Abstract
Motifs of only 1–4 letters can play important roles when present at key locations within macromolecules. Because existing motif-discovery tools typically miss these position-specific short motifs, we developed kpLogo, a probability-based logo tool for integrated detection and visualization of position-specific ultra-short motifs from a set of aligned sequences. kpLogo also overcomes the limitations of conventional motif-visualization tools in handling positional interdependencies and utilizing ranked or weighted sequences increasingly available from high-throughput assays. kpLogo can be found at http://kplogo.wi.mit.edu/.
INTRODUCTION
The specificity of many biological processes relies on the recognition of sequence motifs. Accordingly, sequence-motif analysis, including both discovery and visualization of motifs, has long provided important insights into molecular biology. However, most existing motif-analysis tools have fundamental limitations.
Existing motif-visualization tools, such as WebLogo (1), iceLogo (2) and pLogo (3), usually take a set of aligned sequences as input, calculate the weight (frequency or statistical significance) of each letter at each position, and generate logo plots in which letter heights are scaled relative to their weights. Because each position is considered separately, these tools are unable to model and visualize interdependence among multiple positions and thus cannot resolve motifs that overlap with each other. Moreover, these tools treat each input sequence equally and thus do not support weighted or ranked sequences, which are increasingly available from high-throughput studies, such as in vitro selection (4) and massively parallel reporter assays (5).
In contrast, existing motif-discovery tools exclusively model interdependences between neighboring letters, and some can handle weighted or ranked sequences (6,7). However, unlike motif-visualization tools, which precisely model each position, motif-discovery tools typically ignore positional information and thus miss ultra-short motifs (with lengths 1–4 letters) or other information-poor motifs whose specificities are conferred by both sequence identity and relative position. Examples of such hidden specificity have recently been discovered by high-throughput analyses of interactions that were previously thought to be sequence-independent (4,8,9).
Because of the limitations of existing tools and the strong synergy between motif discovery and visualization, we developed an integrated framework for sensitive detection and visualization of position-specific ultra-short motifs from either weighted or unweighted sequences for which positions have a direct correspondence. The utility of our tool, called kpLogo (k-mer probability logo), is illustrated by three examples. In the first two examples, it provided simple and efficient ways of summarizing and visualizing weighted sequences from a massively parallel reporter assay study of enhancer variants and ranked sequences from a high-throughput screen of CRISPR/Cas9 guide RNAs. In the third example, it was applied to a list of unweighted sequences of human microRNA precursors and identified from sequence alone all four ‘hidden’ ultra-short motifs important for precursor processing, without considering the results of high-throughput experimental datasets from which these four motifs were originally discovered.
MATERIALS AND METHODS
kpLogo overview
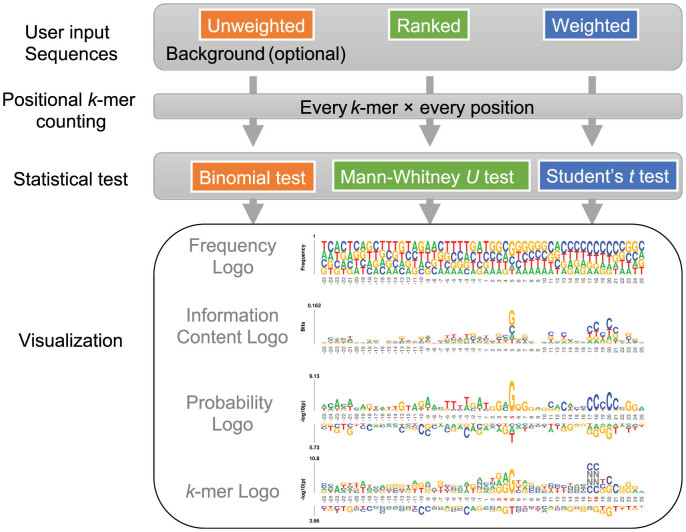
kpLogo extended the framework of pLogo (probability logo), which scales the height of each motif residue to show the statistical significance of its enrichment (3), in two aspects. First, to support ranked and weighted sequences, kpLogo calculates test statistics and their corresponding P values using Mann–Whitney U tests and Student's t tests, respectively. Second, in addition to testing the statistical significance for single letters at each position, kpLogo tests it for all short k-mers starting at each position (default k ≤ 4, allowing degenerate letters). To visualize the results, kpLogo generates a new type of logo plot called the k-mer logo, in which at each position the most significant k-mer is plotted vertically with the total height scaled to its P value (–log10 transformed) or test statistics, as appropriate. Although primarily designed for DNA/RNA sequence analysis, kpLogo also works for protein sequence analysis.
kpLogo web server
kpLogo was developed in C++, and can be used via either a simple command-line interface or a web server. As input, it accepts a list of sequences of identical length (or sequences of different lengths anchored on either their first or last position), which are either unweighted, weighted, or ranked (Figure 1). kpLogo enumerates all possible k-mers of user-specified lengths, evaluates their presence at each position in all input sequences, and reports their enrichment and depletion at each position as determined using an appropriate statistical model (described below). kpLogo tests all k-mers ranging from 1–4 letters by default and can also be configured to test k-mers of other lengths. Degenerate letters can be specified using the IUPAC code. In addition to probability logo and k-mer logo, kpLogo also generates logo plots for monomer frequency and information content.
Figure 1.
Schematic of the kpLogo web server. Users upload or paste sequences that could be unweighted, ranked, or weighted. kpLogo enumerates all possible k-mers of user-specified lengths, counts their frequency at each position in input sequences, and then performs appropriate statistical tests to determine their enrichment and depletion at each position. kpLogo then generates four types of logo plots: frequency logo, information content logo, probability logo, and k-mer logo.
Statistical models
Weighted or ranked sequences
For each position and for every possible k-mer of user-specified size range (default from 1–4) at that position, input sequences are divided into a positive group and a negative group, depending on whether a match to the k-mer can be found at the specific position in the sequence. The weights in the two groups are then compared using the one-sided two-sample Student's t test, or ranks in the two groups are compared using the Mann–Whitney U test (using a z-test approximation).
Unweighted sequences
For each possible k-mer of user-specified size range (default from 1–4) at each position, the one-sided binomial test (using a z-test approximation) is used to evaluate whether the frequency of the k-mer is higher or lower than expected. The expected frequency is determined using one of three background models specified by users to be either the average frequency of the same k-mer across all positions (default), the frequency of the same k-mer at the same position but in a separate set of background sequences or shuffled input sequences that preserve sequence composition, or the frequency calculated from Markov models learned from the input sequences or background sequences.
Logo generation
In each position of a k-mer logo, only the most enriched k-mer and the most depleted k-mer starting at that position are shown above and below the coordinate, respectively. Each k-mer reads vertically from top to bottom, with total height scaled by either its test statistic (either t statistic or z score, depending on the test) or its –log10-transformed P value, depending on the user-defined preference, although in instances in which an absolute P value is too small to be represented in the current computer system (P < 10−324) the analysis and scaling defaults to test statistics. In a probability logo, single letters are stacked on top of each other and each scaled by associated test statistics or P values. Enriched/depleted letters are stacked above/below the coordinates, respectively. In both k-mer logo and probability logo, coordinates of positions with P values smaller than a specified threshold (default 0.05) after Bonferroni correction are highlighted in red. Positions for which the frequency of a single letter exceeds a defined threshold (default 0.75) are designated fixed positions, and coordinates of these positions are highlighted in black. Only the dominant letter is shown at fixed positions, and letters at fixed positions are shown at heights 10% higher than the max height of non-fixed positions.
RESULTS
Weighted sequences
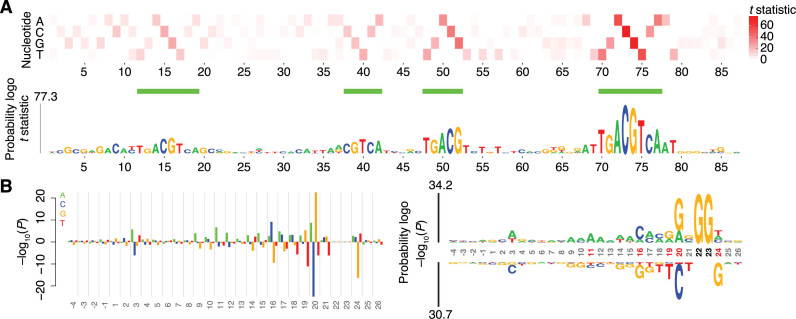
To illustrate the ability of kpLogo to examine weighted sequences, which are currently not handled by widely used tools such as WebLogo and pLogo, we used kpLogo to summarize and visualize the results of a massively parallel reporter assay in which 26,438 variants of the cAMP-responsive enhancer were generated and assayed for activity (5). In the original publication, the relative importance of each nucleotide at each position was visualized by either four bar plots (one for each of the four nucleotides) or a heatmap resembling that of Figure 2A (top). Starting from the list of variant sequences and their activity scores, kpLogo performed Student's t tests to evaluate for each nucleotide at each position whether variants with that nucleotide at that position had higher activity than the rest of variants, and generated a probability logo with more readily visible sequence motifs (Figure 2A, bottom).
Figure 2.
Analysis of weighted and ranked sequences. (A) Comparison of a heatmap (top) and a kpLogo probability logo plot (bottom) for summarizing results of reporter assays for 26,438 variants of the cAMP-responsive enhancer. Both formats visualize t statistics from Student's t tests. Only the top half of the kpLogo probability logo plot is shown. Four consensus CREB sites are highlighted (horizontal green bars). (B) Comparison of a barplot (left) and a kpLogo probability logo plot (right) for depicting the P values from Mann–Whitney U tests of whether guide RNAs with a specific nucleotide at a specific position are more (above 0) or less (below 0) efficient than other guide RNAs. In the probability logo, positions with significant nucleotides (Bonferroni corrected P < 0.01) are highlighted (red coordinates), as are the fixed positions of the GG PAM (black coordinates).
Ranked sequences
We also used kpLogo to analyze and display the results of a study that ranked the efficiency of 1,841 Cas9 guide RNAs (gRNAs) designed to knock out reporter genes (9). The original publication compared the gRNAs in the top quintile of efficiency (an arbitrary cutoff) to the rest, calculating the enrichment and depletion of each base at each position using a binomial test and then plotting the results on a bar plot resembling that of Figure 2B (left). Existing tools would not have been helpful in this analysis, as illustrated by the WebLogo plot generated using the top quintile of gRNA sequences, which did not uncover any visible signal beyond the GG protospacer-adjacent motif (PAM), which did not vary (Supplementary Figure S1A). In contrast, kpLogo could use these ranked data without imposing an arbitrary binary cutoff by performing Mann–Whitney U tests to find nucleotides at each position that were associated with higher or lower efficiency. The kpLogo-generated probability logo was also easier to read compared to a bar plot that graphed the same results (Figure 2B, left versus right). Moreover, the kpLogo-generated k-mer logo uncovered preference for not only mononucleotides but also di-, tri-, and tetra-nucleotide motifs at specific positions within the gRNA (Supplementary Figure S1B).
Unweighted sequences and k-mer logo
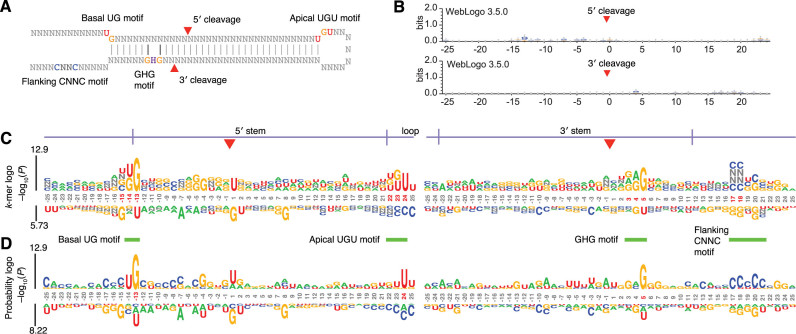
The utility of the k-mer logo for revealing ultra-short position-specific motifs was also illustrated in its analysis of the primary transcripts of human microRNAs (miRNAs). miRNAs are a class of small RNAs that direct the post-transcriptional regulation of most human mRNAs (10). They are processed by endonucleolytic excision from stem-loop regions of primary transcripts known as pri-miRNAs (Figure 3A) (8,11). For about a decade, the only features of pri-miRNAs known to be recognized by the processing machinery were structural (e.g., pairing within the pri-miRNA stem and unstructured RNA in the loop and flanking segments). Consistent with this early understanding of the defining features of pri-miRNAs, no visible sequence signals were observed when human miRNA precursors were anchored at the sites of initial endonucleolytic cleavage and visualized by WebLogo (1) (Figure 3B), nor did motif-discovery tools such as MEME (12) identify any significant motifs within 25 nucleotides (nt) of these cleavage sites. In reality, however, the processing machinery also recognizes position-specific ultra-short motifs. High throughput experimental analyses of many pri-miRNA variants has recently revealed four ultra-short motifs that promote pri-miRNA processing, including a UG motif at the base of the hairpin (14 nt upstream of the 5΄-cleavage site), a UGU motif in the apical loop (22-nt downstream of the 5΄-cleavage site), a mismatched GHG motif (3 bp downstream of the 3΄-cleavage site, H = non-G), and a CNNC motif (17 or 18 nt downstream of the 3΄-cleavage site, N = any nucleotide) (Figure 3A) (4,8). The reason that these motifs had not been identified earlier is that they are partially redundant with each other and most important for pri-miRNAs with suboptimal structural features (8). Therefore, although they are enriched in human pri-miRNAs and have been preferentially conserved in evolution, most of the motifs are each found in fewer than half of the human pri-miRNAs (4,8).
Figure 3.
Analysis of unweighted sequences from human miRNA hairpins. (A) Diagram of a model miRNA hairpin indicating the two cleavage sites and four motifs. (B) WebLogo (version 3.5.0) output using 50 nt centered on each cleavage site. (C) k-mer logo generated by kpLogo using 50 nt centered on the 5΄ cleavage site (left) and the 3΄ cleavage site (right), with k ranging from 1 to 4, allowing the degenerate nucleotide N. The k-mer logo shows the most enriched (above the coordinates) and most depleted (below the coordinates) k-mer starting at each position, with the option of showing instead those ending at each position (Supplementary Figure S2). k-mers read from top to bottom. Positions with significant enrichment or depletion (Bonferroni corrected P < 0.01) are highlighted (red coordinates). (D) Probability logos generated by kpLogo. Nucleotides are scaled by statistical significance (P value) and then stacked on top of each other.
In sharp contrast to existing tools, kpLogo identified all four of these ‘hidden’ motifs in silico starting from just the known human pri-miRNA sequences, without considering any of the experimental results (Figure 3C, highlighted by red coordinates and green horizontal bars). Moreover, no other motif was found to have a P value <0.01 (Figure 3C, Bonferroni corrected, one-sided binomial test), which demonstrated high specificity. Notably, the k-mer logo correctly identified the CNNC motif starting at either position 17 or position 18 (Figure 3C), which would have been incorrectly interpreted as a single CCNCC motif from other types of logos, such as the probability logo shown in Figure 3D, thereby demonstrating the ability of kpLogo to discover and visualize overlapping motifs.
By default, kpLogo assigns each k-mer to the position of its starting (i.e., most 5΄ or most N-terminal) letter and compares the significance of its enrichment to that of all other k-mers assigned to the same position. kpLogo also allows the option of assigning each k-mer to the position of its end letter. Comparing these two schemes using pri-miRNA sequences shows that known motifs sharing a start or end position with a stronger motif can be masked out in one scheme or the other but that the strongest motif among all overlapping ones was identified in both schemes (Supplementary Figure S2).
Running kpLogo on pri-miRNA sequences from other bilaterian species, for which no high-throughput functional data were available, often uncovered motifs resembling those observed in human, as might have been expected from previous analyses examining these motifs in other species (although kpLogo found the motifs in an unbiased analysis, whereas the previous analyses searched specifically for the motifs) (4,8). These previous analyses also found that none of the four motifs were present in nematodes miRNAs (4,8), consistent with the observation that nematode pri-miRNAs are typically not processed when ectopically expressed in human cells (4). To search for motifs that might facilitate the processing of pri-miRNAs in nematodes, we ran kpLogo on a set of 95 Caenorhabditis elegans pri-RNAs that had been previously curated to remove paralogous sequences. The two most prominent motifs were on opposite strands and together formed a paired CC/GGNG motif in the vicinity of the human mismatched GHG motif (Supplementary Figure S3).
DISCUSSION
Our case studies exemplify increasingly common types of settings in which kpLogo is expected to be particularly useful. As illustrated by the analysis of enhancer variants, high-throughput sequencing enables parallel quantitative measurements of thousands-to-millions of variants informative for the understanding of many aspects of nucleic acid binding, processing and function, including endonuclease specificity (4,8,13), affinity landscapes of DNA/RNA-binding proteins (14,15), fitness landscapes of tRNA and snoRNA (16,17), splicing specificity (18), and mRNA 3΄-end processing efficiency and structure (19). Sequencing-based approaches are also widely used to study endogenous sequences that are diverse yet have direct correspondence in positions, typically related to each other due to common binding or processing events. For example, a high-resolution CLIP-seq experiment typically identifies thousands of protein-binding sites across the transcriptome, each captured with an efficiency that corresponds, at least to some degree, with its affinity to the RNA-binding protein. As illustrated by our analysis of pri-miRNAs, RNA-binding proteins typically bind short degenerate motifs, and their function can depend on their placement relative to other binding or processing events. Thus, kpLogo is positioned to summarize those semi-quantitative measurements into visual patterns and facilitate their interpretation.
In summary, kpLogo streamlines sensitive motif discovery with logo-type visualization and enables the discovery of motifs missed by existing tools as well as the generation of sequence-logo plots for ranked or weighted sequences. With the increasing use of high-throughput sequencing for quantitative measurement of a large number of sequence variants, tools like kpLogo will have an expanding role in the discovery and interpretation of patterns hidden in biological sequences.
AVAILABILITY
kpLogo can be found at http://kplogo.wi.mit.edu/. The source code and web code are both available in GitHub: https://github.com/xuebingwu/kpLogo. In addition, kpLogo has been wrapped as a Galaxy tool and available in Galaxy Tool Shed: https://toolshed.g2.bx.psu.edu/view/xuebing/kplogo.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jeff Morgan for testing the server, Kathy Lin and Namita Bisaria for comments on the manuscript, and the Information Technology department at Whitehead Institute for Biomedical Research for hosting the server.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM118135 to D.B.]; X.W. is a Helen Hay Whitney Foundation Fellow; D.B. is an investigator of the Howard Hughes Medical Institute. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Colaert N., Helsens K., Martens L., Vandekerckhove J., Gevaert K.. Improved visualization of protein consensus sequences by iceLogo. Nat. Methods. 2009; 6:786–787. [DOI] [PubMed] [Google Scholar]
- 3. O'Shea J.P., Chou M.F., Quader S.A., Ryan J.K., Church G.M., Schwartz D.. pLogo: a probabilistic approach to visualizing sequence motifs. Nat. Methods. 2013; 10:1211–1212. [DOI] [PubMed] [Google Scholar]
- 4. Auyeung V.C., Ulitsky I., McGeary S.E., Bartel D.P.. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013; 152:844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Melnikov A., Murugan A., Zhang X., Tesileanu T., Wang L., Rogov P., Feizi S., Gnirke A., Callan C.G., Kinney J.B. et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 2012; 30:271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eden E., Lipson D., Yogev S., Yakhini Z.. Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol. 2007; 3:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bussemaker H.J., Li H., Siggia E.D.. Regulatory element detection using correlation with expression. Nat. Genet. 2001; 27:167–174. [DOI] [PubMed] [Google Scholar]
- 8. Fang W., Bartel D.P.. The menu of features that define primary microRNAs and enable de novo design of microRNA genes. Mol. Cell. 2015; 60:131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E.. Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat. Biotechnol. 2014; 32:1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ha M., Kim V.N.. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014; 15:509–524. [DOI] [PubMed] [Google Scholar]
- 12. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pattanayak V., Lin S., Guilinger J.P., Ma E., Doudna J.A., Liu D.R.. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013; 31:839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nutiu R., Friedman R.C., Luo S., Khrebtukova I., Silva D., Li R., Zhang L., Schroth G.P., Burge C.B.. Direct measurement of DNA affinity landscapes on a high-throughput sequencing instrument. Nat. Biotechnol. 2011; 29:659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lambert N., Robertson A., Jangi M., McGeary S., Sharp P.A., Burge C.B.. RNA bind-n-seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell. 2014; 54:887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li C., Qian W., Maclean C.J., Zhang J.. The fitness landscape of a tRNA gene. Science. 2016; 352:837–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Puchta O., Cseke B., Czaja H., Tollervey D., Sanguinetti G., Kudla G.. Network of epistatic interactions within a yeast snoRNA. Science. 2016; 352:840–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rosenberg A.B., Patwardhan R.P., Shendure J., Seelig G.. Learning the sequence determinants of alternative splicing from millions of random sequences. Cell. 2015; 163:698–711. [DOI] [PubMed] [Google Scholar]
- 19. Wu X., Bartel D.P. Widespread influence of 3΄-end structures on mammalian mRNA processingand stability. Cell. 2017; in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.