Abstract
ReFOLD is a novel hybrid refinement server with integrated high performance global and local Accuracy Self Estimates (ASEs). The server attempts to identify and to fix likely errors in user supplied 3D models of proteins via successive rounds of refinement. The server is unique in providing output for multiple alternative refined models in a way that allows users to quickly visualize the key residue locations, which are likely to have been improved. This is important, as global refinement of a full chain model may not always be possible, whereas local regions, or individual domains, can often be much improved. Thus, users may easily compare the specific regions of the alternative refined models in which they are most interested e.g. key interaction sites or domains. ReFOLD was used to generate hundreds of alternative refined models for the CASP12 experiment, boosting our group's performance in the main tertiary structure prediction category. Our successful refinement of initial server models combined with our built-in ASEs were instrumental to our second place ranking on Template Based Modeling (TBM) and Free Modeling (FM)/TBM targets. The ReFOLD server is freely available at: http://www.reading.ac.uk/bioinf/ReFOLD/.
INTRODUCTION
The refinement of 3D models of proteins can be thought of as the ‘end game’ of protein structure prediction (1). Subtle improvements in predictive models are often required to move them the ‘last mile’, beyond the template or fragments upon which they are based, so they more accurately reflect the time averaged observed structures. Comparative protein modelling is now routinely used across the life sciences. However, many biological applications of 3D models are critically dependent on high model accuracy, particularly in key regions, such as binding sites for drug discovery (2). Improving the accuracy of comparative models, beyond the information derived from the template, therefore continues to be one of the pressing problems in structural bioinformatics. However, it has proved difficult to develop reliable and practically useful refinement methods, highlighted by the relatively slow progress seen in CASP (Critical Assessment of Techniques for Protein Structure Prediction). Indeed, the CASP assessors have stated that ‘the endgame problem is going to be at least several orders of magnitude harder than the TBM (Template Based Modeling) problem’ (1).
Refinement was introduced as a new category in CASP7 to encourage development. While quality assessment (QA) methods, such as ModFOLD (3), can accurately identify the magnitude of errors in models and locate where those errors are, refinement methods aim to fix the errors in models. Once an atomic model has been obtained, it may be tweaked to idealize bond geometry and to remove unfavorable contacts that may have been introduced by the initial modelling process. Ironically, in the early years of refinement, often most methods made models worse, rather than improving upon them. In recent years, the situation has been improved, however, it is hard to quantify if improvements between CASPs are being made in refinement, as maintaining the level of ‘target difficulty’ is problematic (1).
Refinement methods can be loosely categorized into the automated server-based programs and non-server-based highly CPU intensive programs, referred to as the ‘human’ refinement groups in the CASP experiments as they allow a large degree of manual intervention in their pipelines. The freely available automated server GalaxyRefine, focuses on rebuilding and repacking of the side chains, before performing overall structure relaxation via molecular dynamic simulation (MDS) (4). Additionally, the KoBaMIN server, relies on minimization of a knowledge-based potential of mean force (5). Another successful fully automated method is the 3Drefine method (6,7), which works in a two-step process involving optimization of the hydrogen bonding network and composite physics and knowledge-based force fields to give atomic-level energy minimization using the MESHI molecular modelling framework (8). The advantage of such automated approaches is that they are easily distributed, accessible and user-friendly. However, successes in improving protein models via automated approaches remains relatively low, with the most successful servers making relatively conservative changes. In contrast, the less well automated approaches, which often make more bold changes to models, have arguably achieved better results.
The human refinement methods commonly use physics based approaches that select structures from Molecular Dynamics (MD)-based ensembles followed by structural averaging, which has led to a high-level of refinement success (9,10).The most successful human refinement group in both CASP10 and CASP11 was the Feig group (1,11). Despite the relatively high performance of such strategies, using highly intensive MDS approaches for refinement has some drawbacks. The refinement procedure used by the Feig group consumed 75,000 core hours (12 days on 265 cores) on the multicore Intel Xenon CPU machines of the day, just to refine a single 3D model for a single protein target (12). Given that each protein structure prediction method will often produce hundreds or thousands of alternative models for each target, using similarly intensive MDS refinement procedures would quickly become problematic to manage in terms of CPU/GPU loads, if it were to be performed on multiple targets and models. This means that MDS refinement protocols, such as those developed by the Feig group, would be less practical to be used routinely for large scale fully automated structure prediction pipelines. Thus, faster, more practical and, ideally, as accurate and consistent, fully automated refinement servers are required. While many automated servers exists, few servers make use of the power of MDS and few adequately evaluate the models and present users with accurate reports of the likely improvement in errors. User feedback on when and, more specifically, where a 3D model has been improved is important to get right and it is often neglected.
We have a good track record in building 3D models (13–15) and estimating the likely errors they may have, recently termed Accuracy Self Estimates or (ASE) (16–18). We have had some success at using quality assessment guided multiple template based modelling to improve upon errors in single template models (19), but this approach requires sequence-structure alignments and template data. The ReFOLD server is our first successful attempt at developing a method for directly fixing likely errors in any user supplied 3D models, using global quality assessment guided refinement. The ReFOLD server also equips users with the ability to identify specific domains or regions in a protein that are likely to be correctly refined, via its accurate per-residue error estimates.
MATERIALS AND METHODS
ReFOLD is the server implementation of the refinement method we initially developed for the CASP12 experiment. The ReFOLD method works using a unique hybrid approach consisting of rapid iterative refinement with i3Drefine (7) and molecular dynamics simulations with NAMD (20), combined with the latest version of our leading model quality estimation method, ModFOLD (3) (http://www.reading.ac.uk/bioinf/ModFOLD/, manuscript for version 6 in preparation). Input models were refined and evaluated over a number of successive stages. This iterative filtering process led to the generation of hundreds of alternative refined models, which were then ranked by quality.
The ReFOLD pipeline consisted of three protocols outlined in the flowchart shown in Supplementary Figure S1. The first protocol simply used a rapid iterative strategy for refinement of starting models, with 20 refinement cycles (iterations) of i3Drefine (7). Although the authors recommend not to run i3Drefine for more than 10 iterations, an optimal number of iterations is not specified, and often models will improve further beyond 10 iterations. Simply, all i3Drefine requires is a starting model, in PDB format, and a given number iterations as input parameters.
The second protocol employed a more complex and CPU/GPU intensive molecular dynamic simulation strategy, using NAMD (20) to refine each starting model. The NAMD protocol that we implemented was inspired by that of Feig and Mirjalili (10), utilizing all atom MD sampling in explicit solvent. Simulations were conducted at 298k under neutralized pH conditions with 1 bar of atmospheric pressure to resemble normal cellular conditions. Weak harmonic restraints with a spring constant of 0.05 kcal/mol/A2 on all C-alpha atoms were added to conserve aspects of the starting model. The CHARMM22/27 (21) force field (for protein systems, the two are equivalent) was used as the parameter file with default TIP3P water model (22). As some proteins are sensitive to ionic conditions in the solvent and PME (Particle Mesh Ewald) (23) was being used, the system was neutralized by adding either Na+ or Cl- ions. Only non-bonded interactions were calculated with bonded interactions excluded; the exclusion parameter was set to exclude up to four pairs of bonded atoms. Electrostatic and van der Waal interactions for these atom pairs were instead calculated using the parameter file (CHARMM27). A 12 Å cut-off for calculating non-bonded interactions (mostly van der Waal's) was applied, as this is the official standard for CHARMM force fields, with the smoothing function switching distance of 10 Å to avoid discontinuity in energy and forces. The pairlistdist function was set to a distance of 14 Å between atom pairs for inclusion in pair lists. All hydrogen bonding was rigidified, using the rigidBonds functions, allowing the time step to be increased to 2 fs. PME was used to calculate full electrostatics and the temperature in the system was controlled through Langevin dynamics (24), which balances random noise with friction to push atoms to the target temperature (298 K). The langevinDamping function was set to 1/ps to give some temperature control without dampening the system to the extent that effects were not seen. Periodic boundary conditions were used to achieve maintenance of conditions such as pressure, density and water box and also enable the use of PME, making the simulation more biologically realistic. The first stage of the MD simulation was 1000 steps of minimization, to lower the potential energy of the system reducing bad initial contacts, high force and temperature regions. This process zeros velocity, so that the reinitvels function returns the system to the desired temperature (298 K). For CASP12, the second protocol was run using multiple short trajectories in place of a single long trajectory; four parallel simulations were run for 2 ns, giving a cumulative simulation time of 8 ns per target.
The 164 refined models generated from the second protocol were assessed and ranked using ModFOLD6_rank method. The third protocol was a combination of the first two approaches, where the top ranked model from the second protocol was further refined using 20 iterations of i3Drefine. Subsequently, all of the refined models generated by each of these protocols and the starting model were pooled and re-ranked again using ModFOLD6_rank and the final top models were identified. Finally, each refined model was evaluated and compared with the original starting model in terms of local and global model quality scores. Static and dynamic graphical outputs were generated using the raw QA scores in order to display the top refined models and estimated improvements in a user friendly manner.
RESULTS AND DISCUSSION
Server inputs and outputs
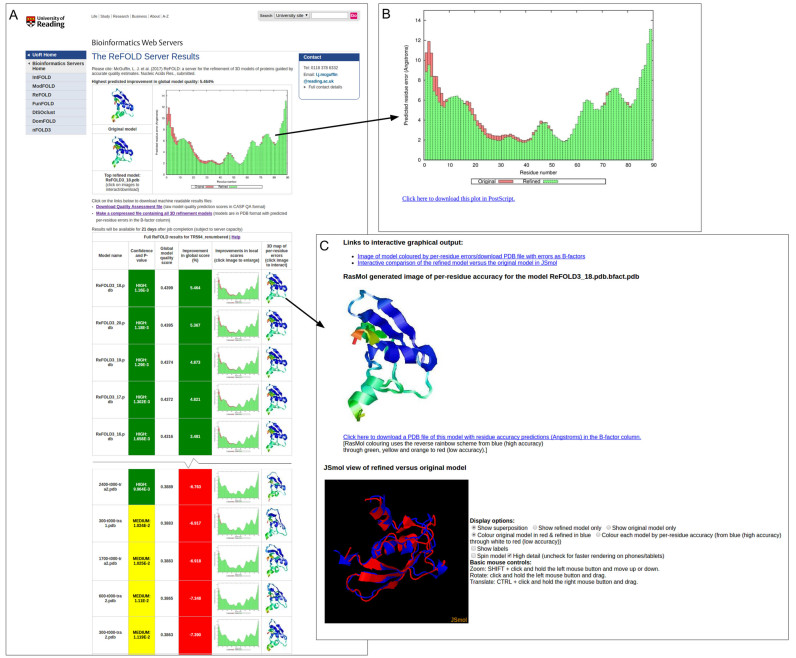
The only required inputs to the ReFOLD server are the amino acid sequence for the target protein and a single 3D model (in PDB format) for refinement. Users may optionally provide a name for their protein sequence and their email address. The ReFOLD server results page provides users with an accurate estimate of the likely percentage improvement in their global quality score based on the top refined model (Figure 1A). In addition, the server is unique in providing output for multiple alternative refined models in a way that allows users to quickly visualize the key residue locations, which are likely to have been improved upon compared to their original model. The results page provides users with a series of per-residue error plots, which demonstrate the reduction in local errors in the refined models compared with the uploaded original (Figure 1B). This is important, as global refinement of a full chain model may not always occur, whereas local regions, or individual domains, may often be much improved. Presenting results to users in this way also gives them the choice to easily compare alternative refined models, allowing them to focus their attention to key interacting residues or specific domains. Users can also click through the images in the table in order to compare the refined and original 3D models interactively in using the JSmol/HTML5 framework (Figure 1C). No plugins are required and, conveniently, interactive results may also be viewed on mobile devices.
Figure 1.
ReFOLD server results for CASP12 refinement target TR594. In CASP12, the McGuffin group used ReFOLD to improve the GDT_TS score from 55.34 → 58.43 (A) Main results page with a summary of the scores for the top refined model with the highest predicted improvement in model quality. The full table of scores for every alternative refined model is displayed below the top hit (truncated here to fit page). Clicking on the images on the main results page allows results to be visualized in more detail and downloaded. (B) Histogram of the local or per-residue ModFOLD6 errors for the top refined model (green bars) compared with the original model. Plots for each alternative refined model may be downloaded. (C) Interactive views of the refined model compared with the original model, which can be manipulated in 3D using the JSmol/HTML5 framework and/or downloaded for local viewing.
Independent benchmarking
Our ReFOLD refinement approach was independently tested by the assessor team in the recent CASP12 experiment, and it was a key factor contributing to our success. During the CASP12 prediction season (May–August 2016), we used ReFOLD to build hundreds of alternative refined models, for both the main tertiary structure prediction and refinement categories. ReFOLD gave us a significant performance boost in the main tertiary structure prediction category, where it enabled us to further improve the quality of some of the very best initial server models. As a result of our high performance, we were invited to speak at the meeting in Gaeta about our template based modelling (TBM) strategy. Our group ranked in 2nd position overall on both the TBM and TBM/FM (Free Modelling) targets according to the assessors’ formula (Supplementary Tables S1 and S2), and we ranked 11th overall on FM targets. Our group also improved the global and local quality scores for many of the starting models provided in the refinement category itself, where we ranked 14th overall (http://predictioncenter.org/casp12/).
The results in Table 1 summarize our CASP12 results for regular targets, where the top server models, selected by ModFOLD_rank, were then further refined with ReFOLD. Arguably, this benchmark represents a more realistic user test case, where each of the starting models have been selected in a fully automated manner and have been generated for full length protein chains. The results in Table 1 show that the majority of models were either improved upon or unchanged according to the GDT_TS (25) and MolProbity (26) scores. There is an average overall improvement in scores (∑ΔGDT_TS = 8.15, ∑ΔMolProbity = –10.16), which are greater than the standard errors, and there is a strong correlation between the ModFOLD6_rank global scores and GDT_TS scores (Pearson's r = 0.8094). The GDT_TS score measures the global positioning of the backbone C-alpha atoms based on multiple superpositions of the predicted and experimental structure. It is promising to see that ReFOLD improves backbone quality, but these changes are relatively small, so significant improvements in GDT_TS are hard to detect on the CASP data set. While it is encouraging that the improvement in cumulative GDT_TS scores is greater than the error, the pairwise t-test on the data in Table 1 does not provide evidence that the improvement in the backbone is statistically significant (P = 0.1196). However, the improvement in the MolProbity full atom scores is statistically significant at the 95% confidence level, according to the pairwise t-test on the data in Table 1 (P = 0.0301). Whereas the GDT_TS score focuses only on the C-alpha atoms, the MolProbity score is an all-atom composite score, which means that it also measures the finer detail of differences in the local errors of the side chains. The MolProbity score denotes the expected resolution with respect to experimental structures, therefore models with lower MolProbity scores are more physically realistic. Figure 2 shows examples where ReFOLD has successfully refined starting models of varying target difficulty (FM, TBM, TBM/FM targets and a refinement target), with improved GDT_TS scores for each.
Table 1. Summary of ReFOLD performance on CASP12 targets for full chains.
| CASP model ID | ModFOLD6_rank | GDT-TS | MolProbity | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Target ID | Starting model | Refined model | Starting model | Refined model | Starting model | Refined model | Diff. | Starting model | Refined model | Diff. |
| T0859 | T0859TS479_1-D1 | T0859TS017_1-D1 | 0.4059 | 0.4189 | 24.78 | 27.66 | 2.88 | 2.88 | 2.7 | –0.18 |
| T0862 | T0862TS183_3-D1 | T0862TS017_1-D1 | 0.3809 | 0.3809 | 58.6 | 58.6 | 0 | 2.66 | 2.66 | 0 |
| T0863 | T0863TS005_1 | T0863TS017_1 | 0.2994 | 0.3024 | 9.62 | 9.54 | –0.08 | 0.93 | 1.61 | 0.68 |
| T0864 | T0864TS479_4-D1 | T0864TS017_1-D1 | 0.3514 | 0.3658 | 20.22 | 19.92 | –0.3 | 3.17 | 1.85 | –1.32 |
| T0866 | T0866TS479_5-D1 | T0866TS017_1-D1 | 0.4049 | 0.425 | 44.71 | 45.91 | 1.2 | 3.44 | 2.45 | –0.99 |
| T0868 | T0868TS479_3-D1 | T0868TS017_1-D1 | 0.4338 | 0.4438 | 57.97 | 60.99 | 3.02 | 2.83 | 0.65 | –2.18 |
| T0869 | T0869TS479_3-D1 | T0869TS017_1-D1 | 0.4288 | 0.432 | 32.21 | 31.97 | –0.24 | 3.17 | 3.79 | 0.62 |
| T0870 | T0870TS183_4-D1 | T0870TS017_1-D1 | 0.4733 | 0.4804 | 33.94 | 32.32 | –1.62 | 2.86 | 1.07 | –1.79 |
| T0872 | T0872TS405_1-D1 | T0872TS017_1-D1 | 0.6467 | 0.6539 | 66.76 | 67.9 | 1.14 | 2.63 | 3.12 | 0.49 |
| T0874 | T0874TS005_2-D1 | T0874TS017_1-D1 | 0.512 | 0.5148 | 46.17 | 46.17 | 0 | 0.92 | 2.1 | 1.18 |
| T0875 | T0875TS005_2-D1 | T0875TS017_1-D1 | 0.5254 | 0.53 | 43.1 | 43.1 | 0 | 1.19 | 1.19 | 0 |
| T0876 | T0876TS220_1-D1 | T0876TS017_1-D1 | 0.4563 | 0.4684 | 48.54 | 49.38 | 0.84 | 1.9 | 2.41 | 0.51 |
| T0878 | T0878TS220_2-D1 | T0878TS017_1-D1 | 0.3544 | 0.364 | 13.01 | 12.5 | –0.51 | 2.66 | 2.09 | –0.57 |
| T0880 | T0880TS405_1 | T0880TS017_1 | 0.3885 | 0.4201 | 10.75 | 11.27 | 0.52 | 3.07 | 2.15 | –0.92 |
| T0882 | T0882TS357_4-D1 | T0882TS017_1-D1 | 0.6089 | 0.6161 | 79.43 | 81.33 | 1.9 | 1.73 | 1.36 | –0.37 |
| T0886 | T0886TS479_1 | T0886TS017_1 | 0.3569 | 0.3758 | 26.96 | 28.27 | 1.31 | 3.19 | 2.91 | –0.28 |
| T0887 | T0887TS220_2-D1 | T0887TS017_1-D1 | 0.4649 | 0.4756 | 40.22 | 38.51 | –1.71 | 1.79 | 1.44 | –0.35 |
| T0888 | T0888TS183_4-D1 | T0888TS017_1-D1 | 0.3677 | 0.3778 | 19.84 | 19.21 | –0.63 | 3 | 3.93 | 0.93 |
| T0890 | T0890TS220_2 | T0890TS017_1 | 0.458 | 0.4653 | 25.53 | 24.73 | –0.8 | 2.27 | 1.17 | –1.1 |
| T0892 | T0892TS479_3 | T0892TS017_1 | 0.4193 | 0.4295 | 37.18 | 38.34 | 1.16 | 3.03 | 2.01 | –1.02 |
| T0894 | T0894TS183_1 | T0894TS017_1 | 0.3434 | 0.3469 | 50.52 | 51.05 | 0.53 | 3.57 | 3.81 | 0.24 |
| T0895 | T0895TS250_5-D1 | T0895TS017_1-D1 | 0.584 | 0.584 | 71.67 | 71.67 | 0 | 2.32 | 2.32 | 0 |
| T0896 | T0896TS220_2 | T0896TS017_1 | 0.3193 | 0.3252 | 19.63 | 20.75 | 1.12 | 1.94 | 1.93 | –0.01 |
| T0897 | T0897TS005_2 | T0897TS017_1 | 0.3141 | 0.3179 | 10.02 | 9.64 | –0.38 | 1.03 | 1.79 | 0.76 |
| T0898 | T0898TS405_1 | T0898TS017_1 | 0.39 | 0.4062 | 28.42 | 28.11 | -0.31 | 3.47 | 3.63 | 0.16 |
| T0899 | T0899TS183_2 | T0899TS017_1 | 0.3994 | 0.4062 | 27.88 | 28.1 | 0.22 | 3.69 | 3.02 | –0.67 |
| T0900 | T0900TS479_1-D1 | T0900TS017_1-D1 | 0.4995 | 0.5129 | 42.65 | 44.85 | 2.2 | 3.13 | 1.05 | –2.08 |
| T0901 | T0901TS479_2 | T0901TS017_1 | 0.4908 | 0.4952 | 37.37 | 37.2 | –0.17 | 3.37 | 3.82 | 0.45 |
| T0904 | T0904TS479_1-D1 | T0904TS017_1-D1 | 0.4628 | 0.4656 | 38.74 | 38.55 | –0.19 | 3.2 | 2.63 | –0.57 |
| T0905 | T0905TS479_2 | T0905TS017_1 | 0.5289 | 0.531 | 35.06 | 34.98 | –0.08 | 3.23 | 2.87 | –0.36 |
| T0907 | T0907TS479_1 | T0907TS017_1 | 0.3736 | 0.391 | 21.32 | 20.55 | –0.77 | 3.41 | 2.82 | –0.59 |
| T0909 | T0909TS251_5-D1 | T0909TS017_1-D1 | 0.44 | 0.4485 | 41.07 | 41.07 | 0 | 3.32 | 2.59 | –0.73 |
| T0911 | T0911TS405_2-D1 | T0911TS017_1-D1 | 0.6009 | 0.6053 | 52.02 | 52.63 | 0.61 | 3.37 | 3.4 | 0.03 |
| T0912 | T0912TS220_1 | T0912TS017_1 | 0.4187 | 0.4237 | 47.66 | 47.16 | -0.5 | 2.36 | 1.87 | –0.49 |
| T0913 | T0913TS479_5-D1 | T0913TS017_1-D1 | 0.5734 | 0.5762 | 61.98 | 62.06 | 0.08 | 3.06 | 3.75 | 0.69 |
| T0914 | T0914TS183_1 | T0914TS017_1 | 0.349 | 0.349 | 16.33 | 16.33 | 0 | 3.71 | 3.71 | 0 |
| T0915 | T0915TS220_1-D1 | T0915TS017_1-D1 | 0.4873 | 0.4988 | 48.86 | 47.4 | –1.46 | 1.85 | 1.94 | 0.09 |
| T0918 | T0918TS183_1 | T0918TS017_1 | 0.3226 | 0.3315 | 14.83 | 15.54 | 0.71 | 3.25 | 2.11 | –1.14 |
| T0923 | T0923TS220_2-D1 | T0923TS017_1-D1 | 0.3305 | 0.3334 | 18.18 | 18.18 | 0 | 1.92 | 1.24 | –0.68 |
| T0941 | T0941TS183_5-D1 | T0941TS017_1-D1 | 0.3366 | 0.3465 | 8.94 | 9.16 | 0.22 | 3.01 | 3.73 | 0.72 |
| T0942 | T0942TS183_1 | T0942TS017_1 | 0.4443 | 0.4497 | 45.54 | 45.22 | -0.32 | 2.78 | 3.4 | 0.62 |
| T0944 | T0944TS220_5-D1 | T0944TS017_1-D1 | 0.6205 | 0.6246 | 72.92 | 73.22 | 0.3 | 1.87 | 1.67 | –0.2 |
| T0945 | T0945TS183_1-D1 | T0945TS017_1-D1 | 0.4438 | 0.4536 | 53.8 | 53.8 | 0 | 3.73 | 3.73 | 0 |
| T0946 | T0946TS479_1 | T0946TS017_1 | 0.4508 | 0.4572 | 45.12 | 44.86 | –0.26 | 3.73 | 3.84 | 0.11 |
| T0947 | T0947TS220_1-D1 | T0947TS017_1-D1 | 0.5149 | 0.5277 | 64.43 | 63.29 | –1.14 | 2.01 | 2.76 | 0.75 |
| T0948 | T0948TS479_1-D1 | T0948TS017_1-D1 | 0.6367 | 0.6368 | 71.98 | 71.64 | –0.34 | 2.69 | 2.09 | –0.6 |
| Total | 1786.48 | 1794.63 | 8.15 | 124.34 | 114.18 | –10.16 | ||||
| Std. Err. | 2.822 | 2.845 | – | 0.1152 | 0.1367 | – | ||||
Mean GDT_TS of starting model = 38.84. ∑ΔGDT_TS = 8.15 (higher scores are better). Mean MolProbity of starting model = 2.70. ∑ΔMolProbity = –10.16 (lower scores are better). Data are from http://www.predictioncenter.org/casp12/.
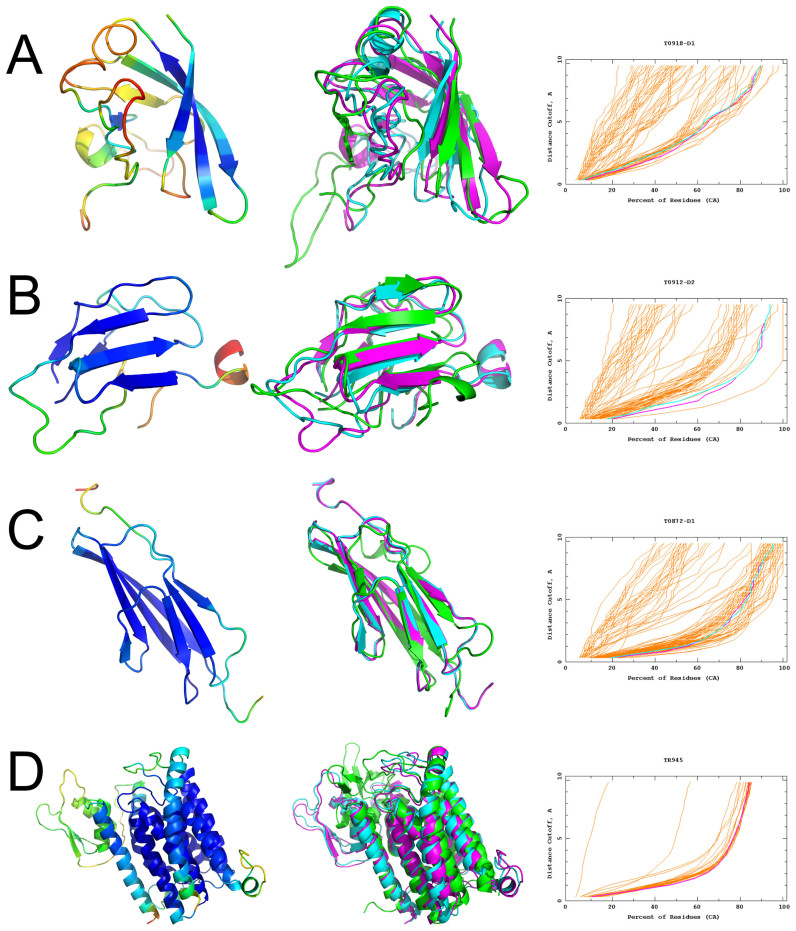
Figure 2.
Four examples of CASP12 targets where using ReFOLD allowed us to fix errors in the top selected server model. Left panels, refined model with ModFOLD6 accuracy self assessment (ASE) displayed using the temperature colour scheme. Middle panels, superposition of the top selected server model (cyan), refined model (magenta) and native structure (green). Right panels, GDT plots comparing top selected server models (cyan) with the ReFOLD refined models (magenta). (A) FM target T0918 domain 1: QUARK_TS1 versus McGuffin_TS1, a GDT_TS improvement from 45.6 to 48.38. (B) FM/TBM target T0912 domain 2: GOAL_TS1 versus McGuffin_TS1, GDT_TS from 62.95 to 65.36. (C) TBM target T0872 domain 1: IntFOLD4_TS1 versus McGuffin_TS1, GDT_TS from 66.76 to 67.9 (D) TR945: starting model versus McGuffin_TS1, GDT_TS from 59.27 to 61.20. Models are rendered using PyMOL (http://www.pymol.org). GDT plots are from http://www.predictioncenter.org/casp12/.
It is clear that the success of refinement is related to the quality of the starting model, when targets are subdivided into domains (Supplementary Tables S3–S5). Dividing targets into domains allows us to pinpoint where the method performance is strongest. The results indicate that the method works better overall on harder FM targets, which have on average lower initial GDT_TS scores. The method is less successful on the assessor selected and edited refinement targets and the TBM domain models (where starting models have, on average, much higher GDT_TS scores) than either the full chain models or FM domain models. Clearly, it is harder to refine models where there is less room for improvement, and likewise it is harder to detect a smaller ΔGDT_TS. In addition, for many of the starting server models, the developers also attempted refinement, which clearly produces a problem of diminishing returns for further refinement. Nevertheless, considering full chain models across regular targets, on average the automatically selected initial models are successfully improved upon by the ReFOLD pipeline.
The time taken to refine a model is dependent on the sequence length. The average time per CASP12 target was ∼10 h for each NAMD run using a quad core desktop (Intel Xeon E5-2407v2) with a standard graphics card (Nvidia GeForce GTX 970). Smaller models were quicker to refine (e.g. 310 min for a 60 residue model) and larger models took longer (e.g. 909 min for a 257 residue model). The other components of the method, including the quality assessment, are run in parallel and will usually take no more than a few extra hours. The ReFOLD pipeline is presently running on a dedicated 56 thread server (Intel Xeon E5-2695 v3) with an Nvidia Tesla K40M GPU card, so most users should expect their results well within 24 h, once they are running.
CONCLUSIONS
The ReFOLD server allows users to attempt to fix errors in their 3D models of proteins and identify improvements with built-in Accuracy Self Estimates. The user friendly, dynamic results pages let users visualise potential improvements for over 200 alternative refined models, at both a global and local level. Providing users with visual comparisons of estimated local improvement allows them to quickly identify those models, which are likely to have been improved upon in a specific region of interest. In addition, the server provides users with a compressed archive all of the generated refined models, which they may rank using their own alternative quality assessment protocols. In the recent CASP12 experiment, the ReFOLD pipeline gave our group a performance boost increasing our cumulative GDT_TS scores and thereby contributing to our high overall rankings.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Malaysian Government (to A.N.S). Funding for open access charge: University of Reading; Malaysian Government.
Conflict of interest statement. None declared.
REFERENCES
- 1. Nugent T., Cozzetto D., Jones D.T.. Evaluation of predictions in the CASP10 model refinement category. Proteins. 2014; 82(Suppl. 2):98–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kryshtafovych A., Fidelis K.. Protein structure prediction and model quality assessment. Drug Discov. Today. 2009; 14:386–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McGuffin L.J., Buenavista M.T., Roche D.B.. The ModFOLD4 server for the quality assessment of 3D protein models. Nucleic Acids Res. 2013; 41:W368–W372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Heo L., Park H., Seok C.. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013; 41:W384–W388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rodrigues J.P., Levitt M., Chopra G.. KoBaMIN: a knowledge-based minimization web server for protein structure refinement. Nucleic Acids Res. 2012; 40:W323–W328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhattacharya D., Cheng J.. 3Drefine: consistent protein structure refinement by optimizing hydrogen bonding network and atomic-level energy minimization. Proteins. 2013; 81:119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhattacharya D., Nowotny J., Cao R., Cheng J.. 3Drefine: an interactive web server for efficient protein structure refinement. Nucleic Acids Res. 2016; 44:W406–W409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kalisman N., Levi A., Maximova T., Reshef D., Zafriri-Lynn S., Gleyzer Y., Keasar C.. MESHI: a new library of Java classes for molecular modeling. Bioinformatics. 2005; 21:3931–3932. [DOI] [PubMed] [Google Scholar]
- 9. Mirjalili V., Noyes K., Feig M.. Physics-based protein structure refinement through multiple molecular dynamics trajectories and structure averaging. Proteins. 2014; 82(Suppl. 2):196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feig M., Mirjalili V.. Protein structure refinement via molecular-dynamics simulations: what works and what does not?. Proteins. 2016; 84(Suppl. 1):282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Modi V., Dunbrack R.L. Jr. Assessment of refinement of template-based models in CASP11. Proteins. 2016; 84(Suppl. 1):260–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mirjalili V., Feig M.. Protein structure refinement through structure selection and averaging from molecular dynamics ensembles. J. Chem. Theory Comput. 2013; 9:1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McGuffin L.J., Atkins J.D., Salehe B.R., Shuid A.N., Roche D.B.. IntFOLD: an integrated server for modelling protein structures and functions from amino acid sequences. Nucleic Acids Res. 2015; 43:W169–W173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McGuffin L.J., Roche D.B.. Automated tertiary structure prediction with accurate local model quality assessment using the IntFOLD-TS method. Proteins. 2011; 79(Suppl. 10):137–146. [DOI] [PubMed] [Google Scholar]
- 15. Roche D.B., Buenavista M.T., Tetchner S.J., McGuffin L.J.. The IntFOLD server: an integrated web resource for protein fold recognition, 3D model quality assessment, intrinsic disorder prediction, domain prediction and ligand binding site prediction. Nucleic Acids Res. 2011; 39:W171–W176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McGuffin L.J. Prediction of global and local model quality in CASP8 using the ModFOLD server. Proteins. 2009; 77(Suppl. 9):185–190. [DOI] [PubMed] [Google Scholar]
- 17. McGuffin L.J. The ModFOLD server for the quality assessment of protein structural models. Bioinformatics. 2008; 24:586–587. [DOI] [PubMed] [Google Scholar]
- 18. Roche D.B., Buenavista M.T., McGuffin L.J.. Assessing the quality of modelled 3D protein structures using the ModFOLD server. Methods Mol. Biol. 2014; 1137:83–103. [DOI] [PubMed] [Google Scholar]
- 19. Buenavista M.T., Roche D.B., McGuffin L.J.. Improvement of 3D protein models using multiple templates guided by single-template model quality assessment. Bioinformatics. 2012; 28:1851–1857. [DOI] [PubMed] [Google Scholar]
- 20. Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kale L., Schulten K.. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005; 26:1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. MacKerell A.D. Jr, Banavali N., Foloppe N.. Development and current status of the CHARMM force field for nucleic acids. Biopolymers. 2000; 56:257–265. [DOI] [PubMed] [Google Scholar]
- 22. Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L.. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983; 79:926–935. [Google Scholar]
- 23. Salomon-Ferrer R., Gotz A.W., Poole D., Le Grand S., Walker R.C.. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 2013; 9:3878–3888. [DOI] [PubMed] [Google Scholar]
- 24. Loncharich R.J., Brooks B.R., Pastor R.W.. Langevin dynamics of peptides: the frictional dependence of isomerization rates of N-acetylalanyl-N'-methylamide. Biopolymers. 1992; 32:523–535. [DOI] [PubMed] [Google Scholar]
- 25. Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003; 31:3370–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.