Abstract
AMMOS2 is an interactive web server for efficient computational refinement of protein–small organic molecule complexes. The AMMOS2 protocol employs atomic-level energy minimization of a large number of experimental or modeled protein–ligand complexes. The web server is based on the previously developed standalone software AMMOS (Automatic Molecular Mechanics Optimization for in silico Screening). AMMOS utilizes the physics-based force field AMMP sp4 and performs optimization of protein–ligand interactions at five levels of flexibility of the protein receptor. The new version 2 of AMMOS implemented in the AMMOS2 web server allows the users to include explicit water molecules and individual metal ions in the protein–ligand complexes during minimization. The web server provides comprehensive analysis of computed energies and interactive visualization of refined protein–ligand complexes. The ligands are ranked by the minimized binding energies allowing the users to perform additional analysis for drug discovery or chemical biology projects. The web server has been extensively tested on 21 diverse protein–ligand complexes. AMMOS2 minimization shows consistent improvement over the initial complex structures in terms of minimized protein–ligand binding energies and water positions optimization. The AMMOS2 web server is freely available without any registration requirement at the URL: http://drugmod.rpbs.univ-paris-diderot.fr/ammosHome.php.
INTRODUCTION
The advances in computational sciences during the last decade enabled extensive use of in silico methods at the interface between chemistry and biology. A modern set of approaches authorizes high-throughput screening computations, prioritization of the hit compounds and different levels of compound optimization (1–3). To date, many online tools have been developed in that direction (1). For instance, free web services for drug likeness and toxicity prediction are available, like Molinspiration (Molinspiration Cheminformatics, for RO5 computations), Aggregator Advisor (to search for molecules that aggregate, http://advisor.bkslab.org/) or FAF-Drugs3 (for compound property calculation and chemical library design) (4). In addition, several web servers performing de novo drug design (e.g. e-LEAD3 (5) or docking of a few ligands (e.g. SwissDock (6), CovalentDock (7)), predicting the binding affinity of a protein–small molecule complex (8), as well as some services for large-scale docking or virtual screening (like iScreen (9), DOCK Blaster (10), USR-VS (11), MTiOpenScreen (12)) have recently been reported. Yet, protein–ligand docking is not a trivial task and its performance strongly depends on the used algorithms and scoring functions, binding site definition, solvation and entropy considerations (13–15). To improve the quality of identified hits/leads and ultimately to assist the overall success of the project, additional refinement of the initially predicted docking/screening results may be required.
Here we present the new web server AMMOS2 dedicated to efficient computational refinement of protein–small organic molecule complexes and web-based visual analysis. The AMMOS2 protocol employs atomic-level energy minimization of a large number of experimental or modeled protein–ligand complex structures that can be generated via a user-chosen docking program or via virtual screening web servers as e.g. MTiOpenScreen (12). Two recent web servers, 3Drefine (16) and KoBaMIN (17), provide refinement of apo protein structures by energy minimization. The AMMOS2 web server is based on the previously developed standalone software AMMOS (Automatic Molecular Mechanics Optimization for in silico Screening) (18) performing minimization of protein–ligand interactions at five different levels of flexibility of the protein receptor. AMMOS has already been rigorously bench marked for virtual screening purposes (19). The new version 2 of AMMOS (20) implemented in the AMMOS2 web server accounts for explicit water molecules and individual metal ions in the protein–ligand complex. Water molecules present at protein–ligand, protein–protein and protein–DNA interfaces play important role as they can form direct hydrogen bonds and ultimately contribute to the stability of the complexes (21,22). Water accommodated in binding sites of proteins is of particular importance for structure-based drug design (23,24). The role and energetics of water displacement upon association have been extensively studied (25,26) and it has been shown that increasing stabilization of water molecules results in an enthalpically more favorable binding (24). Yet, considering water molecules in protein–ligand docking is a challenging task, especially for a large number of ligands like during virtual screening experiments (14,15). A small number of free protein–ligand docking programs (e.g. AutoDock (27), rDOCK, PLANTS (28,29) allow for the incorporation of structural waters. In addition, a few methods are available to predict positions of inter-facial waters important for protein–ligand interactions (for example Fold-X (30), WaterMap (31), WaterDock (32)). Users can easily solvate its protein structure using web-based tools identifying strongly and poorly solvated exposed surfaces on proteins by using, for example, PDB_HYDRO web server, option Run_AquaSol (http://lorentz.immstr.pasteur.fr/pdb_hydro.php) (33). The AMMOS2 web server can be used for the refinement of protein–ligand structures containing water molecules that fill the whole protein or the binding site and for the detection of key waters serving as bridges between the protein and ligand. Thus AMMOS2 server offers valuable solutions to assist structure-based drug design keeping in mind that water molecules mediating protein–ligand interactions are of key importance for identifying high-affinity bioactive molecules (24). In addition, some ligand-binding sites contain metal ions that are important for the interactions with the small molecules. The AMMOS2 server can take into account the presence of metal ions while ions can be added by several online services such as MIB (http://bioinfo.cmu.edu.tw/MIB/) (34) or IonCom (http://zhanglab.ccmb.med.umich.edu/IonCom/) (35).
THE AMMOS2 WEB SERVER
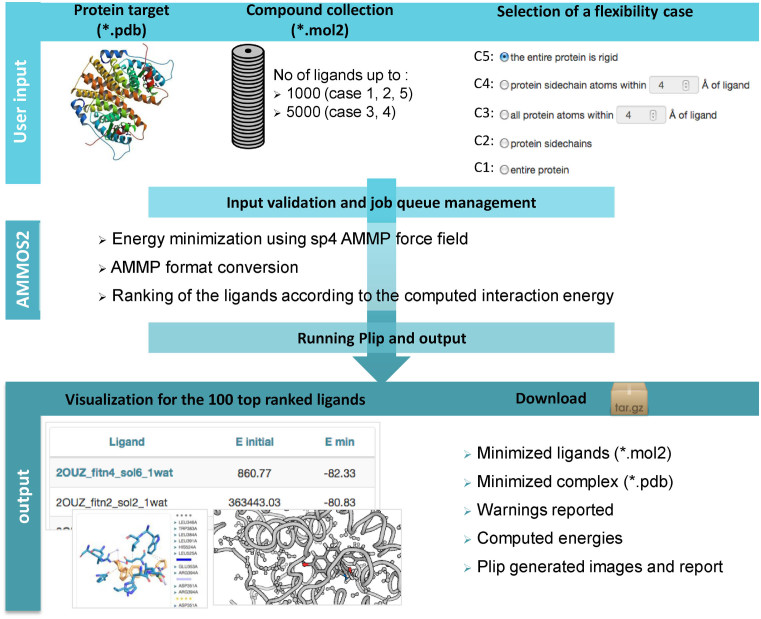
Figure 1 illustrates the overall workflow of the AMMOS2 web server including required inputs, preparation, minimization and ranking procedures. This tool is user-friendly and suitable for non-advanced users. It performs automatic minimization of protein–ligand complexes with different levels of protein flexibility for a large number of pre-docked ligands and ranking according to the minimized protein–ligand interaction energy. AMMOS2 utilizes the physics-based force field AMMP (all-featured molecular mechanics program) sp4 (36,37). The AMMOS2 web server can minimize up to 1000 or 5000 ligands depending on the chosen level of protein flexibility. AMMOS2 permits inclusion of metal ions and explicit water molecules as a part of the receptor during the minimization. Metal ions and explicit water molecules are allowed to move when they are located in the flexible part of the protein receptor. It could help to analyze key water molecules mediating the interactions between the protein and bound ligand.
Figure 1.
Flowchart of AMMOS2 web server workflow depicting interface organization, input, validation, execution and output layers of the server.
Input
AMMOS2 requires as input the protein receptor in pdb format (properly protonated and containing a maximum of 1000 residues) and bound ligands, docked or co-crystallized small organic molecules (with a maximum of 300 atoms), in mol2 format. It performs optimization of the protein–ligand interactions authorizing five levels of flexibility of the protein receptor and keeping ligands flexible—case 1: all atoms of the protein can move (a fully flexible minimization); case 2: all atoms of the protein side chains can move; case 3: all atoms of the protein inside a sphere around the ligand can move; case 4: all atoms of the protein side chains inside a sphere around the ligand can move; case 5: the whole protein is rigid. The accepted radius for cases 3 and 4 is between 4 and 8 Å. A collection of up to 1000 bound ligands is acceptable for cases 1, 2 and 5, while collection of up to 5000 ligands is acceptable for cases 3 and 4. Ligand hydrogen atoms can be assigned by the user or added by AMMOS2.
Output
AMMOS2 returns the coordinates of the minimized ligands, the coordinates of the minimized protein–ligand complex and the minimized interaction energies. AMMOS2 provides an interactive page displaying the 100 top ligands ranked by the minimized protein–ligand binding energies, allowing the users to perform additional analysis. Protein–ligand complex structures before and after minimization are visualized in 3D. An image displaying the predicted minimized protein–ligand interactions in the binding site is generated using the PLIP software (38). The reported interactions include hydrogen bonds, hydrophobic contacts, cation-pi and pi-stacking among others. The user can download the following results: the minimized ligands in mol2 format ranked by the interaction energies, the minimized protein–ligand complex in pdb format, the warning reported during the AMMOS2 minimization, the computed energies and the images and analysis report generated by the PLIP software.
PERFORMANCE OF AMMOS2
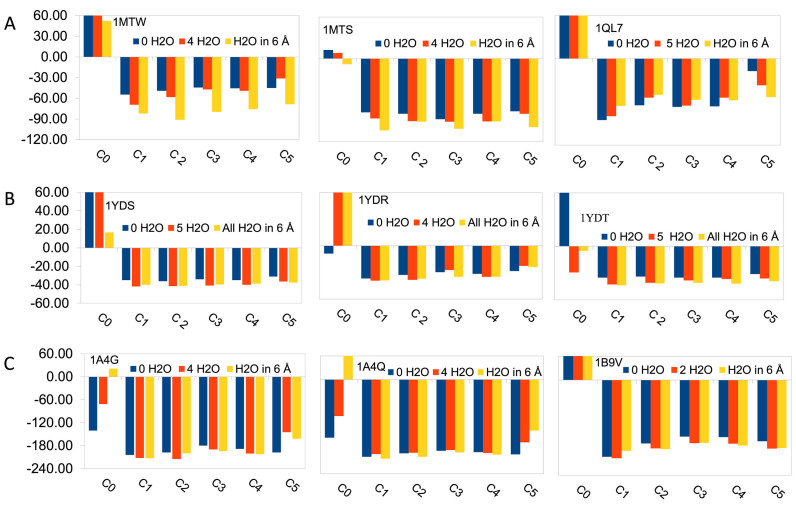
In order to validate the AMMOS2 web server we assessed the performance of the AMMOS2 molecular mechanics minimization on several classes of important therapeutic protein targets (serine proteases, cAMP-dependent protein kinase, carboxypeptidase, matrix metallo proteinase, ribonuclease, neuraminidase and estrogen receptor). We selected 21 extensively checked protein–ligand complexes from the CCDC/Astex Test Set (39). To evaluate the impact of including water molecules on the minimization of protein–ligand complexes pre-docked with AutoDock4.2 (40) we examined: (i) AMMOS2 optimization of the docked protein–ligand complexes without considering any water molecule in the binding site; (ii) AMMOS2 optimization of the docked protein–ligand complexes including selected water molecules located next to the ligands in the X-ray structures, which could be in contacts with the ligand; (iii) AMMOS2 optimization of the docked protein–ligand complexes including all water molecules present in the X-ray structures within a distance of 6 Å of the co-crystallized ligand. For protein flexibility cases 3 and 4 (C3 and C4 in Figure 2) we set a distance of 6 Å around the co-crystallized ligand. Figure 2 and Figure Supplementary S1 (Supplementary data) show the protein–ligand interaction energies after docking (before minimization) and after AMMOS2 minimization for the five different levels of protein flexibility. As seen from the figures, in all cases the interaction energies after AMMOS2 minimization were significantly reduced. In most cases, the higher number of explicit water molecules improved the computed binding energies. Allowing protein flexibility during the minimization resulted in more favorable interaction energies compared to rigid protein minimization (case 5) for most of the proteins. We performed a comparison of energy minimization using AMMOS2 and energy minimization using the free standalone software Chimera for molecular modeling (https://www.cgl.ucsf.edu/chimera/). AMMOS2 minimization improved the binding energies for all used protein–ligand complexes. Chimera minimization improved the binding energies for 14 out of the 21 protein–ligand complexes (Figure S2 in Supplementary data).
Figure 2.
Protein–ligand interaction energies (in kcal/mol) before and after minimization with AMMOS2 for the best docking pose with a different number of water molecules in the binding pocket for trypsin (A), cAMP-dependent protein kinase (B) and neuraminidase (C). C0 notes the energy of the docked complex before minimization, C1 to C5 correspond to the five cases of protein flexibility. The PDB ID of the proteins are given.
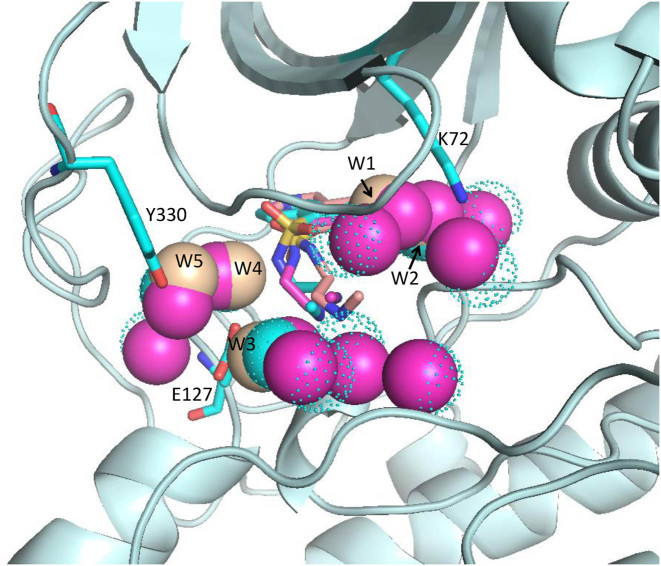
In all our tested proteins containing metal ions, the metal ions remained stable during the AMMOS2 minimization (see for illustration Figure S3 in Supplementary data). We analyzed in more details the water displacement due to protein flexibility in cases 1 and 3 for the best-energy pose of the isoquinolinesulfonyl inhibitor H8 docked into cAMP-dependent protein kinase. We found five water molecules placed close to the ligand and 12 water molecules within a radius of 6 Å of the bound ligand in the crystal structure of the complex (Protein Data Bank PDB ID 1YDS). The distances between the oxygen atoms of the water molecules W1–W5 and key ligand atoms and protein residues after AMMOS2 minimization compared to their X-ray positions are reported in Table 1. It is seen that the AMMOS2 refinement proposes that water molecules W3 and W4 (Figure 3) are the most important for mediating the protein–ligand interaction as remaining in stable hydrogen bonds during the minimization. It may be suggested that W1 is not critical for the interaction as moving by more than 1 Å away from the ligand; similarly, W5 remains located far from the ligand. Thus, the dynamic behavior of water molecules observed with AMMOS2 minimization overcomes the limitation of the ‘snapshot’ X-ray data and allows the user to decide on the importance of structural waters for the stabilization of the interactions between the protein residues and the ligand.
Table 1. Distances between the oxygen atom of water molecules W1–W5 and key ligand atoms and protein residues after AMMOS2 minimization compared to their X-ray positions in the PDB 1YDS complex.
| Water molecules (W) displacement | Distance, in Å | ||||
|---|---|---|---|---|---|
| X-ray | Case 1 (5 Ws) | Case 1 (12 Ws in 6 Å) | Case 3 (5 Ws) | Case 3 (12 Ws in 6 Å) | |
| W1 - O2 ligand | 3.28 | 4.77 | 4.42 | 4.41 | 4.21 |
| W1 - NZ K72 | 3.51 | 3.28 | 2.75 | 2.81 | 2.76 |
| W2 - NZ K72 | 3.81 | 2.89 | 2.71 | 3.41 | 3.5 |
| W3 - N4΄ ligand | 3.28 | 3.8 | 3.9 | 3.52 | 3.85 |
| W3 - OE1 E127 | 3.85 | 3.0 | 3.75 | 3.12 | 3.27 |
| W4 - O1 ligand | 3.75 | 3.14 | 3.68 | 3.64 | 3.43 |
| W4 - N1΄ ligand | 2.37 | 3.67 | 3.76 | 4.7 | 3.45 |
| W4 - OE2 E127 | 2.89 | 2.82 | 2.81 | 2.81 | 2.81 |
| W5 - O1 ligand | 5.66 | 5.69 | 6.18 | 6.17 | 5.78 |
| W5 - OH Y330 | 2.89 | 2.83 | 2.78 | 2.83 | 2.76 |
Figure 3.
Binding site of the catalytic subunit of cAMP-dependent protein kinase in a complex with isoquinolinesulfonyl inhibitor H8. The X-ray structure of the protein and the ligand H8 are shown in cyan cartoon and cyan sticks, respectively; water molecules are shown in cyan dots (PDB ID 1YDS). The ligand H8 and five selected water molecules close to the ligand after AMMOS2 case 1 minimization are shown in salmon. The ligand H8 and all water molecules within 6 Å around the ligand after AMMOS2 case 1 minimization are shown in magenta.
Regarding the speed of performance, AMMOS2 typically requires <1 min for 1 ligand and full protein flexibility computation for a protein of typical length (350 residues). AMMOS2 performed refinement of 1000 ligands in 25 min for case 1 and of 5000 ligands in 90 min for case 3 for a protein containing 356 amino acid residues.
MATERIALS AND METHODS
AMMOS2 method
AMMOS2 performs molecular mechanics of protein–ligand complexes based on the program AMMP (36). The web server parameters are optimized such as to handle a large number of ligands. AMMOS2 employs the program PREAMMP to convert the input protein and ligand files to AMMP format and performs AMMP autolink. AMMP uses a fast multipole algorithm for the efficient calculation of long-range forces thereby allowing evaluation of non-bonded terms without the use of a cutoff radius and increasing the speed, making calculations comparable to a standard treatment with an 8–10 Å radius cutoff (36). The sp4 AMMP force field (36) is developed on the basis of the UFF (Universal Force Field) potential set (41) and the AMBER partial charges (42). Our minimization procedure involves 2 × 500 iterations with conjugate gradient optimization. Finally, all minimized ligands are ranked by the AMMP minimized interaction receptor–ligand energy (18) as the water molecules and metal ions are considered as a part of the receptor.
Implementation
AMMOS2 consists of several programs developed in C and Python, and makes use of the open source programs AMMP and PREAMMP. AMMOS routines written in C ensure the transformation of the input protein and ligands files to a specific ammp format, the rules for the five flexibility cases and at the end of the process they generate the protein in PDB format and the small molecules in mol2 format. The automatization of the procedure described in Figure 1 is accomplished via a Python script. The web server relies on a cluster provided by the RPBS (Ressource Parisienne en Bioinformatique Structurale) platform that has been running since 2009 with about 60 000 connections per year in average. RPBS ensures control program's execution on the server side (storage and resources quota, jobs tracking, etc.). All jobs are kept for 7 days. There is no login requirement and data are not shared with other users.
Preparation of data for performance assessment
All protein and ligand structures were taken from the CCDC/Astex Test Set (39). Selected water molecules and individual metal ions present in the protein–ligand crystal structures were kept as HETATM. The input files preparation and docking analysis were carried out using AutoDockTools4 (40). Grid maps were centered in the bound ligand. Maximal grid sizes were 126 × 126 × 126 Å with a maximal grid spacing of 0.33 Å. Ligand conformational search was performed using Lamarckian genetic algorithm. The protein was kept rigid and all ligand torsion angles were set flexible. Fifty docking runs were performed, with an initial population of 150 random individuals and a maximum number of 25 000 000 energy evaluations. The top three best-scored ligand poses for each complex were taken for AMMOS2 refinement. The protein PDB files subjected for AMMMOS2 web server minimization were protonated with Chimera 1.10.2. The ligands docked with AutoDock4.2 were subjected to energy minimization with AMMOS2 web server with their initial protonation.
The pre-docked ligands for speed performance assessment were generated using virtual screening with the web server MTiOpenScreen for a protein receptor containing 356 amino acid residues (PDB ID: 1YDR). Screening of the diverse chemical compound collection Diverse-lib was performed in a binding site of dimensions 18 × 18 × 18 Å resulted in 2991 poses for 997 compounds. The top ranked 2000 poses were taken for AMMOS2 minimization.
CONCLUSION
The AMMOS2 web server aims at providing the scientific community a free and user-friendly protein–ligand–water minimization tool. AMMOS2 offers the possibility for users to refine a large number of protein–ligand complexes after docking/virtual screening. Including protein flexibility during the minimization with AMMOS2 improves the refinement of the complexes, resulting in more favorable interaction energies. Adding several waters in the binding site also improves the protein–ligand interaction energies. AMMOS2 offers valuable solutions to assist docking and structure-based virtual screening experiments keeping in mind that water molecules mediating protein–ligand interactions are of key importance to design high-affinity hit molecules.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
INSERM institute, Campus France, University Paris Diderot; Bulgarian National Science Fund [DKOST 01/11, NTS/France 01/4]. Funding for open access charge: INSERM UMR-S 973.
Conflict of interest statement. None declared.
REFERENCES
- 1. Villoutreix B.O., Lagorce D., Labbe C.M., Sperandio O., Miteva M.A.. One hundred thousand mouse clicks down the road: selected online resources supporting drug discovery collected over a decade. Drug Discov. Today. 2013; 18:1081–1089. [DOI] [PubMed] [Google Scholar]
- 2. Kar S., Roy K.. How far can virtual screening take us in drug discovery. Expert Opin. Drug. Discov. 2013; 8:245–261. [DOI] [PubMed] [Google Scholar]
- 3. Glaab E. Building a virtual ligand screening pipeline using free software: a survey. Brief Bioinform. 2016; 17:352–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lagorce D., Sperandio O., Baell J.B., Miteva M.A., Villoutreix B.O.. FAF-Drugs3: a web server for compound property calculation and chemical library design. Nucleic Acids Res. 2015; 43:W200–W207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Douguet D. e-LEA3D: a computational-aided drug design web server. Nucleic Acids Res. 2010; 38:W615–W621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grosdidier A., Zoete V., Michielin O.. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011; 39:W270–W277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ouyang X., Zhou S., Ge Z., Li R., Kwoh C.K.. CovalentDock Cloud: a web server for automated covalent docking. Nucleic Acids Res. 2013; 41:W329–W332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pires D.E., Ascher D.B.. CSM-lig: a web server for assessing and comparing protein-small molecule affinities. Nucleic Acids Res. 2016; 44:W557–W561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsai T.Y., Chang K.W., Chen C.Y.. iScreen: world's first cloud-computing web server for virtual screening and de novo drug design based on TCM database@Taiwan. J. Comput. Aided Mol. Des. 2011; 25:525–531. [DOI] [PubMed] [Google Scholar]
- 10. Irwin J.J., Shoichet B.K., Mysinger M.M., Huang N., Colizzi F., Wassam P., Cao Y.. Automated docking screens: a feasibility study. J. Med. Chem. 2009; 52:5712–5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li H., Leung K.S., Wong M.H., Ballester P.J.. USR-VS: a web server for large-scale prospective virtual screening using ultrafast shape recognition techniques. Nucleic Acids Res. 2016; 44:W436–W441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Labbe C.M., Rey J., Lagorce D., Vavrusa M., Becot J., Sperandio O., Villoutreix B.O., Tuffery P., Miteva M.A.. MTiOpenScreen: a web server for structure-based virtual screening. Nucleic Acids Res. 2015; 43:W448–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kalyaanamoorthy S., Chen Y.P.. Structure-based drug design to augment hit discovery. Drug Discov. Today. 2011; 16:831–839. [DOI] [PubMed] [Google Scholar]
- 14. Scior T., Bender A., Tresadern G., Medina-Franco J.L., Martinez-Mayorga K., Langer T., Cuanalo-Contreras K., Agrafiotis D.K.. Recognizing pitfalls in virtual screening: a critical review. J. Chem. Inf. Model. 2012; 52:867–881. [DOI] [PubMed] [Google Scholar]
- 15. Spyrakis F., Cavasotto C.N.. Open challenges in structure-based virtual screening: receptor modeling, target flexibility consideration and active site water molecules description. Arch. Biochem. Biophys. 2015; 583:105–119. [DOI] [PubMed] [Google Scholar]
- 16. Bhattacharya D., Nowotny J., Cao R., Cheng J.. 3Drefine: an interactive web server for efficient protein structure refinement. Nucleic Acids Res. 2016; 44:W406–W409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rodrigues J.P., Levitt M., Chopra G.. KoBaMIN: a knowledge-based minimization web server for protein structure refinement. Nucleic Acids Res. 2012; 40:W323–W328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pencheva T., Lagorce D., Pajeva I., Villoutreix B.O., Miteva M.A.. AMMOS: automated molecular mechanics optimization tool for in silico screening. BMC Bioinform. 2008; 9:438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pencheva T., Soumana O.S., Pajeva I., Miteva M.A.. Post-docking virtual screening of diverse binding pockets: comparative study using DOCK, AMMOS, X-Score and FRED scoring functions. Eur. J. Med. Chem. 2010; 45:2622–2628. [DOI] [PubMed] [Google Scholar]
- 20. Jereva D., Pencheva T., Lagorce D., Desvillechabrol D., Pajeva I., Miteva M.A.. Post-docking optimization of protein-ligand interactions involving water molecules. Asian J. Phys. 2014; 23:745–756. [Google Scholar]
- 21. Li Z., Lazaridis T.. Thermodynamics of buried water clusters at a protein-ligand binding interface. J. Phys. Chem. B. 2006; 110:1464–1475. [DOI] [PubMed] [Google Scholar]
- 22. Parikh H.I., Kellogg G.E.. Intuitive, but not simple: including explicit water molecules in protein-protein docking simulations improves model quality. Proteins. 2014; 82:916–932. [DOI] [PubMed] [Google Scholar]
- 23. de Beer S.B., Vermeulen N.P., Oostenbrink C.. The role of water molecules in computational drug design. Curr. Top Med. Chem. 2010; 10:55–66. [DOI] [PubMed] [Google Scholar]
- 24. Krimmer S.G., Cramer J., Betz M., Fridh V., Karlsson R., Heine A., Klebe G.. Rational design of thermodynamic and kinetic binding profiles by optimizing surface water networks coating protein-bound ligands. J. Med. Chem. 2016; 59:10530–10548. [DOI] [PubMed] [Google Scholar]
- 25. Amadasi A., Surface J.A., Spyrakis F., Cozzini P., Mozzarelli A., Kellogg G.E.. Robust classification of "relevant" water molecules in putative protein binding sites. J. Med. Chem. 2008; 51:1063–1067. [DOI] [PubMed] [Google Scholar]
- 26. Michel J., Tirado-Rives J., Jorgensen W.L.. Energetics of displacing water molecules from protein binding sites: consequences for ligand optimization. J. Am. Chem. Soc. 2009; 131:15403–15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Osterberg F., Morris G.M., Sanner M.F., Olson A.J., Goodsell D.S.. Automated docking to multiple target structures: incorporation of protein mobility and structural water heterogeneity in AutoDock. Proteins. 2002; 46:34–40. [DOI] [PubMed] [Google Scholar]
- 28. Ruiz-Carmona S., Alvarez-Garcia D., Foloppe N., Garmendia-Doval A.B., Juhos S., Schmidtke P., Barril X., Hubbard R.E., Morley S.D.. rDock: a fast, versatile and open source program for docking ligands to proteins and nucleic acids. PLoS Comput. Biol. 2014; 10:e1003571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Korb O., Stutzle T., Exner T.E.. Empirical scoring functions for advanced protein-ligand docking with PLANTS. J. Chem. Inf. Model. 2009; 49:84–96. [DOI] [PubMed] [Google Scholar]
- 30. Schymkowitz J.W., Rousseau F., Martins I.C., Ferkinghoff-Borg J., Stricher F., Serrano L.. Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:10147–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Young T., Abel R., Kim B., Berne B.J., Friesner R.A.. Motifs for molecular recognition exploiting hydrophobic enclosure in protein-ligand binding. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ross G.A., Morris G.M., Biggin P.C.. Rapid and accurate prediction and scoring of water molecules in protein binding sites. PLoS One. 2012; 7:e32036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Azuara C., Lindahl E., Koehl P., Orland H., Delarue M.. PDB_Hydro: incorporating dipolar solvents with variable density in the Poisson-Boltzmann treatment of macromolecule electrostatics. Nucleic Acids Res. 2006; 34:W38–W42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu C.H., Lin Y.F., Lin J.J., Yu C.S.. Prediction of metal ion-binding sites in proteins using the fragment transformation method. PLoS One. 2012; 7:e39252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu X., Dong Q., Yang J., Zhang Y.. Recognizing metal and acid radical ion-binding sites by integrating ab initio modeling with template-based transferals. Bioinformatics. 2016; 32:3260–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weber I.T., Harrison R.W.. Molecular mechanics calculations on Rous sarcoma virus protease with peptide substrates. Protein Sci. 1997; 6:2365–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bagossi P., Zahuczky G., Tözsér J., Weber I., Harrison R.. Improved parameters for generating partial charges: correlation with observed dipole moments. J. Mol. Model. 1999; 5:143–152. [Google Scholar]
- 38. Salentin S., Schreiber S., Haupt V.J., Adasme M.F., Schroeder M.. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015; 43:W443–W447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hartshorn M.J., Verdonk M.L., Chessari G., Brewerton S.C., Mooij W.T., Mortenson P.N., Murray C.W.. Diverse, high-quality test set for the validation of protein-ligand docking performance. J. Med. Chem. 2007; 50:726–741. [DOI] [PubMed] [Google Scholar]
- 40. Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J.. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009; 30:2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rappé A.K., Casewit C.J., Colwell K.S., Goddard W.A. III, Skiff W.M.. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 1992; 114:10024–10035. [Google Scholar]
- 42. Weiner S.J., Kollman P.A., Nguyen D.T., Case D.. An all atom force field for simulations of proteins and nucleic acids. J. Comput. Chem. 1986; 7:230–252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.