Abstract
Functional enrichment analysis has played a key role in the biological interpretation of high-throughput omics data. As a long-standing and widely used web application for functional enrichment analysis, WebGestalt has been constantly updated to satisfy the needs of biologists from different research areas. WebGestalt 2017 supports 12 organisms, 324 gene identifiers from various databases and technology platforms, and 150 937 functional categories from public databases and computational analyses. Omics data with gene identifiers not supported by WebGestalt and functional categories not included in the WebGestalt database can also be uploaded for enrichment analysis. In addition to the Over-Representation Analysis in the previous versions, Gene Set Enrichment Analysis and Network Topology-based Analysis have been added to WebGestalt 2017, providing complementary approaches to the interpretation of high-throughput omics data. The new user-friendly output interface and the GOView tool allow interactive and efficient exploration and comparison of enrichment results. Thus, WebGestalt 2017 enables more comprehensive, powerful, flexible and interactive functional enrichment analysis. It is freely available at http://www.webgestalt.org.
INTRODUCTION
Functional enrichment analysis can help reveal biological insights from high-throughput omics data. It has become indispensable in today's biological studies. WebGestalt (WEB-based Gene SeT AnaLysis Toolkit, http://www.webgestalt.org) is one of the first web tools for functional enrichment analysis. It was initially published in the 2005 NAR (Nucleic Acids Research) Web Server Issue (1), and was updated in the 2013 NAR Web Server Issue (2). Since 2013, WebGestalt has been visited 202 967 times by 81 860 unique users from 142 countries and territories according to Google Analytics, and the two papers have been cited in 1143 scientific papers since 2013 according to Google Scholar. These data clearly demonstrate a substantial and continuously increasing need for web-based functional enrichment analysis.
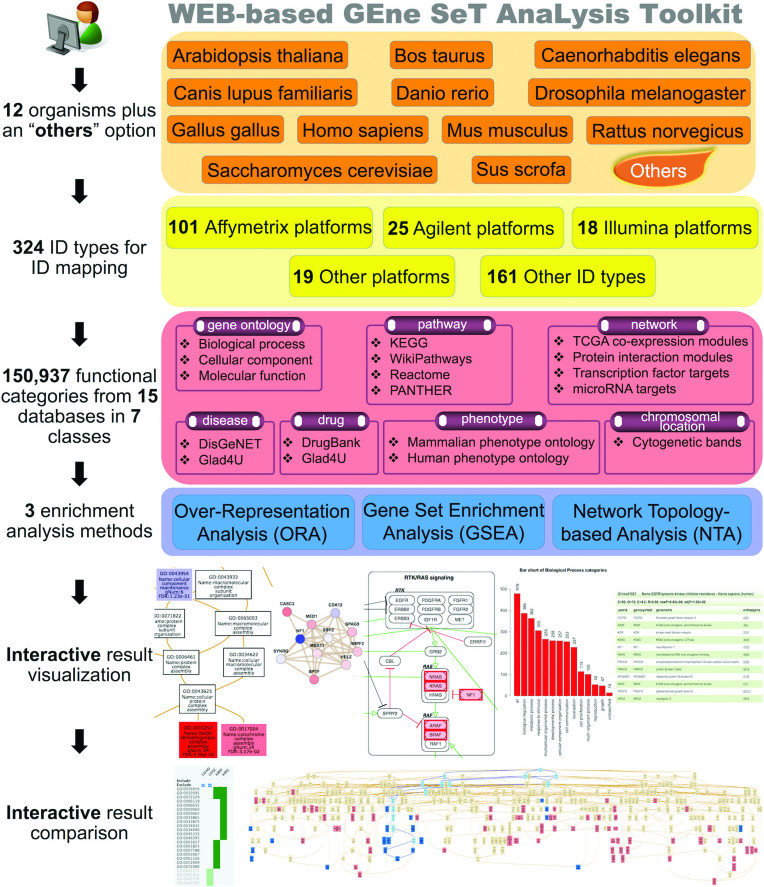
To better serve the needs of biologists from different research areas, we have significantly increased the number of supported organisms, gene identifiers and functional categories in WebGestalt during the past 4 years. The current version of WebGestalt supports 12 organisms, 324 gene identifiers from various databases and technology platforms, and 150 937 functional categories from public databases and computational analyses (Figure 1). Notably, experimental data from organisms or with gene identifiers not covered by the WebGestalt database can also be analyzed in WebGestalt because the current version supports using user-uploaded functional categories for enrichment analysis. There are three well-established and complementary methods for enrichment analysis, including Over-Representation Analysis (ORA) (3), Gene Set Enrichment Analysis (GSEA) (4) and Network Topology-based Analysis (NTA) (5,6). All these methods are now available in WebGestalt, making it a more powerful tool for enrichment analysis. To simplify the process of data uploading and parameter selection, we have consolidated the two-page process in the previous versions into a single, well-organized input page. To facilitate easy exploration and better understanding of the enrichment results, we have revamped the output interface with a user-friendly, tab-based and interactive report. We have also developed a companion tool GOView (http://www.webgestalt.org/GOView) that can help visualize and compare multiple geneontology (GO) enrichment results under the GO Directed Acyclic Graph (DAG) structure (Figure 1). Together, these changes are released as WebGestalt 2017, a more comprehensive, powerful, flexible and interactive web-based enrichment analysis toolkit.
Figure 1.
Summary of the organisms, methods, functional categories, gene identifiers and interactive visualization and comparison features in WebGestalt 2017.
NEW METHODS
WebGestalt 2013 supports only the Fisher's exact test based ORA method for enrichment analysis. Although easy to use and understand, the ORA method has inherent limitations. It requires a threshold to select a subset of genes for the analysis even when measurements for all genes are available, and all selected genes are treated equally in the analysis. Thus, the ORA method may result in information loss. The GSEA method (4) uses data from all genes to maximize information usage; however, it is not appropriate for studies that generate gene lists directly, e.g. gene clustering analysis, cancer genome mutation analysis or pull-down assays in protein complex analyses, etc. Neither ORA nor GSEA accounts for systems-level dependencies and interactions between genes. Thus, methods have been developed to take into consideration topological information of networks in enrichment analysis (5–8).
WebGestalt 2017 has been enhanced to support all the above three methods. The GSEA analysis is performed by the GSEA java jar file downloaded from http://software.broadinstitute.org/gsea/downloads.jsp, whereas the NTA is based on our previously published web application Gene2Net (http://www.gene2net.org). Gene2Net performs random walk-based network propagation (5,6) starting from a list of user uploaded genes. The output can be used to prioritize genes in the uploaded list and to identify new genes of interest.
NEW DATA
Gene Identifier
As shown in Table 1, WebGestalt 2013 supported eight organisms. Four new organisms, including Arabidopsis, cow, chicken and pig, have been added to WebGestalt 2017, expanding the total number of supported organisms to 12. Notably, the addition of the model organism Arabidopsis extends the impact of WebGestalt to the plant research community for the first time.
Table 1. Gene identifier comparison between the two versions of WebGestalt.
| Organism | ID types | |||
|---|---|---|---|---|
| 2013 | 2017 | Changea (%) | ||
| Homo sapiens | 61 | 77 | +26.2% | |
| Mus musculus | 43 | 52 | +20.9% | |
| Rattus norvegicus | 27 | 34 | +25.9% | |
| Canis lupus familiaris | 12 | 19 | +58.3% | |
| Drosophila melanogaster | 13 | 18 | +38.5% | |
| Danio rerio | 18 | 21 | +16.7% | |
| Caenorhabditis elegans | 13 | 29 | +123.1% | |
| Saccharomyces cerevisiae | 14 | 17 | +21.4% | |
| Arabidopsis thaliana | Xb | 9 | NA | |
| Bos taurus | X | 17 | NA | |
| Gallus gallus | X | 17 | NA | |
| Sus scrofa | X | 14 | NA | |
| Others | X | ∞c | NA | |
| Total | 12+∞ | 201 | 324 + ∞ | +61.2% |
aThe percentage of change between the numbers of gene identifiers in version 2013 and 2017 for each organism.
bThe version does not support this organism.
cVersion 2017 allows any kind of data upload.
WebGestalt 2017 has also increased the number of supported gene identifiers from 201 to 324 (+61.2%), with at least 15% increase for each organism (Supplementary Table S1). Specifically, the new version has significantly increased the number of supported Affymetrix platforms from 68 to 101 (+48.5%), Agilent platforms from 18 to 25 (+38.9%) and Illumina platforms from 10 to 18 (+80.0%). These expansions fill the gap between technology platform developments and user's ability to interpret data generated from the new platforms.
Importantly, when the organisms or gene identifiers are not covered by the WebGestalt database, WebGestalt 2017 allows users to upload their own functional categories and experimental data with the same gene identifiers, which provides the maximum flexibility for functional enrichment analysis. In fact, this new development makes it even possible to perform enrichment analysis for non-biological datasets in WebGestalt. Users can also use the WebGestalt forum (https://groups.google.com/forum/#!forum/webgestalt) to deposit and share functional categories and reference lists.
Functional category
Similar with WebGestalt 2013, WebGestalt 2017 also covers seven classes of functional categories, namely GO, pathway, network, phenotype, disease, drug and chromosomal location. However, the total number of categories has increased from 78 612 to 150 937 (+92.0%) in the new version (Table 2).
Table 2. Comparison of the functional categories supported by the old and new versions of WebGestalt.
| Class | New | Database | 2013 | 2017 | Data source | ||
|---|---|---|---|---|---|---|---|
| No. of categoriesa | No. of organismsb | No. of categories | No. of organisms | ||||
| Gene ontology | Biological process | 24 278 | 8 | 29 358 | 12 | http://www.geneontology.org | |
| Cellular component | 3079 | 8 | 4046 | 12 | |||
| Molecular function | 9508 | 8 | 10 539 | 12 | |||
| Pathwayc | KEGG | 390 | 8 | 2677 | 12 | http://rest.kegg.jp/link/has/pathway | |
| WikiPathways | 1018 | 8 | 1345 | 12 | http://www.wikipathways.org | ||
| ✓ | Reactome | X | X | 11 804 | 10 | http://www.reactome.org | |
| ✓ | PANTHER | X | X | 1336 | 10 | http://www.pantherdb.org | |
| Network | ✓ | Hierarchical mRNA co-expression modules | X | X | 30 852 | 1 | Firehose (http://gdac.broadinstitute.org/) |
| Hierarchical protein interaction modules | 1993 | 2 | 2979 | 11 | BioGrid (http://www.thebiogrid.org/) | ||
| MicroRNA target | 884 | 4 | 2210 | 10 | MSigDB (http://software.broadinstitute.org/gsea/misgdb) | ||
| Transcript factor target | 2460 | 4 | 6150 | 10 | MSigDB (http://software.broadinstitute.org/gsea/misgdb) | ||
| Phenotype | Phenotype | 19 023 | 2 | 23 277 | 2 | Human Phenotype Ontology (http://www.human-phenotype-ontology.org) Mammalian Phenotype Ontology (http://www.informatics.jax.org) | |
| Disease | ✓ | DisGeNET | X | X | 7607 | 1 | http://www.disgenet.org |
| GLAD4U | 758 | 1 | 2997 | 1 | http://glad4u.zhang-lab.org | ||
| Drug | ✓ | DrugBank | X | X | 4831 | 1 | http://www.drugbank.ca |
| GLAD4U | 2286 | 1 | 2355 | 1 | http://glad4u.zhang-lab.org | ||
| Chromosomal location | Cytogenetic band | 11 284 | 4 | 6574 | 12 | Entrez Gene (http://www.ncbi.nlm.nih.gov/gene) | |
| Others | ✓ | User-uploaded functional categories | X | X | ∞ | ∞ | |
| Total | 78 612 | 8 | 150 937 + ∞ | 12+∞ | |||
aThe number of categories in each database.
bThe number of organisms supported by each database.
cThe new version of Pathway Commons (http://www.pathwaycommons.org/) focuses on the pathway integration and does not provide the information on individual pathways. Thus, we did not include Pathway Commons in the new version of WebGestalt.
This substantial increase can be partially explained by the increased data content in the data sources already included in WebGestalt 2013 and the increased organism coverage in WebGestalt 2017. For example, the numbers of GO biological process, cellular component and molecular function categories have increased from 24 278, 3079 and 9508 to 29 358, 4046 and 10 539, respectively. In particular, human-related GO biological process, cellular component and molecular function categories increased from 11 763, 1319 and 3897 to 15 248, 1829 and 4624, respectively. As another example, compared to the 1993 protein interaction modules from human and mouse in the previous version, the new version includes 2979 modules derived from the protein–protein interaction networks of 11 organisms downloaded from BioGRID (9).
The increase of functional categories can also be attributed to the addition of new data sources to the WebGestalt database (Table 2). Under the pathway class, two new pathway databases, Reactome (10) and PANTHER (11) have been included in the new version, adding a total of 13 140 pathways. Under the Disease class, DisGeNET (12), a comprehensive disease–gene association database that covers the whole landscape of human diseases and the phenotype–genotype relationships, has been added. DisGeNet contains 2365 diseases with at least two associated genes and 924 diseases with at least five associated genes. Under the drug class, adding the DrugBank database (13) that contains 7607 drugs from Food and Drug Administration (FDA) approved small molecular drugs, FDA-approved biotech drugs, nutraceuticals and experimental drugs will greatly benefit the pharmacogenomics and pharmacoproteomic studies.
One unique addition to the new version is the 33 mRNA co-expression networks and 30 852 corresponding co-expression modules computationally derived from the RNA-Seq datasets across 33 TCGA (The Cancer Genome Atlas) cancer types. Compared with the disease-naive protein–protein interaction networks and protein interaction modules, these co-expression networks and co-expression modules may help reveal novel, cancer type specific molecular mechanisms in cancer systems biology studies. To construct the co-expression network for a cancer type, we first generated 100 bootstrapped data based on the RNASeq data and then built 100 bootstrapped co-expression networks using the K-nearest neighbor method with pair-wise similarities measured by the Spearman's correlation coefficient (6). Following the method described in (14), the 100 bootstrapped networks were combined to construct a consensus co-expression network. To identify co-expression modules from the consensus co-expression network, we used the NetSAM package (15). The co-expression modules are included in the WebGestalt database for functional category enrichment analysis. Moreover, we also included the consensus co-expression networks from all cancer types in the Gene2Net application for network-based analysis as described in the ‘New Methods’ section.
A fundamental improvement in WebGestalt 2017 is to allow users to upload their own functional category databases for enrichment analysis. This not only maximizes the possible functional category coverage but also makes it possible to perform analysis on organisms and gene identifies not covered by the WebGestalt database.
NEW INTERFACE
Both input and output interfaces have been significantly improved in WebGestalt 2017. As shown in Supplementary Figure S1, all input parameters are presented in the same page to reduce the operation time and complexity. This design also facilitates easy submission of multiple analyses. For example, a user can simply change the functional category database while keeping all other parameters the same to perform GO, pathway or other enrichment analyses based on the same uploaded gene list.
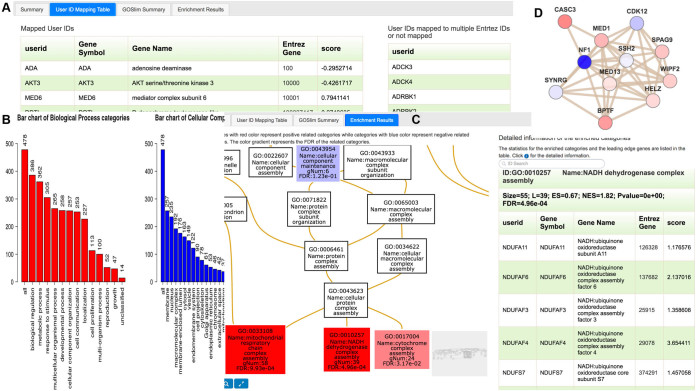
The new user-friendly output interface contains analysis summary, gene identifier (ID) mapping information, GO Slim summary and enrichment results (Figure 2A). We have made several changes from the previous version. First, the GOSlim classification plot is used to show the distribution of uploaded genes across the major branches of the GO Biological Process, Cellular Component and Molecular Function ontologies. The old version used a free version of the ChartDirector tool (http://www.advsofteng.com/) to generate the GOSlim classification plot that cannot be saved as a high-resolution figure for publication. The new version fixed the problem by creating high-resolution plots using R code (Figure 2B). Second, the old version used the graphviz tool (http://www.graphviz.org/) to plot the enriched GO and other enriched DAGs, which are static and come with low resolution. The new version produces interactive DAGs that can be zoomed in or out, and that allow searching and highlighting categories of interest and their related paths (Figure 2C). The categories in the DAGs can be colored based on the direction and FDR values from enrichment analysis. High-resolution figures can be downloaded in different file formats for publication and presentation. Third, because the input to the GSEA analysis contains values for individual genes, the new version can set node colors according to the values of corresponding genes in network visualization (Figure 2D). Finally, output report for each analysis is associated with a unique uniform resource locator and thus can be easily shared.
Figure 2.
Enhanced interfaces and data visualization. (A) Tab-based output interface, (B) GO Slim classification plots, (C) interactive DAG visualization with nodes colored based on the direction and FDR values from enrichment analysis, (D) network visualization with nodes colored based on corresponding gene values.
WebGestalt 2017 also includes a new tool, GOView, for comparing multiple sets of GO enrichment analysis results. Comparison of the enrichment analysis results from different experimental conditions (e.g. time points, drug doses, cancer types, brain regions, experimental platforms, etc.) can help identify unique and common molecular mechanisms across these conditions. This is usually performed by comparing the tabular reports downloaded from the enrichment analysis tools. Although this approach works well for functional categories with a flat structure, it fails to reveal the complex relationships among GO categories. Through the use of tightly coupled, interactive visualization of clickable Venn diagram (or sortable heat map) and the DAG visualization described above, GOView allows easy comparison of multiple GO term lists to identify common and specific biological themes.
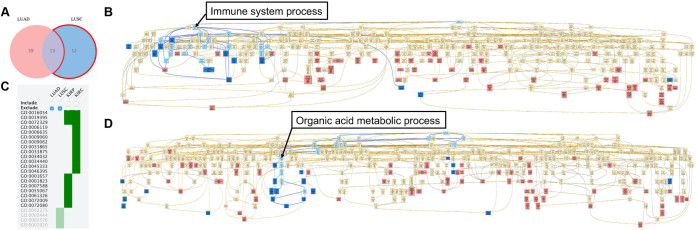
To demonstrate the use of GOView, we downloaded RNA-Seq data from tumors and paired normal samples from the firehose (http://gdac.broadinstitute.org/) for four TCGA cancer types, including lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), kidney renal clear cell carcinoma and kidney renal papillary cell carcinoma. For each cancer type, we calculated the signed–log(P-value) between tumor and normal samples for each gene based on the paired t-test and then identified enriched GO biological processes using the GSEA method with a stringent FDR cut- off of 1e-04. We first compared the enrichment results from the LUAD and LUSC analyses. As shown in the Venn diagram in Figure 3A, 52 and 25 GO biological processes were significantly upregulated in LUAD and LUSC tumor samples, respectively, and all enriched terms (nodes with red background) and their non-enriched parent terms (nodes with yellow background) can be co-visualized in an enriched DAG (Figure 3B). Clicking the LUSC specific part in the Venn diagram (i.e. the section highlighted by a red edge), the color of these 12 terms changed from red to blue in the DAG (Figure 3B). It is obvious that most of the LUSC specific terms are located close to each other in the DAG. The ‘search path’ function in GOView can highlight the ancestors and descendants (nodes with light blue background) of a selected term and related paths (blue edges), which facilitates a quick summary of closely related GO terms. As shown in Figure 3B, most of the LUSC specific terms are related to ‘Immune system process’. Next, we used the sortable heat map in GOView to compare enrichment results from all four cancer types. As shown in Figure 3C, common or unique terms can be identified by checking different combinations of the ‘Include’ or ‘Exclude’ boxes. After selecting the kidney cancer-specific terms in the heat map, corresponding DAG view suggested that many of these terms are the decedents of the term ‘Organic acid metabolic process’ (Figure 3D).
Figure 3.
Comparison of enriched GO terms among different cancer types. (A) Clickable Venn diagram. The clicked part can be highlighted by changing the edge color to red. (B) Interactive DAG visualization. Blue and red nodes in the DAG represent GO terms in the clicked part and other parts of the Venn diagram, respectively. Light blue nodes represent the ancestors and decedents of the selected node (‘Immune system process’ in this case) and blue edges represent the paths connecting these ancestors and decedents. (C) Sortable heat map. ‘Include’ and ‘Exclude’ check boxes are used for GO term selection and the excluded GO terms are blurred at the bottom of the heat map. (D) Selected GO terms in (C) are highlighted in blue in the DAG, and the common ancestor (i.e. organic acid metabolic process) of several highlighted terms were identified.
COMPARISON WITH OTHER TOOLS
Owing to the importance of functional enrichment analysis, many web-based tools have been developed. These tools typically have some overlapping functions but also unique strength. Here we compare WebGestalt 2017 with g:Profiler 2016 (16), Enrichr 2016 (17) and Babelomics 5.0 (18), focusing on the new features introduced in WebGestalt 2017. All of these tools have been updated within the past 4 years and have a substantial user community as determined by the number of citations. As shown in Table 3, three out of these four tools support GSEA. It is worth noting that although g:Profiler can analyze ranked gene lists, its method is based on the hypergeometric statistic and does not consider all genes in the experiment. Moreover, the network enrichment analysis used in g:Profiler is based on the overlapping neighbors of genes in the input list without considering the global network topology. WebGestalt has some advanced visualization features, including the novel DAG-based GO list comparison. Many of the newly added functional categories in WebGestalt are not supported by other tools, especially the computationally derived mRNA co-expression modules from 33 cancer types. WebGestalt provides flexible supports to user-defined functional categories. It is also the only tool that allows the analysis of data from organisms that are not natively included in the tool.
Table 3. A comparison between WebGesalt and three recently published web tools.
| WebGestalt | g:Profiler | Enrichr | Babelomics | |
|---|---|---|---|---|
| Organisms | ||||
| Upload data from other organisms not included in the tool | ✓ | |||
| Enrichment methods | ||||
| Gene Set Enrichment Analysis | ✓ | La | ✓ | |
| Network Topology-based Analysis | ✓ | ✓ | ||
| Functional category | ||||
| TCGA mRNA co-expression module | ✓ | |||
| Reactome | ✓ | ✓ | ✓ | |
| PANTHER | ✓ | ✓ | ||
| DisGeNET | ✓ | |||
| DrugBank | ✓ | |||
| Upload functional categories not included in the tool | ✓ | Lb | ||
| Visualization | ||||
| GO Slim summary | ✓ | |||
| Interactive DAG visualization | ✓ | ✓ | ||
| Network visualization with a color gradient | ✓ | |||
| Comparison | ||||
| GO list comparison under GO DAG | ✓ | |||
aAlthough g:Profiler can analyze ranked gene lists, its method is based on the hypergeometric statistic and does not consider all genes in the experiment.
bBabelomics can only allow uploading the databases from the existing organisms and only support gene symbol as the gene identifier.
DISCUSSION AND CONCLUSION
The new version of WebGestalt has significantly increased the coverage of organisms, gene identifiers and functional categories and supports three types of enrichment analysis methods. Moreover, functional categories and experimental data with gene identifiers not included in the WebGestalt database can be uploaded for enrichment analysis. The new user-friendly HTML report and the GOView comparison tool provide the most interactive and efficient methods to explore and compare enrichment results. Thus, WebGestalt is becoming an even more comprehensive, powerful, flexible and interactive gene set enrichment analysis tool kit.
There are hundreds of functional enrichment analysis tools, but most of them, including WebGestalt, fail to update their data content in a timely manner. One major challenge lies in the difficulty in keeping all gene identifiers and functional categories up-to-date. To make this process more efficient, we are working on an R package that will enable automatic downloading and updating of all data in WebGestalt. With this new tool, we should be able to update the WebGestalt database more frequently.
Despite all exciting new features introduced in WebGestalt 2017, more work remains to be done. One important area for future development is to support batch analysis and to allow easy integration into third-party applications. To this end, we have started developing an R package that will share the same input, analysis methods and output with the web version. Another area for future development is more advanced network and pathway topology-based analysis. For example, many pathways in the pathway databases have directed (e.g. gene A phosphorylates gene B) and signed (e.g. inhibition or activation) edges, but such information is ignored in our current analyses. We will evaluate and implement new methods that can leverage this type of information (19) in the future. In summary, we will continuously update and expand our data content, improve our enrichment analysis methods and eventually make WebGestalt available to the broad user community through both web-based and programmatic interfaces.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute (NCI) [U24 CA210954]; Leidos Biomedical Research, Inc. [15X038]; Cancer Prevention & Research Institutes of Texas Scholarship (CPRIT) [RR160027 to B.Z.]; McNair Medical Institute Scholarship (to B.Z.). Funding for open access charge: NCI [U24 CA210954]; CPRIT [RR160027].
Conflict of interest statement. None declared.
REFERENCES
- 1. Zhang B., Kirov S., Snoddy J.. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005; 33:W741–W748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang J., Duncan D., Shi Z., Zhang B.. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013; 41:W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Khatri P., Sirota M., Butte A.J.. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput. Biol. 2012; 8:e1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang J., Qian J., Hoeksema M.D., Zou Y., Espinosa A.V., Rahman S.M., Zhang B., Massion P.P.. Integrative genomics analysis identifies candidate drivers at 3q26-29 amplicon in squamous cell carcinoma of the lung. Clin. Cancer Res. 2013; 19:5580–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang J., Ma Z., Carr S.A., Mertins P., Zhang H., Zhang Z., Chan D.W., Ellis M.J., Townsend R.R., Smith R.D. et al. . Proteome profiling outperforms transcriptome profiling for coexpression based gene function prediction. Mol. Cell. Proteomics. 2017; 16:121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kohler S., Bauer S., Horn D., Robinson P.N.. Walking the interactome for prioritization of candidate disease genes. Am. J. Hum. Genet. 2008; 82:949–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vandin F., Clay P., Upfal E., Raphael B.J.. Discovery of mutated subnetworks associated with clinical data in cancer. Pac. Symp. Biocomput. 2012; 2012:55–66. [PubMed] [Google Scholar]
- 9. Chatr-Aryamontri A., Oughtred R., Boucher L., Rust J., Chang C., Kolas N.K., O’Donnell L., Oster S., Theesfeld C., Sellam A. et al. . The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017; 45:D369–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S. et al. . The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016; 44:D481–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mi H., Poudel S., Muruganujan A., Casagrande J.T., Thomas P.D.. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016; 44:D336–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pinero J., Bravo A., Queralt-Rosinach N., Gutierrez-Sacristan A., Deu-Pons J., Centeno E., Garcia-Garcia J., Sanz F., Furlong L.I.. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017; 45:D833–D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wishart D.S., Knox C., Guo A.C., Shrivastava S., Hassanali M., Stothard P., Chang Z., Woolsey J.. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006; 34:D668–D672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Margolin A.A., Wang K., Lim W.K., Kustagi M., Nemenman I., Califano A.. Reverse engineering cellular networks. Nat. Protoc. 2006; 1:662–671. [DOI] [PubMed] [Google Scholar]
- 15. Shi Z., Wang J., Zhang B.. NetGestalt: integrating multidimensional omics data over biological networks. Nat. Methods. 2013; 10:597–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reimand J., Arak T., Adler P., Kolberg L., Reisberg S., Peterson H., Vilo J.. g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016; 44:W83–W89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. et al. . Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016; 44:W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alonso R., Salavert F., Garcia-Garcia F., Carbonell-Caballero J., Bleda M., Garcia-Alonso L., Sanchis-Juan A., Perez-Gil D., Marin-Garcia P., Sanchez R. et al. . Babelomics 5.0: functional interpretation for new generations of genomic data. Nucleic Acids Res. 2015; 43:W117–W121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mitrea C., Taghavi Z., Bokanizad B., Hanoudi S., Tagett R., Donato M., Voichita C., Draghici S.. Methods and approaches in the topology-based analysis of biological pathways. Front. Physiol. 2013; 4:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.