-Armi (1:1000) (5), α-Nelf-E (1:500; NELF subunit (30)), α-Xmas-2 (1:1000; TREX-2/AMEX subunit (28)), α-Thoc5 (1:1000; TREX subunit (29)), α-Elys (1:1500; this work) and mouse monoclonal α-beta Actin (1:3000; Abcam, ab8224).
Immunostaining
OSCs in the growth phase were seeded on coverslips for 1 h. After rinsing with PBS, cells were fixed in 4% formaldehyde in PBS for 20 min at room temperature, washed in PBS three times for 5 min, permeabilized with PBTX (PBS with 0.1% Tween-20 and 0.3% Triton X-100) for 20 min, blocked with PBTX containing 3% normal goat serum (Invitrogen) for 1 h at room temperature, incubated with primary antibody in PBTX with 3% normal goat serum for 3 h at room temperature, washed in PBTX three times for 10 min, incubated with secondary antibodies in PBTX with 3% normal goat serum overnight at 4°C and then washed in PBTX three times for 10 min in a dark chamber. Coverslips were mounted with a drop of SlowFade Gold Antifade reagent (Invitrogen) containing DAPI. As primary antibodies we used rabbit polyclonal α-Piwi (1:300; Santa Cruz Biotechnology, sc-98264), mouse monoclonal α-NPC (Mab414, 1:300; ab24609, Abcam), rabbit polyclonal α-Xmas-2 (1:300) (28), rabbit polyclonal α-Thoc5 (1:300) (29), rabbit polyclonal α-Elys (1:600; this work), rabbit polyclonal α-Pol II (1:500; ab5408 (4H8), Abcam) and guinea pig polyclonal α-LBR (1:1000) (31). As a secondary antibodies we used Alexa Fluor 546-conjugated goat α-rabbit IgG (Invitrogen) or Alexa Fluor 488-conjugated goat α-mouse IgG (Invitrogen), or Alexa Fluor 633-conjugated goat α-guinea pig IgG (Invitrogen).
Sequencing and bioinformatics analysis of Drosophila genome
High molecular-weight genomic DNA was isolated from the F1 offspring of the Mos1-Cre × Dam-Piwi crossed flies according to the standard procedure (32). A paired-end library of the fragmented genomic DNA (∼250-bp fragment size) was prepared according to the Illumina standard protocol and sequenced on the Illumina HiSeq 2000 in the Laboratory of Evolutionary Genomics, Faculty of Bioengineering and Bioinformatics, Lomonosov Moscow State University. In total, 8.6 million of 101-nt paired-end reads with 9-fold coverage of euchromatic genome were obtained. Insertion sites of TEs were identified by aligning the reads with the consensus sequences of TEs (http://www.fruitfly.org/p_disrupt/TE.html) and the annotated TE insertion sites in the FlyBase (r.5.57) reference genome using the ‘McClintock meta-pipeline’ (https://github.com/bergmanlab/mcclintock). Only the non-redundant, unambiguous TE insertions, identified by ‘TEMP’, ‘TE-locate’, ‘RE-locate’ or ‘ngs_te_mapper’ software with default settings, were taken for further consideration. Totally, 2839 known and 1238 novel TE insertion sites were identified. The TE insertion and Piwi domain were considered as neighbors if Piwi domain overlapped −500 to +500-bp genomic region around TE insertion site.
Bioinformatics analysis of DamID-seq and microarray DamID data
Two biological replicates of Dam-only or Dam-Piwi DamID-seq reads were adapter clipped, mapped to the dm3/R5 genomic assembly by ‘bowtie2’ (33) and counted by ‘HTSeq-count’ software (34) on the GATC-GATC fragments or the ∼300-bp genomic bins, corresponding to genome positions of microarray Nup98 data, covering the nonrepetitive Drosophila genome (35). The resulting counts of Dam-Piwi samples were converted to reads per million mapped reads (RPM), normalized to RPM values of the Dam-only samples and log2-transformed. The domain calling was performed with the two-state Hidden Markov Model (HMM) algorithm (the code is available upon request). The domains were merged if the distance between them was less than 500 bp. The domains overlapped in both biological replicates were used for further analysis.
Nup98_NPC-tethered (Nup98_NPC) or Nup98_nucleoplasmic (Nup98_nucl) DamID microarray data for Kc167 cells (35) were retrieved from Gene Expression Omnibus (GEO) NCBI (GSM479750 and GSM479755) and the domain calling was done by two-state HMM similar to the Piwi domain identification. Piwi and Nup98 domains were further repeat-masked against the known and the newly identified TEs. Piwi domain scores were calculated as an average log2(Dam-Piwi/Dam) values across the corresponding Piwi domains. In cases where several Piwi domains overlap the gene, the highest Piwi score was ascribed. Piwi and Nup98_NPC domains were ranked according to their scores and divided into 10 groups, each containing ∼1000 or ∼700 domains, respectively. For the estimation of overlap between different domain types (Piwi and Nup98_NPC, Piwi and Nup98_nucl, Piwi and protein-coding genes or their TSSs), the UCSC Genome Browser intersection option was employed. For the calculation of Piwi/Nup98_NPC intersected domain scores, represented in Figure 2C, first, the average scores for the Nup98_NPC subdomains, intersected with each group of ranked by scores Piwi domains, were calculated, and then, these scores were compared with the calculated average scores for the intersected regions in each group of Piwi domains. For gene annotation, the Ensembl 78 release was used. To generate average Piwi log2(Dam-Piwi/Dam) profile around TSSs, the profiles within the regions of −1.5 to +1.5 kb around TSSs of all genes were averaged using ‘deepTools2’ (36), regardless of whether they are in or out of Piwi domains. To generate Piwi-DamID profiles for the TE families, the DamID-seq reads were mapped onto 100-nt bins of TEs consensus sequences, counted, converted to RPM, normalized to RPM values of the Dam-only samples and log2-transformed. The profiles were smoothed by applying the cubic spline interpolation. More than 2-fold enrichment was considered as significant.
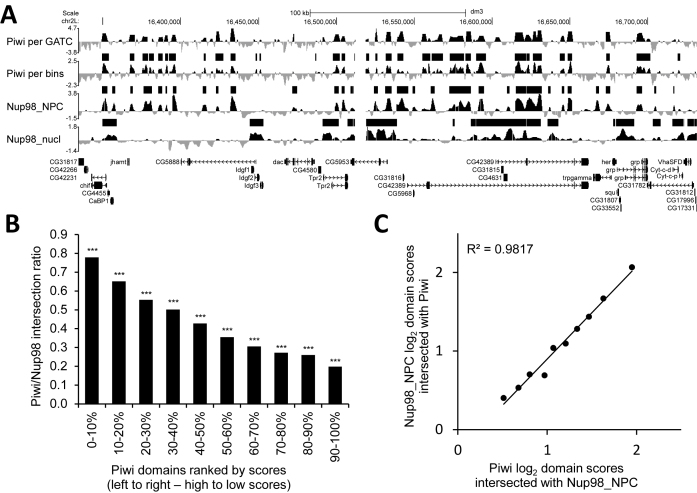
Figure 2.
Piwi-interacting domains significantly overlap with Nup98_NPC domains. (A) A snapshot from the UCSC Genome Browser shows log2(Dam-X/Dam) profiles for Piwi (per GATC-GATC fragments and per ∼300-bp bins), Nup98_NPC, Nup98_nucl, as well as their enriched domains, determined by HMM (black rectangles above the profiles). Below are FlyBase gene models. (B) Ratio of Piwi domains, ranked by scores, intersected with Nup98_NPC domains. ***P < 0.001, **P < 0.01. The permutation test was used for estimation of a probability of occasional domain intersections. (C) Piwi and Nup98_NPC log2 average scores for the Piwi/Nup98_NPC intersected domains. For the calculation of these scores, first, the average scores for the Nup98_NPC domains, intersected with each group of ranked by scores Piwi domains, were calculated and then, these scores were compared with the average scores for the intersected regions in each group of Piwi domains.
Bioinformatics analysis of RIP-seq
Adapter sequences were removed from the reads using FlexBar tool (37). Sequencing data for ‘mock’, Piwi RIP or Piwi RIP after actinomycin D treatment were mapped to the dm3/R5 genome assembly by ‘bowtie2’ (33). For the exonic read counting, the ‘TopHat’ (38) and ‘Cufflinks’ (39) software was applied. To obtain values of reads per kilobase per million mapped reads (RPKM), read counts were normalized on the library size (including rRNA read counts for correct inter-sample comparisons) and on the length of corresponding exon regions. For RIP analysis of TEs, reads were mapped onto TE consensus sequences (http://www.fruitfly.org/p_disrupt/TE.html) using ‘bowtie’ (v.1.0.0), allowing up to three mismatches. The corresponding RPKM values were calculated using ‘bamutils’ software (v.0.5.7; (40)).
Bioinformatics analysis of ChIP-seq and GRO-seq data
The Pol II pausing (stalling) index was calculated for the set of non-overlapping and having more than 500-bp in length genes from the Ensembl 78 release (n = 4108), using Pol II ChIP-seq or GRO-seq data for OSCs retrieved from GEO NCBI database (GSE41729) (7), or Pol II ChIP-seq data for Kc167 cells (GSE48512) (41), similar to Zeitlinger et al. (42). The ChIP-seq and GRO-seq reads were mapped relative to the most distal TSS onto −200 to +200-bp region (promoter, TSS) and +201 to the gene end region (gene body, GB) binned into 100-bp intervals. The number of single-mapped and de-duplexed reads in each interval were counted and the pausing index was calculated by formula: PI = (TSS_IP/GB_IP)/(TSS_input/GB_input), where TSS_IP and TSS_input are the maximal RPM-normalized signals at the promoter bins for IP and input, respectively, and GB_IP and GB_input are the median RPM-normalized signals at the GB bins for IP and input, respectively. Based on the GRO-seq data, the pausing index was calculated as the ratio of maximal signal at promoter bins to the median signal at the GB bins. The pausing index was considered significant only for the genes with Pol II-enriched regions at promoters, identified by the MACS2 software (v.2.1.1, Q-value cutoff 10−5, https://pypi.python.org/pypi/MACS2). Genes were considered overlapped, if their promoters (most distal TSSs) overlapped with Piwi domains, or if at least 1-bp of their bodies or promoters overlapped with Nup98 domains.
Bioinformatics analysis of RNA-seq data
The processed RNA-seq data for egfp-RNAi-treated or piwi-RNAi-treated OSCs were retrieved from the GEO NCBI GSE47006 (12) and compared with the Piwi domains and Piwi RIP data in MS Excel.
GO analysis
The gene ontology (GO) analysis was performed using the ‘topGO R’/Bioconductor package (version 2.22.0) with the elim algorithm and a P = 0.05 (Fisher's exact test) and Q = 0.05 (Benjamini–Hochberg correction) as the cut-offs. All the terms with the evidence codes inferred from electronic annotation, inferred from reviewed computational analysis or no biological data available were removed before the analysis.
Statistical analysis
For P-value estimation the Mann–Whitney (M–W) U-test was used for comparison of two sample distributions, or the permutation test (n = 1000) for the comparison of an overlap between two domain sets.
Data deposition
Raw and processed NGS data for Piwi-DamID and Piwi RIP were deposited in the NCBI GEO under the accession number GSE87346; NGS data of Drosophila genome used for the Piwi-DamID were deposited in the NCBI SRA Database (SRR4296946).
RESULTS
Whole-genome Piwi-DamID mapping in somatic cells of Drosophila ovaries
To clarify the mechanism of nascent RNA target recognition by Piwi, we generated whole-genome interaction map with Piwi in Drosophila ovaries using DamID technique, capturing both constant and transient protein–DNA interactions, including spatial proximity between protein and DNA without their direct binding (43,44). We applied a tissue-specific DamID-seq approach (23,45,46), illustrated in Figure 1A. Transgenic flies, carrying Dam or Dam-Piwi fusion coding regions, separated from the hsp70 core promoter of pUAST vector (47) by transcriptional stop-cassette flanked by loxP sites, were obtained by φC31-mediated integration at the attP40 site (20,21). The stop-cassette efficiently blocks any transcription, allowing Dam or Dam-Piwi expression only in the tissue where the stop-cassette has been removed by site-specific recombination at the loxP sites. We crossed females, expressing Cre-recombinase in oocytes and early embryos under the control of Mos1 promoter (line #766 from the Bloomington Drosophila Stock Center, devoid of the balancer chromosomes) (48), with the males, carrying Dam-Piwi or Dam constructs. The stop-cassette removal in a large fraction of embryonic cells results in the female offspring, mosaically expressing Dam or Dam-Piwi. We dissected ∼200 ovaries (in two biological replicates) from 0–4 days old females, isolated genomic DNA and performed standard DamID procedure (25), including DpnI digestion, adaptor ligation, DpnII digestion, followed by PCR-amplification of methylated fragments (Figure 1A and B). We note that in order to obtain similar quantity of PCR-amplified methylated DNA fragments as the output it was necessary to carry out 14 cycles of PCR for Dam × Mos1-Cre samples and 19 cycles of PCR for Dam-Piwi × Mos1-Cre samples (Figure 1B). However, in case of Dam-Piwi × Mos1-Cre this was still the amplification of specifically methylated genomic fragments, because the negative control samples (Dam-Piwi without Mos1-Cre or without DpnI digestion) gave substantially less PCR-amplified products (Figure 1C). As the levels of expression of Dam and Dam-Piwi constructs are very similar (Supplementary Figure S3), we propose that ∼30-fold lower methylation in Dam-Piwi × Mos1-Cre relative to Dam × Mos1-Cre ovaries may be explained by the assumption that only a small Piwi fraction has an access to DNA, whereas the majority of Piwi molecules are somehow isolated from the contacts with DNA (see ‘Discussion’ section).
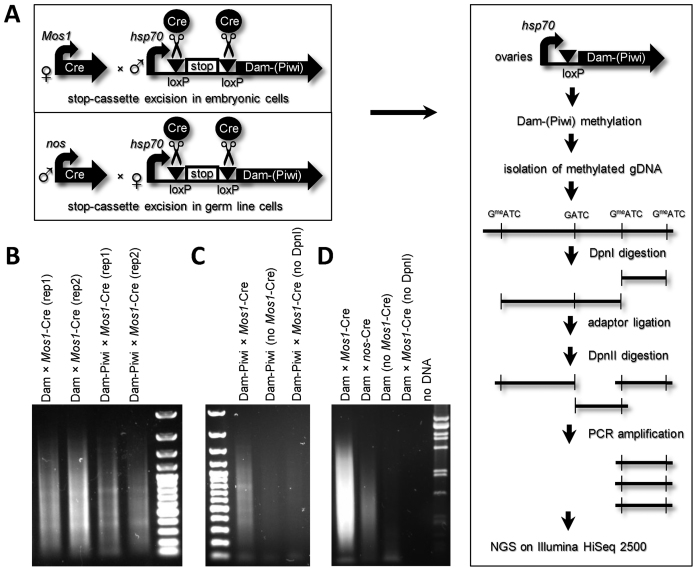
Figure 1.
Experimental design of tissue-specific Piwi-DamID in Drosophila ovaries. (A) Scheme, depicting DamID-seq procedure. (B) PCR-amplification of methylated fragments results in the smear on the gel, characteristic of the correct DamID procedure. (C) The PCR-amplification of Mos1-Cre × Dam-Piwi sample produces more DNA product, than in the negative controls (Dam-Piwi without Mos1-Cre or Mos1-Cre × Dam-Piwi without DpnI digestion), thus indicating the selective amplification of methylated fragments. (D) PCR-amplification of methylated fragments produces substantially less DNA product of samples with nos-Cre germ line-specific stop-cassette removal, than in the case of its removal in all cells (Mos1-Cre).
The equal quantities of PCR-amplified fragments were further subjected to NGS on Illumina HiSeq 2500, resulting in 28–33 million 101-nt single-end reads per sample. Unique reads were mapped on the GATC–GATC fragments of the Drosophila dm3/R5 reference genome, giving genomic profiles that were highly correlated between replicates (Spearman's correlation coefficients, ρ = 0.96 for Dam and ρ = 0.89 for Dam-Piwi, Supplementary Figure S4). Dam-Piwi signals were normalized to Dam-only signals to correct for local chromatin accessibility, and log2(Dam-Piwi/Dam) profiles per GATC fragment were generated.
We also attempted to perform Piwi-DamID specifically in the female germ-line cells by crossing males, expressing Cre-recombinase under the control of germ-line specific nanos promoter, with the females carrying Dam-Piwi or Dam constructs (Figure 1A). In this case, the stop-cassette was removed only in the germ line cells of the offspring flies. However, the PCR-amplification of methylated fragments on the isolated genomic DNA from ovaries of this offspring was significantly less efficient than after DamID procedure in the whole ovaries (Figure 1D, compare Dam × Mos1-Cre and Dam × nos-Cre). This is likely due to the failure of hsp70 core promoter to work efficiently in the female germ line (47). These data imply that Piwi-DamID mapping in whole ovaries was performed mainly in the somatic cells of the ovaries.
Piwi-interacting domains significantly overlap with NPC-tethered genomic regions
Previously, the genomic regions interacting with nucleoporin Nup98 were identified in embryonic Drosophila Kc167 cells using the DamID technique (35). In higher eukaryotes many nucleoporins (including Nup98) may be found not only at the NPCs but also in the nucleoplasmic fraction. The authors (35) succeeded in separating the Nup98-interacting regions into those tethered to NPCs (Nup98_NPC) and those of the nucleoplasmic Nup98 fraction (Nup98_nucl). A visual comparison of our Piwi DamID profile, generated from the somatic cells of ovaries, with the Nup98_NPC DamID profile for Kc167 cells revealed a high degree of similarity (Figure 2A). To compare these profiles genome-wide, we first remapped Piwi-DamID-seq reads to ∼300 bp bins, corresponding to the genome positions of microarray oligonucleotides that were used for Nup98-DamID mapping and generated log2(Dam-Piwi/Dam) profiles per bin. Then we employed a two-state HMM algorithm and in the somatic cells of Drosophila ovaries identified ∼10800 Piwi-interacting domains with a 2.1 kb median size, covering ∼27% of the euchromatic genome (Supplementary Table S1). Using the same algorithm, we identified ∼7100 Nup98_NPC (2.1 kb median size) and ∼3900 Nup98_nucl (3.6 kb median size) domains in Kc167 cells (Supplementary Table S1). We found moderate but significant correlation between Piwi and Nup98_NPC log2 profiles (Pearson's correlation coefficient, R = 0.48, P = 0), but only low correlation between Piwi and Nup98_nucl profiles (R = 0.12). Consistent with these correlations, Piwi-interacting domains significantly overlapped with the Nup98_NPC domains (43.8% of Piwi domains overlap (by coverage) with 68.5% Nup98_NPC domains; P = 10−5, permutation test), but not with the Nup98_nucl domains (13.8% of Piwi domains overlap; P = 0.98, permutation test). We divided Piwi domains into 10 groups according to their ranked average enrichment scores (see ‘Materials and Methods’ section for details). The degree of intersection between Piwi and Nup98_NPC domains progressively increased with the higher enrichment scores of Piwi domains, approaching ∼80% for the top 10% Piwi-interacting domains (Figure 2B and Supplementary Table S1). High overlap between Piwi and Nup98_NPC domains is consistent with the models, when either Piwi and NPCs independently bind the same genomic regions, or alternatively, Piwi molecules bound to NPCs as ‘passengers’ appear in close proximity to the genomic regions tethered to NPCs. We note that Piwi is absent in S2 (27) and, according to our data, in Kc167 (Supplementary Figure S5) embryonic cell cultures (although the low level of Piwi was recently reported for Kc167 cells (49)), thus indicating the absence of cooperative interactions of Piwi and NPCs with genomic sites, and NPC-tethering to these sites without Piwi assistance. Moreover, the linear regression between Piwi and Nup98_NPC enrichment scores in the regions of their overlap (Figure 2C) strongly supports the Piwi ‘passenger’ model because only bound to NPC Piwi may interact with each genomic site with the same ‘strength’ as the NPC does.
Recently, about one hundred of Piwi-bound genomic regions were revealed in Drosophila ovaries using ChIP (18). We found that the genomic positions of only 12 out of 42 sites, shared by 3 ChIP replicates, overlapped with the Piwi domains, determined by DamID (P = 0.69, permutation test). We, thus, conclude that Piwi may be stably bound with DNA only at several genomic sites, whilst at many more genomic locations Piwi interacts with DNA more transiently.
Together these data indicate that in somatic cells of Drosophila ovaries a small fraction of Piwi is likely to be linked to the NPCs and these Piwi molecules can transiently interact with the genomic regions tethered to NPCs.
Piwi-interacting domains overlap with many protein-coding genes
We next explored the interaction of Piwi with protein-coding genes. Piwi domains overlap with 36% of the protein-coding genes residing within the euchromatin, mostly with the TSSs of these genes (67%) (Figure 3A and Supplementary Table S2). Genes, interacting with Piwi, are on average three times larger (median length 4.8 kb), than the non-interacting genes (P = 0, M–W U-test), with the most high-scoring Piwi domains overlapping with the longest Drosophila genes (median length 24.5 kb, Supplementary Figure S6). Interestingly, high-scoring Piwi domains preferentially overlap with gene bodies or intergenic regions, whilst low-scoring Piwi domains overlap mostly with promoter regions (i.e. the most distal TSSs) (Figure 3B). Moreover, the average profile of Piwi at promoters peaks ∼100 bp downstream of their TSSs (Figure 3C).
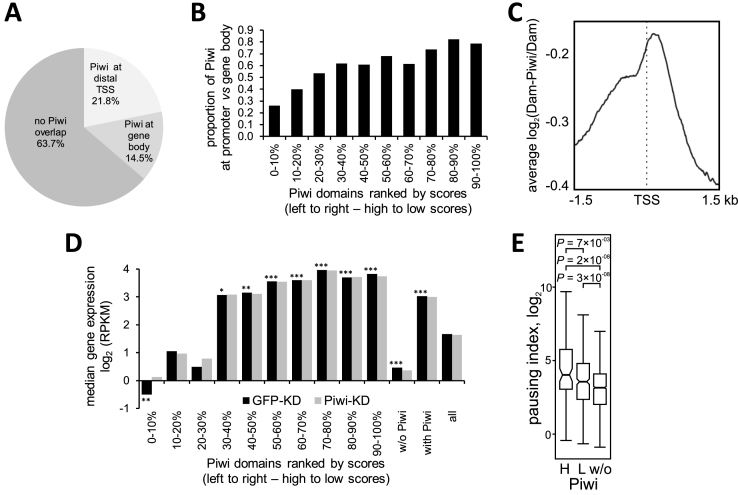
Figure 3.
Piwi domains overlap with protein-coding genes, demonstrating various levels of expression. (A) Pie chart for the Piwi domains, overlapping with protein-coding genes. (B) The ratio of promoter-overlapped to gene-overlapped Piwi domains, sorted by scores. (C) Averaged log2(Dam-Piwi/Dam) profile around the most distal TSSs for overall gene set. (D) Median expression levels in OSCs according to (12) for the genes, overlapping with Piwi domains (sorted by scores). Black bars – control cells (with gfp RNAi), gray bars – cells with piwi RNAi. ***P < 0.001, **P < 0.01, *P < 0.05. M–W U-test was used for the pairwise comparison of expression levels of the genes, overlapping with each Piwi domain subgroup, with the overall gene set (all). Significance is indicated for control cells (with GFP-KD) only. No significant difference was revealed between expression of corresponding gene sets in control and Piwi-KD cells. (E) Box plots showing Pol II-pausing indexes, calculated on the basis of Pol II ChIP-seq data in OSCs (7), for the genes, overlapping with the first three groups of high-scoring Piwi domains (H), or with the other low-scoring Piwi domains (L), or non-overlapping with Piwi domains (w/o). M–W U-test was used for the pairwise comparison of expression levels of the genes in high-scoring or low-scoring Piwi domains, with the genes outside Piwi domains.
To test whether Piwi-interacting genes are regulated by Piwi directly, we compared Piwi-DamID domains with the RNA-seq data from (12) for control and Piwi-depleted OSCs. In control cells, Piwi-interacting genes appear to be expressed at significantly higher levels (P = 2 × 10−29, M–W U-test), than those genes which do not interact with Piwi (Figure 3D and Supplementary Table S2). However, genes, overlapping with the top 30% high-scoring Piwi domains, exhibit significantly lower (P = 2 × 10−3, M–W U-test) expression levels compared to the overall gene set, and the genes, overlapping with the top 10% of Piwi domains, display the lowest expression (Figure 3D and Supplementary Table S2). Conversely, the median expression of genes in the low-scoring Piwi domains is significantly higher than median expression in the overall gene set (Figure 3D and Supplementary Table S2). A similar effect is observed for the genes overlapping with Piwi domains at the most distal TSSs (Supplementary Figure S7 and Table S2). These data indicate that Piwi interacts with both low-expressed/silent genes as well as actively-expressed genes, albeit with different strength/frequency of interaction. Importantly, the knock-down of Piwi in OSCs did not notably affect (P = 0.7, M–W U-test) expression of protein-coding genes, interacting with Piwi (Figure 3D and Supplementary Figure S7) and ∼900 genes, which have changed their expression more than 2-fold after Piwi knock-down in OSCs (441 upregulated and 445 downregulated) (12), were equally represented among Piwi-interacting (7%) and Piwi-non-interacting (7%) genes. This indicates that in OSCs Piwi interacts with the genes of all expression levels and these interactions do not result in the silencing of the majority of these genes.
To examine the characteristic features of protein-coding genes interacting with Piwi, we calculated the Pol II pausing (stalling) index (42,50), as the ratio of Pol II occupancy at the promoter (i.e. distal TSS) to its occupancy at the GB, using Pol II ChIP-seq data or GRO-seq for OSCs (7). We found that Pol II at the promoters, overlapping with either high-scoring or low-scoring Piwi domains, exhibited substantially higher median values of pausing indexes, than at the promoters without Piwi (Figure 3E; Supplementary Figure S8A and Table S2). Therefore, genes interacting with Piwi appeared to contain mostly paused Pol II at their promoters. We then assessed, whether Pol II pausing is a unique feature of Piwi-interacting genes, or it is typical for the NPC-interacting genes as well. Based on the Pol II ChIP-seq data for Kc167 cells (41), we calculated the Pol II-pausing index for the genes localized in the Nup98_NPC domains and found strongly increased pausing indexes for the genes in these domains as compared to the genes outside these domains, with especially high pausing in the high-scoring Nup98_NPC domains (Supplementary Figure S8B). Thus, the increased pausing index of Piwi-interacting genes may reflect tight contacts of Piwi as the NPC ‘passenger’ with the NPC-tethered genomic regions.
Pol II pausing is a characteristic of developmentally regulated genes, including transcription factors and components of signal transduction (reviewed in (51)). Accordingly, GO analysis indicates that amongst the genes, interacting with either high- or low-scoring Piwi domains, are those enriched in G-protein coupled receptor signaling pathway (60% genes in corresponding GO category), signal transduction (49% genes) and regulation of transcription (46% genes) (Supplementary Table S2). Earlier, the developmental genes were found to be enriched in the Nup98-interacting domains (35), emphasizing the link between Piwi and NPCs.
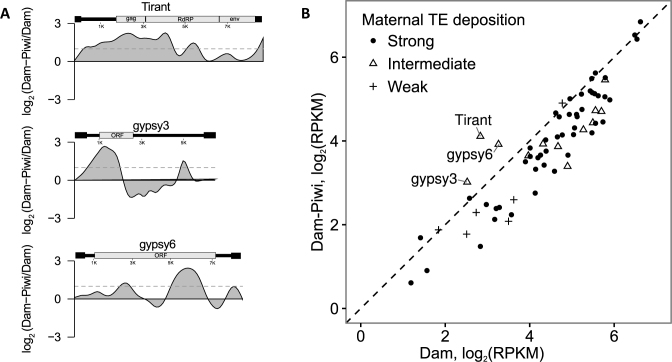
Piwi does not significantly associate with TE DNA
To determine Piwi-DamID interactions with the TEs, we mapped sequencing reads from Dam-Piwi and Dam samples onto all Drosophila TE consensus sequences, divided onto ∼100-bp bins and then compiled the log2(Dam-Piwi/Dam) profiles for the TE families. As a result, we found that Piwi were locally enriched at the specific regions of only 13 TE families (Figure 4A and Supplementary Figure S9), with the majority of TE families as a whole being devoid of DamID interactions (Figure 4B). Moreover, we did not observe selective enrichment of Piwi over TE families, known to be regulated by Piwi specifically in OSCs, as compared to other TEs. However, it was still possible that Piwi is enriched at the individual TE copies, scattered in the euchromatin. To test this idea, we sequenced the genome of the fly strains, that were used for Piwi-DamID crossing and identified TE insertion sites (see ‘Materials and Methods’ section for details). Using this technique, we identified the 2839 TE insertion sites annotated in the reference genome and a further 1238 novel euchromatic TE insertion sites (Supplementary Table S3). We then compared their genomic positions with that of Piwi domains and found no significant association between TE insertion sites and Piwi domains (27% TE insertions were within 0.5 kb from the corresponding Piwi domains, P = 1, permutation test). These data indicate that Piwi targeting to chromatin is not mediated by abundant piRNAs against TEs present in somatic ovarian cells. Thus, the perfect complementarity between piRNA and TE transcript does not lead to long-term Piwi association with the corresponding chromosomal region, supporting the idea that Piwi is required only for target recognition, but not for the subsequent silencing (14,15).
Figure 4.
According to DamID-seq, Piwi is enriched in the minor fraction of TE families. (A) TE families with the log2 (Dam-Piwi/Dam) values, exceeding the threshold (the complete list is provided on the Supplementary Figure S9). The structure of corresponding TEs is depicted above the profile. (B) Scatter plot showing TE families with overall Dam-Piwi over Dam enrichment (positioned to the left of diagonal), classified according to strong, intermediate or weak maternal piRNA deposition into embryos (2).
Piwi binds many genic and TE transcripts in OSCs
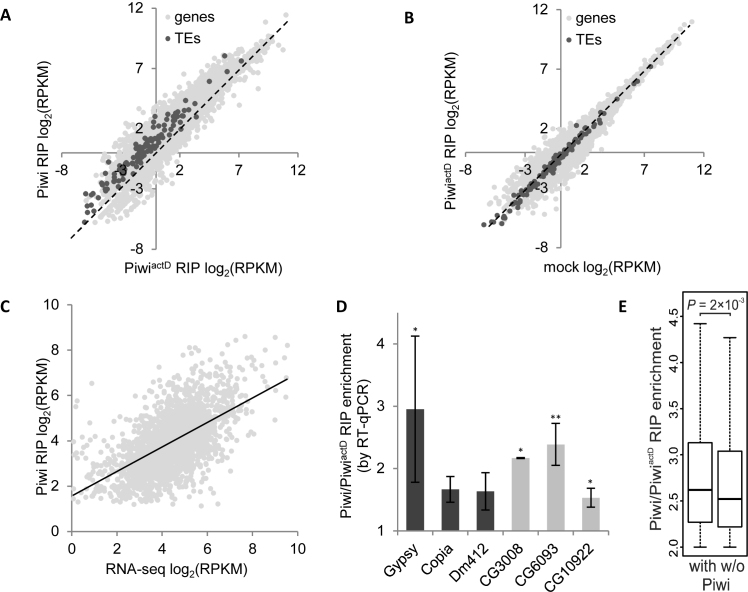
To characterize the pool of nuclear RNAs, bound by Piwi, we immunoprecipitated (without cross-linking) the RNA/Piwi complexes from OSC extracts (i.e. performed RNA-immunoprecipitation with subsequent NGS, RIP-seq) (reviewed in (26)). We carried out ‘mock’ RIP without antibodies, Piwi RIP (with α-Piwi antibodies) and PiwiactD RIP after 1-h treatment of cells with actinomycin D, the inhibitor of Pol I and Pol II (52). The efficacy of transcription blockage by actinomycin D was confirmed by the incorporation of 5-ethynyl uridine into the nascent transcripts with their subsequent ligation with the fluorescence dye in Click-iT reaction (Supplementary Figure S1A). For RIP assay, RNA was extracted, converted to cDNA using random primers and subjected to NGS. We counted the RPKM values for the exons of protein-coding genes as well as for TEs (Supplementary Table S4) and found that many of them were enriched in Piwi as compared to PiwiactD RIP (median enrichment 1.74 for the protein-coding genes and 2.34 for the TEs; Figure 5A and Supplementary Table S4) or as compared to ‘mock’ (Supplementary Figure S10A). At the same time, the log2 RPKM values for ‘mock’ and PiwiactD RIP samples were strongly correlated (Pearson's correlation coefficient, R = 0.97 for the genes and R = 0.99 for the TEs, P = 0). Accordingly, no systematic difference was revealed between the latter two samples (median ‘mock’/PiwiactD ratio 1.14 for the genes and 1.27 for the TEs; Figure 5B). As ‘mock’ RPKM values reflect the background RNA binding to Protein G Dynabeads in the cell extracts, we concluded that actinomycin D treatment leads to the loss of Piwi binding to transcripts due to efficient blockage of nascent transcription and the depletion of pool of nuclear RNAs.
Figure 5.
According to RIP-seq, Piwi promiscuously binds nuclear transcripts. Scatter plots demonstrating moderate correlation between Piwi and PiwiactD (A) and high correlation between PiwiactD and ‘mock’ (B) log2 RPKM RIP values for the protein-coding genes (light gray dots) or TEs (dark gray dots). Most dots are located to the left of diagonal (dotted line) in (A) or close to diagonal in (B). (C) Scatter plot showing the dependence of Piwi RIP log2 RPKMs and RNA-seq log2 RPKMs (12) in OSCs. Linear regression line is indicated. (D) Piwi/PiwiactD RIP enrichment by RT-qPCR for TEs (black bars) and genes (gray bars) normalized on 28S rRNA. Error bars correspond to standard deviation from three biological replicates. **P < 0.01, *P < 0.05 (Student's two-tailed t-test). (E) Box plots showing the distribution of Piwi/PiwiactD RIP enrichment values for the RIP-enriched gene set, dependent on the presence (with) or absence (w/o) of Piwi domains. The median is represented by a horizontal line, the whiskers indicate the maximum and minimum value, the outliers are omitted. The comparison of distributions was done by M–W U-test.
We considered transcripts with RPKM values ≥ 1 in Piwi and PiwiactD RIPs and with Piwi/PiwiactD ratio ≥ 2 to be significantly enriched in Piwi RIP. Approximately 2000 genes appeared to be significantly enriched in Piwi RIP, thus indicating the in vivo Piwi binding to their transcripts in the OSC nuclei. Importantly, Piwi RIP-seq log2 RPKM values for significantly RIP-enriched genes positively correlated (Pearson's correlation coefficient, R = 0.57; P = 0) with the steady state transcript abundance of these genes, determined by RNA-seq (12) (Figure 5C and Supplementary Table S4). Moreover, similar correlation is seen for the TEs (Pearson's correlation coefficient, R = 0.67; P = 0.007), indicating that Piwi binds to TE transcripts without notable preference compared to transcripts of protein-coding genes. Considering that median Piwi/PiwiactD RIP enrichment for TEs only slightly exceeds that for the protein-coding genes (Figure 5A), we conclude that Piwi binds nuclear transcripts in a ‘promiscuous’ fashion and that this binding has minimal dependence on the presence of the bulk of piRNAs complementary to TEs in OSCs.
To verify our Piwi RIP-seq results we carried out Piwi RIP and PiwiactD RIP experiments using another α-Piwi antibody. RT-qPCR analysis of several randomly selected genes and TEs with various expression levels confirmed their ∼1.5- to 3-fold enrichment in Piwi RIP relative to PiwiactD RIP after normalization on the 28S rRNA (Figure 5D).
We then assessed, whether there is a correlation between Piwi RIP-seq and Piwi-DamID data. For the protein-coding genes, resided in Piwi domains, we did not find any correlation between RIP-seq log2(Piwi/PiwiactD) enrichment values and log2(Dam-Piwi/Dam) enrichment scores (Pearson's correlation coefficient, R = 0.01, P = 0.56; Supplementary Figure S10B). However, for the set of significantly RIP-enriched nascent transcripts, the Piwi to PiwiactD RIP ratios appeared to be slightly higher for the genes, residing in Piwi domains, than for the genes lacking Piwi (median enrichment 2.62 versus 2.52 for the genes in or out of Piwi domains, respectively, P = 2 × 10−3, M–W U-test; Figure 5E and Supplementary Table S4). The same trend was observed, when transcripts with low (but not zero) RPKM values in RIPs were included (median 1.85 versus 1.77, P = 0.04, M–W U-test; Supplementary Figure S10C).
Together this indicates that most nuclear transcripts are bound by Piwi roughly proportionally to their abundance, but there is some binding preference for genes, localized in Piwi domains.
Piwi interacts with the NPC component Elys, as well as with the subunits of mRNA export complexes TREX and TREX-2 that are partially colocalized with NPCs
To test Piwi association with promoter-bound protein complexes, we carried out a coimmunoprecipitation from OSC protein extracts by α-Piwi antibodies. As a positive control we detected in the Piwi-immunoprecipitate the Armi protein (Figure 6A), known to interact with Piwi (4,5). However, we were unable to reveal Piwi interactions with Pol II and its associated pausing factor NELF (53) (Supplementary Figure S11).
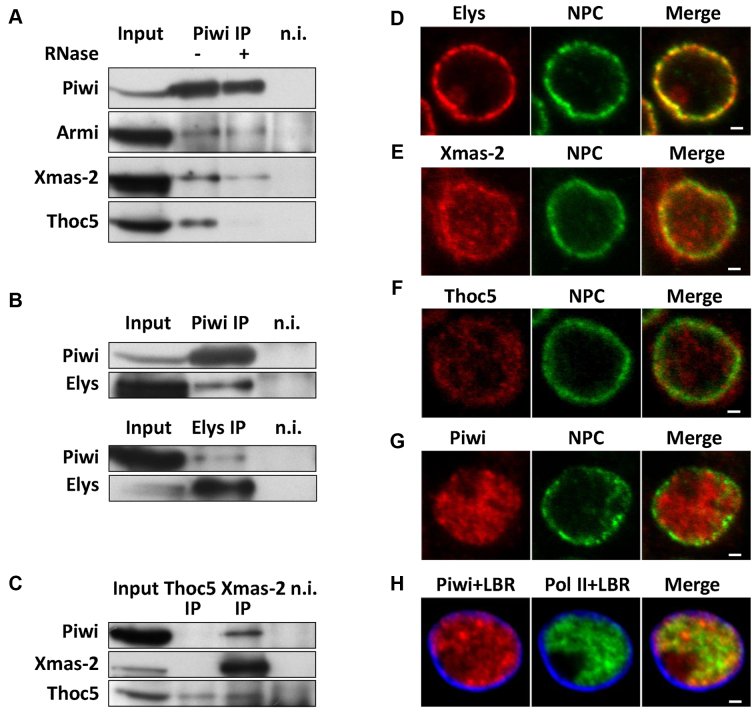
Figure 6.
Piwi is coimmunoprecipitated with the component of NPC Elys, as well as with the subunits of mRNA export complexes TREX and TREX-2 that are partially colocalized with NPCs. (A) Western-blot analysis of proteins, coimmunoprecipitated with α-Piwi antibodies from OSC extracts in the presence (+) or absence (−) of RNase A, probed by α-Piwi, α-Armi, α-Xmas-2 or α-Thoc5 antibodies (n.i. – non-immune serum, 5% of input was loaded). (B) Western-blot analysis of proteins, coimmunoprecipitated with α-Piwi (upper panel) or α-Elys (lower panel) antibodies from OSC extracts, probed by α-Piwi or α-Elys antibodies. (C) Western-blot analysis of proteins, coimmunoprecipitated with α-Xmas-2 or α-Thoc5 antibodies from OSC extracts, probed by α-Piwi, α-Xmas-2 or α-Thoc5 antibodies. (D–G) Double immunostaining of OSCs by Mab414 (NPC, green) together with α-Elys (red) (D), α-Xmas-2 (red) (E), α-Thoc5 (red) (F) or α-Piwi (red) (G) antibodies. (H) Triple immunostaining of OSCs with α-Piwi (red), α-Pol II (green) and α-LBR (blue). Note that unlike Pol II, Piwi does stain nucleolus. Scale bar 1 μm.
We further examined an association of Piwi with NPCs. We generated rabbit polyclonal antibodies against Drosophila Elys/Mel-28, which is the known component of NPCs in mammals, Xenopus and nematode (54–57), as well as in Drosophila (our unpublished data). Elys was clearly coimmunoprecipitated with Piwi from OSC extracts (Figure 6B) indicating the direct physical interaction between Piwi and NPCs. Previously, the Xmas-2 subunit of TREX-2/AMEX mRNA export complex (28) and the Thoc5 subunit of TREX mRNA export complex (29,58, reviewed in 59) were shown to colocalize with NPCs, and the Xmas-2 to coimmunoprecipitate with NPCs in S2 cells (28,29). Importantly, both Xmas-2 and Thoc5 were found in the Piwi-immunoprecipitate from OSC extracts (Figure 6A) and Piwi was reciprocally revealed in the Xmas-2-immunoprecipitate (Figure 6C). Of note, we did not detect Piwi, as well as Xmas-2 in the Thoc5-immunoprecipitate (Figure 6C). However, we believe that this is due to the inability of α-Thoc5 antibodies to efficiently capture the native, non-denatured Thoc5 protein. To address, whether Piwi association with the TREX and TREX-2 complexes is mediated by RNA, we treated cell extracts with the RNase A and found notable signal loss for the Thoc5, but not for the Xmas-2 (Figure 6A). We thus conclude that Piwi is associated with the Xmas-2 via protein–protein interactions, while with the Thoc5 mostly, but not exclusively, via RNA.
We stained OSCs with α-Elys, α-Xmas-2 or α-Thoc5 antibodies together with the Mab414 antibodies, recognizing FG-repeat nucleoporins and observed their almost complete (for Elys), or partial (for Xmas-2 or Thoc5) colocalization with NPCs, particularly at the nuclear rim (Figure 6D–F). Therefore, Piwi binding to Elys, Xmas-2 and, less likely, to Thoc5 might provide the link for Piwi targeting to NPC-tethered genomic regions. We next examined colocalization of Piwi with NPCs by immunostaining of OSCs with antibodies against Piwi and NPCs or Elys. Piwi was widespread in the nucleoplasm of OSCs, whereas Piwi and NPCs (or Piwi and Elys) were only faintly colocalized at the nuclear rim (Figure 6G and Supplementary Figure S12), implying that a small Piwi fraction is located at the NPCs. We further examined whether the widespread localization of Piwi in the nucleus corresponds to the sites of active transcription. Coimmunostaining of OSCs with α-Piwi, α-Pol II and (in order to visualize the nuclear envelope) with α-LBR antibodies shows only partial colocalization of Piwi and Pol II at chromatin (Figure 6H, Pearson's correlation coefficient, R = 0.23+/−0.14 for the ImageJ analysis of 3 immunostaining experiments, with 10 nuclei in each experiment). These results support the hypothesis of promiscuous Piwi binding to messenger ribonucleoprotein particles (mRNPs), as Thoc5 and Xmas-2 are their components and pointed to a small pool of Piwi, interacting with chromatin at NPCs.
DISCUSSION
Using DamID technique, we revealed for the first time multiple Piwi-interacting domains in the Drosophila genome and demonstrated their high correspondence to the NPC-tethered genomic regions, including gene promoters with highly paused Pol II. More than a third of protein-coding genes were found in Piwi domains. Large, tissue-specific genes, which are mostly silent or weakly expressed, reside in the high-scoring Piwi domains, whereas the actively expressed genes occupy low-scoring Piwi domains, which still partially overlap with NPC domains. The scores of Piwi domains strongly correlate with the scores of intersected NPC domains (Figure 2C) favoring the model that Piwi molecules linked to NPCs interact with the attached to NPCs chromatin carrying these genes and TEs.
We confirmed the association of Piwi with NPCs by coimmunoprecipitation of Piwi with the component of NPCs Elys/Mel-28 (54–57; reviewed in (60)) and with the Xmas-2 subunit of mRNA transcription and export complex 2 (TREX-2; reviewed in (61)); the latter was shown to colocalize and physically interact with the NPCs in Drosophila, yeast and mammals (28,62,63). However, our coimmunostaining experiments show very faint Piwi presence at the nuclear rim. Together with the ∼30-fold lower methylation level by Dam-Piwi as compared to Dam, this indicates that only a small fraction of nuclear Piwi may contact DNA at nuclear pores.
As many genes and TEs reside out of Piwi domains, then how does Piwi get in touch with the chromatin of these genes for their scanning? Looking at separate Dam-Piwi and Dam profiles (Supplementary Figure S4), one can see that besides the Dam-Piwi peaks, that are weaker or absent in Dam-only profile (these peaks give the Piwi-enriched HMM domains after normalization on the Dam data), there are many peaks of comparable to Dam height. In fact this implies, that Dam-Piwi (and therefore, Piwi) may have an access to DNA at many more sites than that tethered to NPCs. The access of Piwi to these sites is likely of stochastic nature because Dam-Piwi methylation there does not exceed methylation by freely diffusing Dam.
Two scenarios may be considered to explain how Piwi induces the silencing of its targets. Either binding of many Piwi molecules to the transcript, or Piwi's detection of the perfect complementarity between piRNA and transcript coupled with the conformational changes of Piwi/piRNA complex may be necessary to trigger the repression. Our RIP-seq data as well as the CLIP-seq data on OSS cells of Sytnikova et al. (64) show that Piwi binds to numerous nuclear transcripts, including nascent transcripts, without notable preference towards TEs, demonstrating that this binding is only weakly directed by the bulk of piRNAs with the perfect complementarity against TEs. Importantly, this Piwi binding as well as the Piwi localization close to the NPC-attached chromatin does not lead to the silencing of nearby genes. These findings support the necessity of the perfect base-pairing between piRNAs and their targets for the induction of silencing in agreement with the data of (13–15) indicating that Piwi's presence on the reporter gene is not sufficient for its repression.
What may be the mechanism of Piwi promiscuous binding with RNA? Piwi may associate with RNA indirectly, for example, via RNA-binding ability of its partner Asterix/dmGtsf1 (11,12) or via the protein components of mRNPs. However, we favor the hypothesis of direct Piwi binding with RNA via mostly imperfect base-pairing of transcripts with piRNAs. This idea is supported by the results on Piwi's closest Drosophila homolog Aubergine promiscuously interacting with multiple genic transcripts based on their incomplete (7–13 nt) complementarity with piRNAs (65). Piwi-binding activity in Drosophila cells may be similar to that of Miwi, which directly binds most mRNAs in mouse pachytene spermatocytes and thus stabilizes the mRNAs essential for further spermiogenesis (66). The promiscuous binding of XIWI and XILI with numerous genic transcripts without perfect complementarity to piRNAs was also recently described in Xenopus (67).
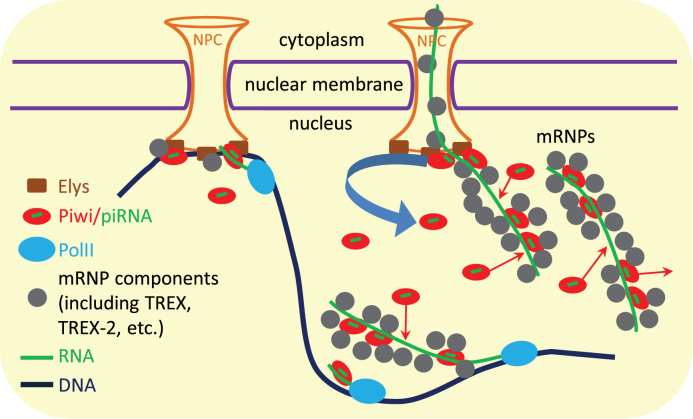
We thus suggest that a small pool of Piwi linked to NPCs, as well as more abundant nucleoplasmic pool of Piwi, interact with the majority of newly synthesized transcripts in the nucleus performing their scanning (Figure 7). If Piwi detects the perfect complementarity between Piwi-loaded piRNAs and the nascent TE transcripts, it initiates the transcriptional silencing of the corresponding TEs (14,15). However, if Piwi reveals the imperfect complementarity in the genic and TE transcripts, it does not trigger silencing but still remains bound to RNA in the mRNPs after their detachment from the sites of transcription. Piwi's association with mRNPs is supported by the coimmunoprecipitation of Piwi with the Thoc5 and the Xmas-2 mRNPs components, as well as by the weak colocalization of Piwi with the chromatin (identified by α-Pol II staining, Figure 6H). We assume that the majority of Piwi molecules in the nucleus are shielded from the contacts with DNA in the mRNPs, by analogy with the nascent transcripts that are protected by mRNP proteins from the interactions with single-stranded DNA in the R-loops (reviewed in (68)). We further suggest that Piwi is released from mRNPs at nuclear pores due to its binding to NPCs via Xmas-2, Elys or other NPC components. Linked to NPCs Piwi interacts with the attached to NPCs chromatin and, as a result, slightly more efficiently associates (Figure 5E) and probably, scans the transcripts of nearby genes and TEs, whereas some Piwi molecules are detached from the NPCs and scan the nascent transcripts at distant to NPCs genomic locations. Weak difference in the strength of Piwi association with RNA between NPC-tethered and nucleoplasmic genes and TEs (Figure 5E) may be explained by the assumption that many Piwi interactions with mRNPs occur in the nucleoplasm after these RNPs have left the transcription sites. These interactions may mask the effect of preferable Piwi binding with the nascent transcripts of genes and TEs localized at NPCs.
Figure 7.
Working model of Piwi circuit in the nucleus. Piwi stochastically interacts with the nascent transcripts of genes and TEs scanning them for the complementarity with loaded piRNAs. While capturing the perfect complementarity, Piwi triggers the transcriptional silencing of TEs. However, while revealing the imperfect complementarity, Piwi remains bound with the transcripts in the mRNPs after their detachment from the sites of transcription and continues to bind and to dissociate from mRNPs in the nucleoplasm. mRNP components may isolate the RNA-bound Piwi molecules from contacts with DNA. Piwi is released from the passing through nuclear pore mRNPs by binding to NPCs (via Xmas-2, Elys or other NPC components). Therefore, different pools of Piwi may exist in the nucleus. The smallest pool is composed from transiently linking to NPCs Piwi molecules, which interact with the attached to NPCs chromatin and, as a result, slightly more efficiently bind and, probably, scan the transcripts of nearby genes and TEs. The released from NPCs, more abundant Piwi fraction moves by Brownian motion to the chromatin positioned inside the nucleoplasm, where Piwi may scan the nascent transcripts and may bind with mRNPs. Finally, the most abundant Piwi fraction is represented by already bound to mRNPs Piwi.
In summary, in Drosophila OSCs, we revealed local Piwi enrichment on genomic regions tethered to NPCs, including promoters with highly paused Pol II. This Piwi enrichment does not result in the repression of protein-coding genes, but slightly enhances the ability of Piwi to bind with nascent transcripts, likely in the search of the complementarity with piRNAs. It is currently unclear whether Piwi's presence at NPCs, as well as Piwi's binding with numerous nuclear transcripts is a side effect of scanning mechanism or it has any biological consequence. Therefore, the suggested model of Piwi circuit in the nucleus may be considered as a framework for future studies aiming to unravel the mechanism of Piwi functioning.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Maria Logacheva and Aleksey Penin (Laboratory of Evolutionary Genomics, Faculty of Bioengineering and Bioinformatics, Lomonosov Moscow State University) for genomic and Piwi RIP sample preparation and NGS, Mikiko Siomi for α-Piwi and α-Armi antibodies, as well as for the OSC culture, Ramesh Pillai for α-Piwi antibody, Sophia Georgieva for α-Nelf-E, α-Thoc5 and α-Xmas-2 antibodies, Georg Krohne for α-LBR antibody, Haifan Lin for providing Piwi ChIP genomic positions, the Bloomington Drosophila Stock Center for fly stocks and Drosophila Genomics Resource Center for Piwi and Elys cDNA clones.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Science Foundation [14-14-01076]; Grants for Russian Leading Scientific Schools [7231.2016.4]; Russian Foundation for Basic Research [16-34-60176 to S.S.R., in part]; Russian Fundamental Scientific Research project [0310-2016-0005]; Skoltech Fellowship for Systems Biology (to S.S.R.). Funding for open access charge: Grants for Russian Leading Scientific Schools [7231.2016.4]
Conflict of interest statement. None declared.
REFERENCES
- 1. Czech B., Hannon G.J.. One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem. Sci. 2016; 41:324–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Malone C.D., Brennecke J., Dus M., Stark A., McCombie W.R., Sachidanandam R., Hannon G.J.. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009; 137:522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Haase A.D., Fenoglio S., Muerdter F., Guzzardo P.M., Czech B., Pappin D.J., Chen C., Gordon A., Hannon G.J.. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 2010; 24:2499–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Olivieri D., Sykora M.M., Sachidanandam R., Mechtler K., Brennecke J.. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010; 29:3301–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saito K., Ishizu H., Komai M., Kotani H., Kawamura Y., Nishida K.M., Siomi H., Siomi M.C.. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010; 24:2493–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang S.H., Elgin S.C.R.. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:21164–21169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sienski G., Dönertas D., Brennecke J.. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012; 151:964–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rozhkov N.V., Hammell M., Hannon G.J.. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013; 27:400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Le Thomas A., Rogers A.K., Webster A., Marinov G.K., Liao S.E., Perkins E.M., Hur J.K., Aravin A.A., Toth K.F.. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013; 27:390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klenov M.S., Lavrov S.A., Korbut A.P., Stolyarenko A.D., Yakushev E.Y., Reuter M., Pillai R.S., Gvozdev V.A.. Impact of nuclear Piwi elimination on chromatin state in Drosophila melanogaster ovaries. Nucleic Acids Res. 2014; 42:6208–6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Donertas D., Sienski G., Brennecke J.. Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev. 2013; 27:1693–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohtani H., Iwasaki Y.W., Shibuya A., Siomi H., Siomi M.C., Saito K.. DmGTSF1 is necessary for Piwi-piRISC-mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev. 2013; 27:1656–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Post C., Clark J.P., Sytnikova Y.A., Chirn G.-W., Lau N.C.. The capacity of target silencing by Drosophila PIWI and piRNAs. RNA. 2014; 20:1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu Y., Gu J., Jin Y., Luo Y., Preall J.B., Ma J., Czech B., Hannon G.J.. Panoramix enforces piRNA-dependent cotranscriptional silencing. Science. 2015; 350:339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sienski G., Batki J., Senti K.-A., Dönertas D., Tirian L., Meixner K., Brennecke J.. Silencio/CG9754 connects the Piwi-piRNA complex to the cellular heterochromatin machinery. Genes Dev. 2015; 29:2258–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moshkovich N., Lei E.P.. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet. 2010; 6:e1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang X.A., Yin H., Sweeney S., Raha D., Snyder M., Lin H.. A major epigenetic programming mechanism guided by piRNAs. Dev. Cell. 2013; 24:502–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peng J.C., Valouev A., Liu N., Lin H.. Piwi maintains germline stem cells and oogenesis in Drosophila through negative regulation of Polycomb group proteins. Nat. Genet. 2016; 48:283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marinov G.K., Wang J., Handler D., Wold B.J., Weng Z., Hannon G.J., Aravin A.A., Zamore P.D., Brennecke J., Toth K.F.. Pitfalls of mapping high-throughput sequencing data to repetitive sequences: Piwi's genomic targets still not identified. Dev. Cell. 2015; 32:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Markstein M., Pitsouli C., Villalta C., Celniker S.E., Perrimon N.. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 2008; 40:476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bischof J., Maeda R.K., Hediger M., Karch F., Basler K.. An optimized transgenesis system for Drosophila using germ-line-specific φC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Niki Y., Yamaguchi T., Mahowald A.P.. Establishment of stable cell lines of Drosophila germ-line stem cells. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:16325–16330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maksimov D.A., Koryakov D.E., Belyakin S.N.. Developmental variation of the SUUR protein binding correlates with gene regulation and specific chromatin types in D. melanogaster. Chromosoma. 2014; 123:253–264. [DOI] [PubMed] [Google Scholar]
- 24. Laktionov P.P., White-Cooper H., Maksimov D.A., Beliakin S.N.. Transcription factor comr acts as a direct activator in the genetic program controlling spermatogenesis in D. melanogaster. Mol. Biol. (Mosk). 48:153–165. [PubMed] [Google Scholar]
- 25. Vogel M.J., Peric-Hupkes D., van Steensel B.. Detection of in vivo protein–DNA interactions using DamID in mammalian cells. Nat. Protoc. 2007; 2:1467–1478. [DOI] [PubMed] [Google Scholar]
- 26. Gagliardi M., Matarazzo M.R.. RIP: RNA immunoprecipitation. Methods Mol. Biol. 2016; 1480:73–86. [DOI] [PubMed] [Google Scholar]
- 27. Saito K., Nishida K.M, Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C.. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006; 20:2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kurshakova M.M., Krasnov A.N., Kopytova D. V, Shidlovskii Y. V, Nikolenko J. V, Nabirochkina E.N., Spehner D., Schultz P., Tora L., Georgieva S.G.. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J. 2007; 26:4956–4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kopytova D. V., Orlova A. V., Krasnov A.N., Gurskiy D.Y., Nikolenko J. V., Nabirochkina E.N., Shidlovskii Y. V., Georgieva S.G.. Multifunctional factor ENY2 is associated with the THO complex and promotes its recruitment onto nascent mRNA. Genes Dev. 2010; 24:86–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu C.-H. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 2003; 17:1402–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wagner N., Weber D., Seitz S., Krohne G.. The lamin B receptor of Drosophila melanogaster. J. Cell Sci. 2004; 117:2015–2028. [DOI] [PubMed] [Google Scholar]
- 32. Maniatis T., Fritsch E.F., Sambrook J.. Molecular Cloning: a Laboratory Manual. 1982; NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 33. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anders S., Pyl P.T., Huber W.. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kalverda B., Pickersgill H., Shloma V. V., Fornerod M.. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010; 140:360–371. [DOI] [PubMed] [Google Scholar]
- 36. Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dodt M., Roehr J., Ahmed R., Dieterich C.. FLEXBAR – flexible barcode and adapter processing for next-generation sequencing platforms. Biology. 2012; 1:895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Trapnell C., Pachter L., Salzberg S.L.. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Breese M.R., Liu Y.. NGSUtils: a software suite for analyzing and manipulating next-generation sequencing datasets. Bioinformatics. 2013; 29:494–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roy S., Ernst J., Kharchenko P. V., Kheradpour P., Negre N., Eaton M.L., Landolin J.M., Bristow C.A., Ma L., Lin M.F. et al. . Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010; 330:1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zeitlinger J., Stark A., Kellis M., Hong J.-W., Nechaev S., Adelman K., Levine M., Young R.A.. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007; 39:1512–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van Steensel B., Henikoff S.. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000; 18:424–428. [DOI] [PubMed] [Google Scholar]
- 44. Greil F., Moorman C., van Steensel B.. DamID: mapping of in vivo protein-genome interactions using tethered DNA adenine methyltransferase. Methods Enzymol. 2006; 410:342–359. [DOI] [PubMed] [Google Scholar]
- 45. Luo S.D., Shi G.W., Baker B.S.. Direct targets of the D. melanogaster DSXF protein and the evolution of sexual development. Development. 2011; 138:2761–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pindyurin A.V., Pagie L., Kozhevnikova E.N., van Arensbergen J., van Steensel B.. Inducible DamID systems for genomic mapping of chromatin proteins in Drosophila. Nucleic Acids Res. 2016; 44:5646–5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brand A.H., Perrimon N.. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993; 415:401–415. [DOI] [PubMed] [Google Scholar]
- 48. Siegal M.L., Hartl D.L.. Transgene coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics. 1996; 144:715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vrettos N., Maragkakis M., Alexiou P., Mourelatos Z.. Kc167, a widely used Drosophila cell line, contains an active primary piRNA pathway. RNA. 2016; 23:108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gilchrist D.A., Dos Santos G., Fargo D.C., Xie B., Gao Y., Li L., Adelman K.. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010; 143:540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gaertner B., Zeitlinger J.. RNA polymerase II pausing during development. Development. 2014; 141:1179–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bensaude O. Inhibiting eukaryotic transcription. Which compound to choose? How to evaluate its activity?. Transcription. 2011; 2:103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yamaguchi Y., Takagi T., Wada T., Yano K., Furuya A., Sugimoto S., Hasegawa J., Handa H.. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999; 97:41–51. [DOI] [PubMed] [Google Scholar]
- 54. Rasala B.A., Orjalo A.V., Shen Z., Briggs S., Forbes D.J.. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:17801–17806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Franz C., Walczak R., Yavuz S., Santarella R., Gentzel M., Askjaer P., Galy V., Hetzer M., Mattaj I.W., Antonin W.. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007; 8:165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Galy V., Askjaer P., Franz C., López-Iglesias C., Mattaj I.W.. MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Curr. Biol. 2006; 16:1748–1756. [DOI] [PubMed] [Google Scholar]
- 57. Bilokapic S., Schwartz T.U.. Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:15241–15246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rehwinkel J., Herold A., Gari K., Köcher T., Rode M., Ciccarelli F.L., Wilm M., Izaurralde E.. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat. Struct. Mol. Biol. 2004; 11:558–566. [DOI] [PubMed] [Google Scholar]
- 59. Heath C.G., Viphakone N., Wilson S.A.. The role of TREX in gene expression and disease. Biochem. J. 2016; 473:2911–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zierhut C., Funabiki H.. Nucleosome functions in spindle assembly and nuclear envelope formation. Bioessays. 2015; 37:1074–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wickramasinghe V.O., Laskey R.A.. Control of mammalian gene expression by selective mRNA export. Nat. Rev. Mol. Cell Biol. 2015; 16:431–442. [DOI] [PubMed] [Google Scholar]
- 62. Fischer T. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002; 21:5843–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Umlauf D., Bonnet J., Waharte F., Fournier M., Stierle M., Fischer B., Brino L., Devys D., Tora L.. The human TREX-2 complex is stably associated with the nuclear pore basket. J. Cell Sci. 2013; 126:2656–2667. [DOI] [PubMed] [Google Scholar]
- 64. Sytnikova Y.A., Rahman R., Chirn G., Clark J.P., Lau N.C.. Transposable element dynamics and PIWI regulation impacts lncRNA and gene expression diversity in Drosophila ovarian cell cultures. Genome Res. 2014; 24:1977–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vourekas A., Alexiou P., Vrettos N., Maragkakis M., Mourelatos Z.. Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm. Nature. 2016; 531:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vourekas A., Zheng Q., Alexiou P., Maragkakis M., Kirino Y., Gregory B.D., Mourelatos Z.. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat. Struct. Mol. Biol. 2012; 19:773–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Toombs J.A., Sytnikova Y.A., Chirn G.W., Ang I., Lau N.C., Blower M.D.. Xenopus Piwi proteins interact with a broad proportion of the oocyte transcriptome. RNA. 2017; 23:504–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Santos-Pereira J.M., Aguilera A.. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 2015; 16:583–597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.