Abstract
We have developed the web application GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html) for the rapid and accurate annotation of organellar genome sequences, in particular chloroplast genomes. In contrast to existing tools, GeSeq combines batch processing with a fully customizable reference sequence selection of organellar genome records from NCBI and/or references uploaded by the user. For the annotation of chloroplast genomes, the application additionally provides an integrated database of manually curated reference sequences. GeSeq identifies genes or other feature-encoding regions by BLAT-based homology searches and additionally, by profile HMM searches for protein and rRNA coding genes and two de novo predictors for tRNA genes. These unique features enable the user to conveniently compare the annotations of different state-of-the-art methods, thus supporting high-quality annotations. The main output of GeSeq is a GenBank file that usually requires only little curation and is instantly visualized by OGDRAW. GeSeq also offers a variety of optional additional outputs that facilitate downstream analyzes, for example comparative genomic or phylogenetic studies.
INTRODUCTION
Next-generation sequencing (NGS) technologies have led to a burst in the availability of organellar genome sequences (1). However, sequence annotation is still a major bottleneck. Four tools for (semi-)automated annotation of organellar genomes are currently available, but all of these programs suffer from limited customizability of reference sequences by the user. This does not only prevent annotation of a subset of organellar genomes or custom features, but also precludes the user from taking advantage of any proprietary high-quality references. Mitofy (2) was developed for the annotation of plant mitochondrial sequences only, whereas CpGAVAS (3) and Verdant (4) solely annotate chloroplast genomes. DOGMA (5) annotates chloroplast and animal mitochondrial sequences, but no plant mitochondrial genomes. In addition to restrictions in reference selection, none of these tools provide a manually curated reference sequences database, take RNA editing into account or allow the user to directly compare annotations based on different methods like BLAST/BLAT and HMMER and different de novo tRNA prediction algorithms.
Here, we present GeSeq, a web-based annotation tool for the annotation of organellar sequences. The program was developed for plants. With an appropriate reference set, however, it also annotates mitochondria genomes from non-green species, such as mammals. Even entire plasmid collections can be annotated using GeSeq, although we have not tested the tool rigorously outside the plant lineage. GeSeq produces high-quality annotations in short runtime, is highly customizable and accepts batch submissions. GeSeq's functions are integrated in an easy-to-use GUI and with its high flexibility it will meet the demands of most annotation jobs. For high-quality annotation of chloroplast genomes, GeSeq is equipped with manually curated reference sequences and a corresponding profile hidden Markov model (profile HMM) database for chloroplast protein and rRNA-coding genes.
Aside of the ‘classical’ annotation job, the annotation of a complete organellar genomes or genome segments, GeSeq can be easily adjusted to annotate multiple sequences derived from NGS contigs, generate codon-based alignments for specific genes or, when supplemented with additional references, used for any other (smaller) DNA sequence. This allows, for instance, quick annotations for verifying contigs of de novo assemblies and/or to map primer-binding sites for gap closure.
RESULTS AND DISCUSSION
Implementation
GeSeq is written in Java and PHP. It features parallel (multi-threaded) job processing and user management. The front end is a user-friendly web interface implemented in Javascript (Supplementary Figure S1A). It was tested with the current versions of Firefox, Chrome, Safari, Internet Explorer and Edge. GeSeq uses the third-party software tRNAscan-SE v1.3.1 (6), ARAGORN v1.2.36 (7), BLAT (Standalone BLAT v.35 × 1; 8), OGDRAW v1.2 (9,10), TranslatorX v1.1 (11), MUSCLE v3.8.31 (12,13) and HMMER (14). The GeSeq application is part of the CHLOROBOX toolbox (https://chlorobox.mpimp-golm.mpg.de/index.html) hosted at the Max Planck Institute of Molecular Plant Physiology (Potsdam-Golm, Germany). With ‘GenBank 2 Sequin’ (https://chlorobox.mpimp-golm.mpg.de/GenBank2Sequin.html), the CHLOROBOX offers an accompanying program to convert revised GenBank files generated by GeSeq into the Sequin or BankIt format required for NCBI/EMBL/DDBJ database submission.
Sequence submission
Nucleic acid sequences for annotation must be provided in (multi-)FASTA format (http://blast.ncbi.nlm.nih.gov/blastcgihelp.shtml). All sequence letters not complying with the IUPAC code (15), including gaps, will be removed and ‘U's of RNA sequences converted to ‘T's. Each input sequence for annotation is treated as independent job. Activation of ‘Circular sequence(s)’ enables annotation of genes or features that span the ends of the submitted linear sequence. GeSeq will simulate a circular sequence by appending a copy of up to the first 10 000 bp to the end of the submitted sequence.
Reference selection
GeSeq offers the user to freely select or upload the most appropriate reference sequences for each annotation project and provides a manually curated reference sequences set for chloroplast genomes.
NCBI Reference Sequence (NCBI RefSeq)
GeSeq is equipped with a local copy of the organelle genome records of the NCBI RefSeq project (http://www.ncbi.nlm.nih.gov/genome/organelle/). The hosted NCBI RefSeq records are monthly updated and visualized as a phylogenetic tree, searchable by free text (Supplementary Figure S1B). However, we strongly encourage the user to ascertain the annotation quality of the NCBI references before use (see below).
MPI-MP chloroplast reference set
During this work, we frequently encountered apparent annotation errors in NCBI RefSeq entries. These include unexpected truncations or extensions of genes or likely misannotated exon–intron borders. In order to provide GeSeq with a high-quality reference set of plastid sequences, we selected the complete plastid genomes of 34 plant species spanning the full taxonomic range from mosses to seed plants, with a focus on the latter (Supplementary Table S1). We then generated multiple alignments for each protein and rRNA-coding gene and manually curated these alignments (for examples, see Figure 1). Since for many of the 34 species, no experimental validation of gene or exon–intron annotation was available, manual curation was guided by data from organisms intensely studied with respect to chloroplast gene expression, like Arabidopsis thaliana, Nicotiana tabacum, Oenothera elata and Zea mays.
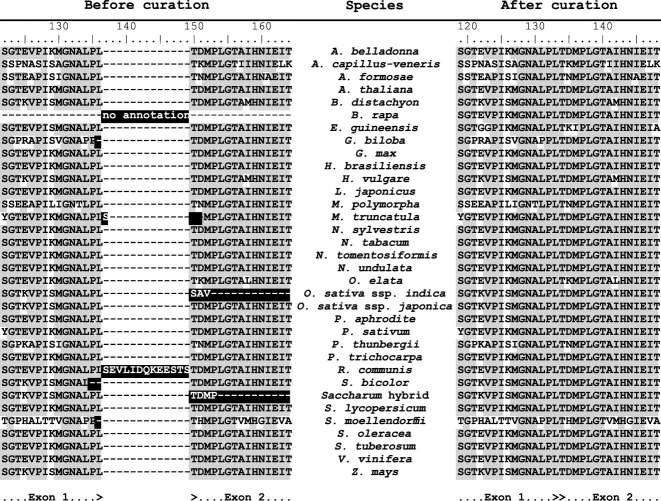
Figure 1.
Examples for a likely mis-annotated gene in some (provisional) NCBI Reference Sequence (NCBI RefSeq) entries encountered during this work. Exon–exon junction of the chloroplast rpl2 gene as translated from NCBI RefSeq CDS entries (left) and comparison with the corresponding entries in the GeSeq internal reference sequences database (right; see Supplementary Table S1 for GeSeq names and GenBank IDs). Suspected annotation errors are labeled in black. Some NCBI RefSeq records lack annotation of given genes (‘no annotation’, cf. Brassica rapa) or introns resulting in no or truncated CDS (Oryza sativa ssp. indica and Saccharum), respectively. Inconsistent exon borders result in likely erroneous insertions or deletions of one or several codons (Ginkgo biloba, Medicago truncatula, Ricinus communis, Sorghum bicolor and Selaginella moellendorffii).
Users are invited to donate references sets, for example for mammalian or fugal mitochondrial genomes, or algal chloroplasts.
Custom references
In addition to the above described server-hosted references, the user can upload (multi-)FASTA or (multi-)GenBank files containing custom reference sets. This does not only enable the user to take advantage of proprietary high-quality references, but also to add custom annotations like origins of replication or ‘foreign’ features like primer or protein-binding sites.
Annotation pipeline
The GeSeq core annotation pipeline is based on a BLAT-driven best-match approach. This is complemented by additional profile HMM searches for protein and rRNA-coding genes and de novo prediction of tRNA genes (Figure 2). We decided in favor of a best-match and against a (more sensitive) profile HMMs-based approach for the core annotation pipeline because we prioritized a free selection of reference sequences by the user. Generation of high-quality profile HMMs requires high-quality multiple alignments, ideally manually curated (http://hmmer.org/documentation.html), which is incompatible with a highly flexible and readily extendable reference selection system. In order to still take advantage of profile HMM searches, we built profile HMM databases for plastid protein and rRNA-coding genes (see above). Thus, the user can benefit from both free reference selection and profile HMMs searches. We selected BLAT as sequence similarity search tool, because it runs fast and provides an accurate annotation of exon–intron borders.
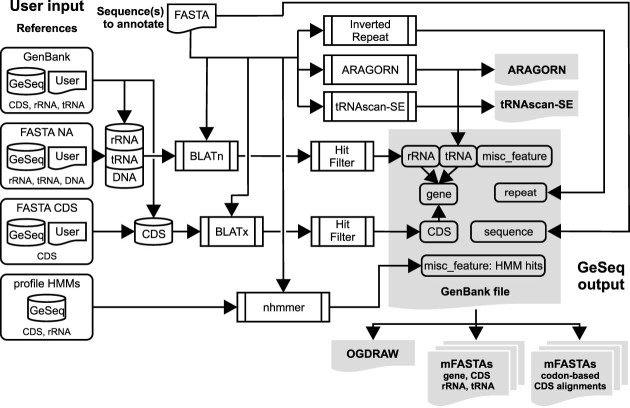
Figure 2.
GeSeq annotation pipeline. The user provides nucleic acid FASTA sequence(s) for annotation and selects or provides reference nucleic acid sequences in GenBank or FASTA format (‘User Input’). Based on the selected or uploaded reference sequences (‘References’), GeSeq builds a non-protein-coding (rRNA, tRNA and DNA) and a protein-coding (CDS) BLAT database, carries out standard (‘BLATn’) and translated BLAT (‘BLATx’) searches, respectively, and filters the hits (‘Hit Filter’). GeSeq annotates from the filtered hits the classes rRNA, tRNA, CDS and gene (‘gene entries’). ‘Gene entries’ result from tRNA, rRNA and CDS hits, and include introns (if present). DNA hits are annotated as ‘misc_features’ (as shown here) or, alternatively, as ‘primer_bind’ if invoked by the user (see text for details). In addition, the user can activate an nhmmer search by selecting profile HMMs of CDS and rRNA sequences (currently chloroplast only) as references. All profile HMM hits are annotated as misc_features to support manual curation. Optionally, the user can invoke ARAGORN or tRNAscan-SE for de novo annotation of tRNAs and a self-BLATn search for the detection of the inverted repeat (IR) pair typically found in chloroplast genomes. The minimum GeSeq output (all output files are labeled in gray) is a GenBank file that contains all annotations and its interpretation by OGDRAW for a quick evaluation. Additionally, the user can choose additional optional outputs, including separate multi-FASTA files (‘mFASTAs’) containing the annotated sequences belonging to the classes gene, CDS, rRNA and tRNA. If several sequences were uploaded for annotation in the same job, also combined mFASTAs for all annotated sequences of the four classes are offered for download and optionally, codon-based alignments can be produced for all annotated CDS sequences with or without the selected or uploaded GenBank references.
BLAT reference sequence databases
GeSeq generates two databases for each annotation job: a protein-coding (CDS) and a non-protein-coding (NA) database. The latter may contain rRNA, tRNA and other nucleic acid sequences. GeSeq does not accept protein sequences. The program parses all selected or uploaded GenBank files for CDS, tRNA and rRNA entries, extracts these features and adds them to the corresponding database. The identifiers of extracted sequences are generated by the concatenation of the GenBank qualifier ‘/gene’ with the source GenBank IDs or accession number separated by an underscore (e.g. ‘psaA_AJ271079’).
Sequences of selected or uploaded CDS or NA FASTA files are added to the appropriate database. Uploaded NA FASTA sequences are flagged as tRNA, rRNA or primer sequences if the FASTA file name starts with ‘tRNA_’, ‘rRNA_’ or ‘primer_’ respectively, and hits will be annotated accordingly (otherwise, hits will be annotated as ‘misc_features’). The sequence identifiers are kept and we recommend the syntax: ‘>gene/feature name_source’ (e.g. ‘>psaA_Oelata’ for the psaA gene from Oenothera elata). This nomenclature is required for hit filtering and prevents redundant annotation of the same gene or feature by multiple references (see ‘BLAT hit filtering’ below).
BLAT hit filtering
Standard (‘BLATn’) and translated BLAT (‘BLATx’) searches are executed with default parameters (unless the user changes the BLAT minimum identity threshold in GeSeq's GUI) using the collected sequences of the NA and the CDS databases, respectively (Figure 2). Activation of the ‘find short matches’ option changes the parameters of the standard BLAT search to allow the detection of short (at least 12 bases) identical matches. This enables the detection of short regulatory elements, like promoters, origins of replication or primer-binding sites, if provided by the user in a custom FASTA file. Then, GeSeq applies a best-match hit filtering to the BLAT outputs in order to avoid multiple annotations of the same gene or feature by different references. For each gene or feature name, GeSeq retains only the hit(s) with the highest score for annotation. In addition, GeSeq retains the second-best, non-overlapping hit(s) for each gene or feature. Such secondary hits will be annotated as ‘gene-’ or ‘feature-fragment(s)’ and usually represent gene fragments resulting from recombination events at direct or inverted repeats (IRs) that are frequently found in plant organellar genomes (1,16). Trans-spliced genes (such as the chloroplast gene rps12) are also annotated that way.
HMM search for protein and rRNA-coding genes
In addition to the BLAT-based core annotation pipeline, a profile HMM search for chloroplast protein and rRNA-coding genes can be invoked. For this, we generated profile HMMs for the manually curated reference set of chloroplast CDS and rRNA sequences and GeSeq executes a standard nhmmer search (Figure 2). The resulting hits (envelope coordinates) will be shown as ‘misc_features’ in the output GenBank file. Thus, the user can easily compare the annotations by BLAT with profile HMM hits (Figure 3).
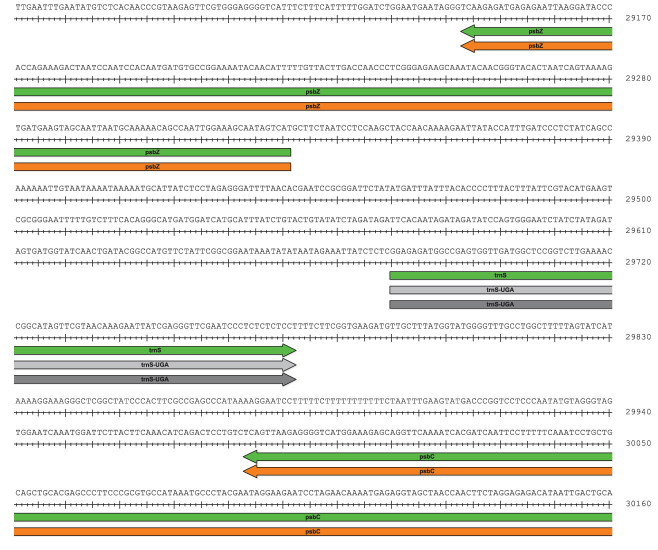
Figure 3.
Examples of protein-coding and tRNA gene annotations (green arrows) by GeSeq with high-quality settings visualized by SeqBuilder.v13.0.0 (DNASTAR, Madison, WI, USA). The coordinates of the chloroplast protein-coding genes psbZ and psbC (only 3΄ end shown) as annotated by BLATx exactly match to their corresponding profile HMM hits (orange arrows). The tRNA gene trnS which is annotated in-between the two protein-coding genes by BLATn is confirmed by both de novo tRNA predictors tRNAscan-SE and Aragorn (light and dark gray arrows, respectively). See Supplementary Figure S2B and (C) for the corresponding GenBank file excerpt.
De novo prediction of tRNA genes
tRNA genes can be additionally predicted de novo by the third-party tools tRNAscan-SE and ARAGORN (Figure 3). The default parameters are set for the discovery of tRNAs in seed plant chloroplast genomes. Since several tRNA species contain introns, tRNAscan-SE runs in COVE mode (17), allowing the detection of long introns. In general, search parameters of both tools can be flexibly adjusted to many purposes. Often, the genes for the initiator tRNA trnfM-CAU and tRNAs with modified anticodons [such as trnI-CAU in chloroplast genomes; (18)] remain unannotated or can be misannotated by one of the two tRNA discovery programs. The presence of these tRNAs can be verified by the implemented BLATn search for tRNAs in GeSeq.
Identification of the chloroplast Inverted Repeat (IR) pair
GeSeq can annotate the IR pair typically found in chloroplast genomes. If the option ‘Annotate plastid IR’ is activated, the longest, identical IR pair within a submitted sequence is identified from a self-BLATn search (Figure 2).
Assembly of GenBank files
The GenBank file generated by GeSeq lists all settings and references used for annotation in the header section (Supplementary Figure S2A). Annotations of genes, rRNAs, tRNAs and CDS are written according to the filtered BLATn and BLATx outputs, including exon–intron positions. This works well for the vast majority of spliced genes. All annotations based on BLAT contain information about the annotated gene and the BLAT hit parameters in ‘/note’ entries (Supplementary Figure S2B). CDS translations are predicted if (i) the length of the annotated CDS is dividable by three, (ii) the CDS starts with ATG, GTG, ACG or TTG, and (iii) the CDS ends with TAG, TGA, TAA, CAG, CGA or CAA. Note that ACG, CAG, CGA and CAA are included, because these triplets can potentially be converted into canonical translational start or stop codons by C-to-U RNA editing which occurs in plant organelles (19). If ARAGORN or tRNAscan-SE were invoked, predicted tRNAs are annotated accordingly (Supplementary Figure S2C). If ‘Annotate plastid IR’ was enabled, the detected IRs are annotated as ‘repeat_regions’ (Supplementary Figure S2D). Hits based on un-flagged uploaded FASTA files (see above under ‘BLAT reference sequences databases’) or profile HMM hits are annotated as ‘misc_features’.
GeSeq output
The minimal output of GeSeq consists of a GenBank file for review, manual inspection and editing by the user in a third-party genome browser and its visualization by OGDRAW. For downstream analyses or annotation verification, additional outputs can be provided: First, tRNAs identified by tRNAscan-SE and ARAGON are displayed in separate files. Second, GeSeq can optionally generate multi-FASTA files containing all genes, CDSs, rRNAs and tRNAs found in the sequence(s) submitted for annotation. Third, GeSeq can provide the user with codon-based alignments by TranslatorX and MUSCLE for each CDS identified by GeSeq, optionally including the selected reference sequences, for phylogenetic or other analyses.
EVALUATION
GeSeq is a web application for the annotation of organelle genomes. During its development, we have focused on achieving a high annotation quality while maintaining maximum flexibility.
During extensive testing of GeSeq for the annotation of organelle genomes, we noticed that GeSeq fails to detect extremely short exons (i.e. <8 bp long first exons of the chloroplast genes petB and petD) and occasionally produces an over-annotation of introns in rapidly evolving genes, like the chloroplast giant open reading frames ycf1 and ycf2 (20). The latter can be easily compensated by decreasing the BLAT protein search identity value. Extraordinarily short exons of few base pairs cannot be detected by a translated BLAT search. To accomplish this, a dedicated pipeline for the detection of such short exons would be required, which would need to include surrounding non-coding bases. However, we decided to stick to a general and uniform annotation approach that is exclusively based on translated searches which are less prone to errors caused by elevated variation occurring at (or biases acting on) non-coding positions or synonymous codon positions. Therefore, these few exceptional exons may require manual curation of the resulting GenBank file during review by the user and we suggest referring to the corresponding hit of the HMMER search for chloroplast genes or by additionally uploading a custom NA FASTA reference file containing the relevant complete gene sequences.
The high flexibility and versatility of GeSeq facilitates various kinds of annotation jobs, some of which are outlined below:
Quick annotation of chloroplast genome sequence: we strongly recommend using GeSeq's chloroplast reference set as default. If less genes are annotated than expected and/or if genes are fragmented, selecting or uploading one or several additional NCBI RefSeq files is appropriate. Ideally, these references should be of high annotation quality and represent the most closely related taxa. With a runtime of about 6 s per vascular chloroplast genome (Supplementary Table S2), GeSeq is currently the fastest tool available. This enables the user to comfortably select appropriate parameters and references by fast trial-and-error test annotations.
High-quality annotation of complete chloroplast genomes: for high-quality annotations, we suggest, in addition to (i), to enable HMMER profile search, tRNA-ScanSE and ARAGORN. This will result in a multi-annotation of protein and rRNA-coding genes by two (BLATx and HMMER) and of tRNA genes by three (BLATn, tRNAscan-SE and ARAGORN) different methods (Figure 3). In addition, we recommend to run a quick annotation and use it as template for manual curation.
Annotation of NGS-derived contigs: NGS-derived contigs from a single-sequencing project should be submitted as individual job. All settings for high-quality annotation should be selected and, in addition, the ‘Generate multi-FASTAs’ option invoked. GeSeq will provide ‘global’ multi-FASTA files for all recognized gene classes (see also Supplementary Figure S1). The complete set of result files can be conveniently downloaded as single zip file.
Extraction of specific gene sequences from many species: GeSeq can generate codon-based alignments for all submitted sequences by calling TranslatorX. If more than 50 sequences are submitted, we recommend adjusting the annotation parameters to the target gene(s) to minimize the runtime (e.g. if alignments of protein coding genes are to be generated, third-party tRNA annotators are not required).
CONCLUSION
GeSeq is a fast web application that generates high-quality annotations in a default mode using our curated reference gene set (typically >97% of genes and coding regions are correctly annotated). Its power rests on its flexibility: GeSeq allows the user to provide custom reference sequences in GenBank or (multi-)FASTA format. This feature represents a particularly critical improvement over existing tools because it enables the user to upload proprietary, most up-to-date or novel feature-containing reference sets. Furthermore, during development of GeSeq, a strong emphasis was placed on annotation quality. GeSeq urges the user to curate gene annotations that failed or are particularly tricky (like tiny exons or rapidly evolving genes) and can show independent annotations by the best-matching reference sequences and its corresponding profile HMM hit or third party tRNA annotators. Although this may demand a final manual curation by the user (usually just for a few genes), we consider this an important step in order to achieve optimal annotation quality and provide high-quality sequence data files to the public databases.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank the IT Service Team of the Max Planck Institute of Molecular Plant Physiology for excellent technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) [GR 4193/1-1 to S.G., TI 605/5-1 to M.T.]; Human Frontier Science Program (HFSP) [RGP0005/2013 to R.B.]; Max Planck Society (MPG). Funding for open access charge: MPG.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ruhlman T.A., Jansen R.K.. Maliga P. Chloroplast Biotechnology: Methods and Protocols, Methods in Molecular Biology. 2014; 1132:NY: Springer; 3–38. [DOI] [PubMed] [Google Scholar]
- 2. Alverson A.J., Wei X., Rice D.W., Stern D.B., Barry K., Palmer J.D.. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 2010; 27:1436–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu C., Shi L., Zhu Y., Chen H., Zhang J., Lin X., Guan X.. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 2012; 13:715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McKain M.R., Hartsock R.H., Wohl M.M., Kellogg E.A.. Verdant: automated annotation, alignment, and phylogenetic analysis of whole chloroplast genomes. Bioinformatics. 2017; 33:130–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wyman S.K., Jansen R.K., Boore J.L.. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004; 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- 6. Lowe T.M., Eddy S.R.. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997; 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Laslett D., Canback B.. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004; 32:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kent W.J. BLAT - The BLAST-like alignment tool. Genome Res. 2002; 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lohse M., Drechsel O., Bock R.. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007; 52:267–274. [DOI] [PubMed] [Google Scholar]
- 10. Lohse M., Drechsel O., Kahlau S., Bock R.. OrganellarGenomeDRAW - a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013; 41:W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Abascal F., Zardoya R., Telford M.J.. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010; 38:W7–W13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004; 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wheeler T.J., Eddy S.R.. nhmmer: DNA homology search with profile HMMs. Bioinformatics. 2013; 29:2487–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. 1985; 13:3021–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sloan D.B. One ring to rule them all? Genome sequencing provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytol. 2013; 200:978–985. [DOI] [PubMed] [Google Scholar]
- 17. Eddy S.R., Durbin R.. RNA sequence analysis using covariance models. Nucleic Acids Res. 1994; 22:2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alkatib S., Fleischmann T.T., Scharff L.B., Bock R.. Evolutionary constraints on the plastid tRNA set decoding methionine and isoleucine. Nucleic Acids Res. 2012; 40:6713–6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takenaka M., Zehrmann A., Verbitskiy D., Härtel B., Brennicke A.. RNA editing in plants and its evolution. Annu. Rev. Genet. 2013; 47:335–352. [DOI] [PubMed] [Google Scholar]
- 20. Drescher A., Ruf S., Calsa T. Jr, Carrer H., Bock R.. The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 2000; 22:97–104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.