Abstract
The widespread application of next-generation sequencing technologies has revolutionized microbiome research by enabling high-throughput profiling of the genetic contents of microbial communities. How to analyze the resulting large complex datasets remains a key challenge in current microbiome studies. Over the past decade, powerful computational pipelines and robust protocols have been established to enable efficient raw data processing and annotation. The focus has shifted toward downstream statistical analysis and functional interpretation. Here, we introduce MicrobiomeAnalyst, a user-friendly tool that integrates recent progress in statistics and visualization techniques, coupled with novel knowledge bases, to enable comprehensive analysis of common data outputs produced from microbiome studies. MicrobiomeAnalyst contains four modules - the Marker Data Profiling module offers various options for community profiling, comparative analysis and functional prediction based on 16S rRNA marker gene data; the Shotgun Data Profiling module supports exploratory data analysis, functional profiling and metabolic network visualization of shotgun metagenomics or metatranscriptomics data; the Taxon Set Enrichment Analysis module helps interpret taxonomic signatures via enrichment analysis against >300 taxon sets manually curated from literature and public databases; finally, the Projection with Public Data module allows users to visually explore their data with a public reference data for pattern discovery and biological insights. MicrobiomeAnalyst is freely available at http://www.microbiomeanalyst.ca.
INTRODUCTION
The past decade has seen an immense growth in the number of studies that aim to characterize the structures, functions and dynamics of host-associated microbial communities (microbiota) within the context of host development, pathophysiology, diet and environment perturbations (1,2). These studies have revealed a wide array of important roles that the microbiota play in human and animal health. Due to drastic reduction in costs and its high-throughput capacity, next-generation sequencing has become the preferred method to study the collective genetic contents of microbial communities (microbiome). Currently, microbiome datasets are mainly generated using one of the three common sequencing strategies including marker gene (i.e. 16S rRNA) survey to characterize microbial community compositions, shotgun metagenomics to study their functional potentials, and shotgun metatranscriptomics to identify those actively expressed genes. These studies usually generate datasets that are both large (with regard to data size) and complex (with regard to data structure), posing substantial ‘big data’ challenges in downstream data analysis.
The initial computational effort in microbiome data analysis focused on raw sequence processing, clustering and annotation. This led to the development of several powerful tool suites such as MEGAN, MG-RAST, mothur and QIIME (3–6), which together helped to establish the essential pipelines and procedures for processing raw reads generated from microbiome studies. Given the ever-increasing data sizes and computational costs, raw data processing is now typically handled at the same sequencing center following standardized protocols. These procedures produce a key summary table containing feature (Operational Taxonomic Units (OTUs), taxa or genes) abundance information across samples, along with various annotations and sample metadata. The Biological Observation Matrix (BIOM) file was recently developed to store all these types of information to facilitate the interoperability of existing bioinformatics tools and future meta-analyses (7). For most researchers, their primary challenge in data analysis is how to make sense of the abundance tables or BIOM files within the context of different experimental factors or study conditions.
Microbiome data analysis can be placed into four general categories: (i) taxonomic profiling - to characterize community compositions based on methods developed in ecology such as alpha-diversity (within-sample diversity) or beta-diversity (between-sample diversity); (ii) functional profiling - to assign genes into different functional groups (i.e. metabolic pathways or biological processes) to understand their collective functional capacities; (iii) comparative analysis - to identify features that are significantly different among conditions under study and (iv) meta-analysis - to integrate user data with public data or knowledge accumulated for improved statistical power or biological understanding. The first two categories are now relatively straightforward to perform, while the last two categories still remain very challenging and become the focus of intense research efforts.
Microbiome abundance data presents several unique challenges including sparsity (containing many zeros), vast differences in sequencing depth, and large variance in distributions (over-dispersion) (8). These unique characteristics have made it inappropriate to directly apply methods developed in other omics fields to perform comparative analysis on microbiome data. As a result, non-parametric permutation-based methods are often employed for identification of significant features in microbiome data (9,10). Although robust, the main limitations of such approaches are the lack of statistical power and the inability to model confounding factors to accommodate complex experimental designs. To deal with uneven sequencing depth, researchers often resort to two common approaches: rescaling the total reads in each sample to a constant sum (using proportions), or resampling the reads in each sample to an equal amount (rarefying). The former will lead to typical issues associated with compositional data (11), and the latter may lead to the loss of important information. In general, it is statistically more appropriate to develop suitable statistical models for sparse count data to accommodate differences in sequence depth, or to develop strategies to transform data to have distributions that fit the models assumed by other well-established algorithms. There has been significant progress towards these directions in recent years. For instance, the metagenomeSeq algorithm integrates cumulative-sum scaling (CSS) method and a statistical model based on the zero-inflated Gaussian (ZIG) distribution to improve the power for differential abundance analysis of microbiome data (12). It has also been shown that, following proper data normalization, the methods developed for RNAseq such as edgeR and DESeq2 perform similarly to or better than many other algorithms developed specifically for microbiome data (13–15). To account for compositional data, different data transformation approaches have been proposed such as the centered log-ratio (CLR) transformation (16).
The majority of these recent methods have been implemented as R packages. In particular, the phyloseq package has been developed to provide a unified framework to allow R users to explore different statistical algorithms for microbiome data analysis (17). Although powerful and flexible, learning R programming and the underlying statistics can be demanding for most clinicians and bench researchers. There is an urgent demand for user-friendly tools that support these recent approaches for comprehensive statistical analysis of microbiome data. In addition, with the increasing number of public datasets and our growing knowledge about microbiome, it is now possible to perform meta-analyses to reveal larger pictures or novel insights beyond a single study, such as using compatible public datasets for contextualizing new experiments (18), pooling new data with existing cohorts for increased power (19), or comparing microbial signatures with those reported from other studies (20).
To address these gaps as well as to meet new requests arising from current microbiome data analysis, we have developed MicrobiomeAnalyst, a web-based program to allow clinical and basic scientists to easily perform exploratory analysis on common abundance profiles and taxonomic signatures generated from microbiome studies. The key features of MicrobiomeAnalyst include:
Support for a wide array of common as well as advanced methods for taxonomic diversity analysis, functional profiling, visualization and significance testing;
Comprehensive support for various data filtering and transformation methods coupled with well-established as well as more recent algorithms for differential abundance analysis;
A powerful, fully-featured metabolic network visualization framework for intuitive exploration of results from functional profiling;
Support for meta-analysis with compatible public datasets for context reference and pattern discovery using 3D visual analytics;
Enrichment analysis based on >300 taxon sets manually collected from literature and public databases.
MicrobiomeAnalyst also contains a comprehensive list of frequently asked questions (FAQs) and tutorials to help researchers easily navigate different analysis tasks. Collectively, these features consist of comprehensive tool suites for microbiome data analysis. MicrobiomeAnalyst is freely available at http://www.microbiomeanalyst.ca.
PROGRAM DESCRIPTION AND METHODS
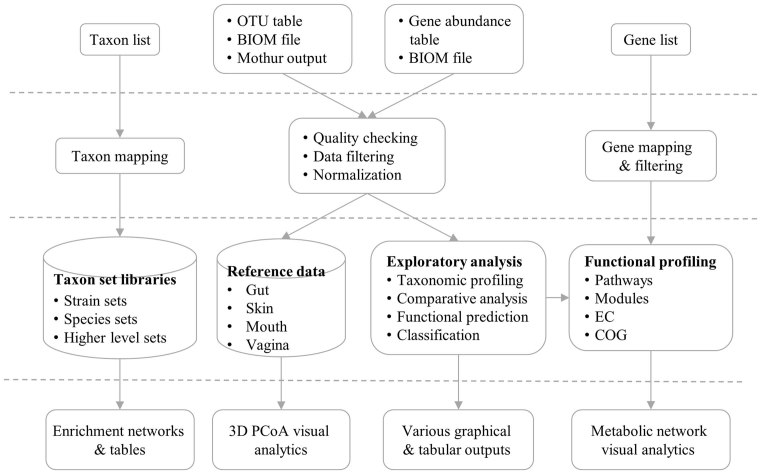
MicrobiomeAnalyst is comprised of four modules. The first is the Marker Data Profiling (MDP) module that is designed for analysis of 16S rRNA marker gene survey data. The second is the Shotgun Data Profiling (SDP) module that contains functions for analyzing metagenomics or metatranscriptomics data. The third module, the Taxon Set Enrichment Analysis (TSEA), is designed to test whether there are biologically or ecologically meaningful patterns from a given list of taxa of interest. Finally, the Projection with Public Data (PPD) module allows users to visually compare their data with our collection of curated public datasets for novel patterns or biological insights. Figure 1 summarizes the overall design and the flowchart of MicrobiomeAnalyst. Different modules provide a variety of options and procedures to help users to complete their tasks. We advise users to start with our tutorials and to first try our example data to become familiarized with the basic features and main steps.
Figure 1.
MicrobiomeAnalyst flow chart. MicrobiomeAnalyst accepts taxa/gene lists, OTU/gene abundance tables, or BIOM files. Three consecutive steps are performed - data processing, data analysis, and result exploration. The associated web interface offers a rich set of options, and produces various tables and graphics to allow users to intuitively navigate the data analysis tasks.
Data upload and processing
Overview of data inputs
The four modules (MDP, SDP, TSEA and PPD) are represented as four interactive circular buttons on the homepage of MicrobiomeAnalyst. Users must choose a module based on their data types. The MDP and PPD are designed for 16S rRNA maker gene survey data. Users need to provide a taxon or OTU abundance table together with a sample metadata file containing group information. The files can be uploaded as a tab delimited text (.txt) or in comma separated values (.csv). MicrobiomeAnalyst also accepts BIOM files as well as the common output files from the mothur software package. The SDP module requires the same formats for data input except that the features should be genes annotated by KEGG Orthology (KO), Enzyme Commission (EC) numbers or Cluster of Orthologous Groups (COG) IDs. For more details, users can go to the corresponding FAQs and tutorials, or download our test examples for inspection.
Data filtering
By default, features containing all zeros or only appear in a single sample are excluded in downstream analyses based on technical, statistical and biological considerations. In particular, features with very low counts in very few samples cannot be distinguished from sequencing errors, and significant differences in features characterized by low abundance or rare occurrence are difficult to interpret with respect to their general importance in the whole community. This ‘minimally cleaned’ data is reserved for various alpha diversity analyses in which the primary goal is to understand individual sample diversity. For all other data analysis, further data filtering is necessary. By default, features are filtered based on their abundance levels and sample prevalence. Users can also filter low-count features using a minimum count cutoff based on their mean or median values. If the primary goal is comparative analysis, users should exclude features that exhibit low variance based on their inter-quantile ranges, standard deviations or coefficient of variations. These features are very unlikely to be significant in the comparative analysis. Filtering those uninformative features can ameliorate the data sparsity issue, as well as improve statistical power by reducing the issue of multiple testing in downstream analysis.
Data normalization
After data filtering, users need to perform data normalization in order to make more meaningful comparisons. MicrobiomeAnalyst offers three types of data normalization - scaling, transformation and rarefying, based on various options implemented in phyloseq. The normalized data is used for visual data exploration including beta-diversity and clustering analysis. It is also used for those comparative analysis methods without a known preference for certain normalization procedures, such as univariate statistics and linear discriminant analysis effect size (LEfSe). Other comparative analyses will use their own specific normalization methods. For instance, the cumulative sum scaling (CSS) normalization is used for metagenomeSeq, and the trimmed mean of M-values (TMM) normalization is applied for edgeR. MicrobiomeAnalyst also allows users to perform data rarefying. Recent studies have suggested that this procedure may still be necessary when the library sizes are vastly different (i.e. differing more than 10 folds) (21). The function supports rarefaction curve analysis to allow users to visually assess the sequencing depth with regard to the number of OTUs detected.
Community profiling
Taxonomic diversity profiling
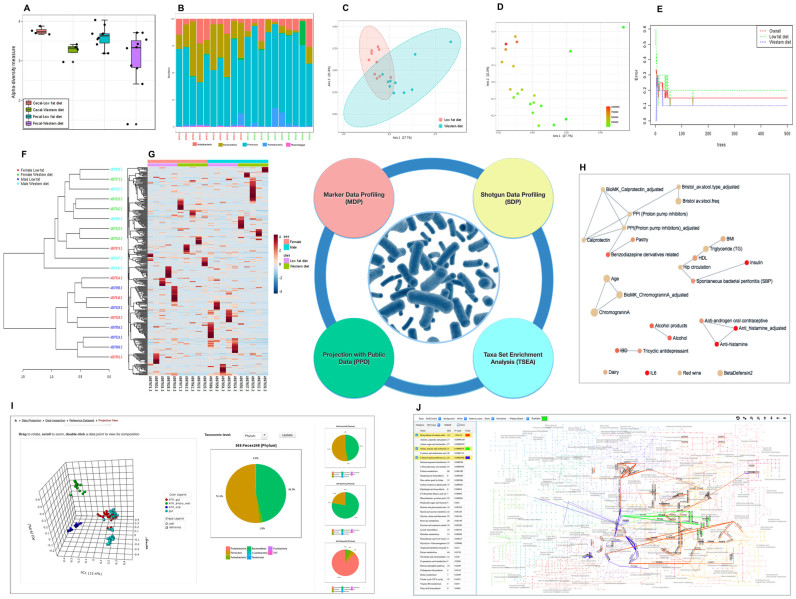
The community diversity profiling was implemented based primarily on the R phyloseq and vegan packages (17,22). The analysis can be performed at different taxonomic levels based on the available annotations. The alpha-diversity analysis function currently supports six common diversity measures. The results are plotted across samples and are also summarized as box plots for each group (Figure 2A). The corresponding statistical significance is estimated automatically using either parametric or non-parametric tests based on user selection. Users can also visualize abundance profiles at different taxonomic levels using a stacked area or stacked bar plot (Figure 2B). The beta-diversity analysis supports five common distance measures. The results are presented as both 2D and 3D ordination plots based on principal coordinate analysis (PCoA) or non-metric multidimensional scaling (NMDS). The corresponding statistical significance is assessed using one of the three statistical methods with Permutational Multivariate Analysis of Variance (PERMANOVA) as the default option. To help identify patterns or gain biological insight, the samples displayed on PCoA or NMDS plots can be colored based on the metadata (default), their alpha diversity measures, or the abundance levels of a particular feature they contain. The last option has often been used to show the potential association between the metadata and a specific feature whose abundance levels (shown as color gradients) vary in the same (or opposite) direction as the separation patterns according to the metadata (Figure 2C and D).
Figure 2.
Example outputs from MicrobiomeAnalyst. (A) A box plot summary of the Shannon diversity index across different groups. (B) A stacked bar chart showing Phylum level abundance profiles across samples. Sample names in red and green colors indicate mice fed a low fat diet (LFD) and a western-style diet (WSD), respectively. (C) A PCoA plot with sample colors based on different diets. (D) The same PCoA plot with color gradients based on the abundance levels of family Bacteroidaceae. (E) A graphical summary of the classification performance on different diets using the Random Forests algorithm. (F) A dendrogram showing the clustering of samples with colors based on diet and sex. (G) A clustered heatmap showing the variation of taxonomic abundance with regard to diet and sex. (H) An interactive network summarizing enriched taxon sets from TSEA. (I) A 3D PCoA plot from the PPD module, with the taxonomic composition of the currently selected sample shown in the middle and the session history on the right. (J) A screenshot showing functional enrichment analysis and visualization within the global metabolic network.
Predicting metabolic potentials and profiling functional diversity
Based on their phylogenetic distances or sequence similarities to those from the microbes whose whole genomes have been sequenced and annotated, the 16S rRNA data can be used to infer the metabolic potentials of the corresponding microbial species. In particular, the PICRUSt is used for Greengenes annotated data (23) and the Tax4Fun is used for data annotated by SILVA database (24). The result is a table containing relative KO abundance levels. The KO profiles obtained from predictions or actually measured from shotgun metagenomics or metatranscriptomics can be used for functional diversity profiling based on the KEGG (pathways, modules or EC categories) or COG annotation systems. Since one KO or COG can be assigned to multiple functional groups, MicrobiomeAnalyst offers different approaches to deal with this issue, including simple sum, normalized sum, or weighted sum methods. The results are presented as a stacked area plot organized by experimental factors to help visualize patterns of variations across different conditions. The underlying abundance table is available for download.
Comparative analysis
Differential abundance analysis
This section allows users to perform formal statistical comparisons to identify features that are significantly different. For marker gene data, the OTU tables can be collapsed to higher levels based on their taxonomic assignments before conducting differential analysis. Although this procedure can reduce data sparsity, a large proportion of OTUs will usually be placed into one ‘Not_Assigned’ bin, making it very challenging for biological interpretation. MicrobiomeAnalyst supports both common parametric and non-parametric univariate analysis, as well as more recent approaches such as metagenomeSeq, edgeR and DESeq2 (15). The results from the differential analysis are displayed as a numerical table. Users can click the ‘Details’ icon to see a box plot summary of any feature of interest. Since different statistical models sometimes produce P values that can be vastly different, it is advisable to compare results from multiple methods and to visualize the features to gain more confidence. By default, MicrobiomeAnalyst will display a maximum of 500 top features according to their P values. The rows containing significant features (if present) are automatically highlighted in orange. This implementation allows users to easily focus on the features of interest while minimizing the chance of missing important ones. For shotgun data, significant KOs can be mapped to metabolic networks for enrichment analysis and visual exploration.
Biomarker identification and classification
This section provides two well-established methods - LEfSe (10) and Random Forests (25). The former has been developed specifically for microbiome data to help identify robust and biologically relevant features for biomarker discovery; while the latter is a generic non-parametric machine learning algorithm which has been shown to perform well in many recent microbiome data analyses and classifications (26–28). In particular, LEfSe uses the non-parametric Kruskal-Wallis rank sum test to detect features with significant differential abundance in different groups, followed by linear discriminant analysis to evaluate the effect size of those significant features. Users can select significant features using a combination of P value and effect size. The Random Forests algorithm uses an ensemble of classification trees (forest), with class prediction based on the majority vote of the ensemble. As the forest is built, it can provide an unbiased estimate of the classification errors by aggregating cross-validation results using bootstrapped samples. In addition, the algorithm also measures the importance of each feature based on the increase of the classification errors when it is permuted. A graphical output is generated to summarize its classification performance with regard to increasing number of trees (Figure 2E).
Other features
MicrobiomeAnalyst also provides other useful features for visual comparison and clustering analysis. Users can generate stacked bar or stacked area plots to view the abundance profiles across samples at different taxonomic levels. The interactive pie chart summarizes the taxonomic compositions for a selected group. Users can click a particular section of interest to further investigate its compositions at a lower taxonomic level. MicrobiomeAnalyst also provides comprehensive support for the widely used hierarchical clustering coupled with dendrograms and heatmaps. Figure 2F and G shows the example outputs from these two functions. All graphical outputs can be downloaded as either Portable Document Format (PDF) or Scalable Vector Graphics (SVG) files for publication purposes.
Taxon set enrichment analysis (TSEA)
Taxon set collection
The taxon sets have been collected from literature and public databases using a combination of text mining and manual curation. The 105 strain sets were obtained mainly from the Genomes Online Database (GOLD) database (29) and the Pathosystems Resource Integration Center (PATRIC) (30), organized primarily based on their phenotypic traits. The 174 species sets were manually collected from over 60 literature publications, organized based on their associations with various host physiological and biochemical measures, disease states or life style factors. Finally, the 40 higher level taxon sets were obtained from the MicroPattern website (20). These taxon sets were manually annotated to improve name readability with links to their original databases or publications.
Enrichment analysis and interpretation
The goal of this analysis is to determine whether members in a particular taxon set are represented more frequently within the user-uploaded list of taxa than expected by random chance. Such a list can be those significant features identified in differential abundance analysis, or those exhibiting similar behaviors based on clustering analysis. The enrichment analysis is calculated using hypergeometric tests. The results are presented as an interactive network (Figure 2H) at the top and a detailed result table at the bottom of the page. The enrichment network offers a high-level overview of important taxon sets and their relationships (31). Each node represents a taxon set with its color based on the P value, and its size based on the number of hits. Two taxon sets are connected by an edge if the number of their shared hits is >20% of the total number of their combined taxa. Users can manually drag and drop a node to improve the layout. Double clicking a node will display the members in the taxon set with those hits highlighted in red.
Projection with public data (PPD)
This module allows users to visually explore their 16S rRNA data within the context of a compatible public dataset. Such comparisons have been increasingly used to reveal microbiome compositional differences in different developmental stages (18) or across different populations (32). The public datasets were collected from the Qiita database (http://qiita.microbio.me) by selecting those well-annotated 16S rRNA marker gene datasets collected from different body sites in human, mouse and cow. The key metadata (sequencing platforms, regions targeted by the primers and associated publications) are displayed to help users choose a suitable dataset. In order to achieve meaningful comparisons, MicrobiomeAnalyst requires that there must be at least 20% OTU overlap between the user data and the selected public reference data.
The results are presented as an interactive 3D PCoA plot (Figure 2I) with node colors based on different experimental factors and node shapes representing different datasets. Users can intuitively rotate (mouse dragging), zoom in and out (mouse scrolling), or directly click on any node (sample) of interest to view its taxonomic composition. The view history is displayed on the right. By comparing the compositions of nodes in distinctive clusters, users can easily identify key taxa underlying the separation patterns. Unlike alpha diversity, beta diversity is mainly affected by those abundant taxa that are shared across samples. Normalization (including data rarefying) tends to have very little effect on data clustering patterns, which has been confirmed in a recent large-scale benchmarking test (33). These observations have been applied in the PPD module to help save computing time, in which the default PCoA is computed from the top 20% most abundant taxa using the Bray-Curtis index distance measure. Users can choose to use the complete dataset or other distance measures to explore the cluster patterns.
Metabolic network visualization
For shotgun data, users can perform enrichment analysis and visually explore the results within a metabolic network. The framework has been developed based on the KEGG global metabolic network using the KEGGscape (34) followed by manual curation. A screenshot of the metabolic network view is shown in Figure 2J. It is composed of three main components - the central network visualization area, the toolbar at the top, and the pathway table on the left. The network is displayed at the central area, with nodes and edges representing metabolites and enzymatic reactions, respectively. In the KEGG layout, certain reactions are represented multiple times at different places to reduce cluttering. A KO is assigned to one or several edges if it encodes the corresponding enzyme. Double click on an edge will show the corresponding reaction information (KO and compounds). Users can use the mouse scroll to zoom in and out of the network. The top toolbar contains functions for common tasks such as changing the background color, switching the view style, specifying a highlighting color, or downloading the current network view as images. The left panel displays the names of the metabolic pathways or modules ranked by their enrichment P values. Clicking on a name will highlight its KO members (edges) within the network, with the edge thicknesses reflecting their abundance levels.
USE CASE
To illustrate the utility of MicrobiomeAnalyst, we conducted a gut microbiome study on the effects of two different diets - a low fat diet (LFD) or a western-style diet (WSD), using male and female wild-type (C57BL/6) mice born in the same family. Fecal samples and cecal contents were collected after 10 weeks on the diets, and DNA extracted from the samples were used to generate 16S rRNA gene libraries. Raw reads processing and taxonomy assignment were performed using the MG-RAST pipeline (4). The BIOM file was then uploaded to the MDP module of MicrobiomeAnalyst. We first compared libraries from fecal samples and cecal contents. According to Shannon alpha-diversity index, the cecal samples displayed higher diversity than fecal samples (Figure 2A); however, both the fecal and cecal samples showed a consistent decrease in microbial diversity when mice were consuming the WSD as compared to the LFD. Additional analyses were carried out using the data from the fecal samples to represent the gut microbiota. As shown in Figure 2B, the abundance of Bacteroidetes phylum was lower, while the abundance of Firmicutes and Proteobacteria phyla were both higher, in mice consuming the WSD as compared to the LFD, an expected effect of the WSD on the gut microbiota (35). Furthermore, the PCoA plot (Figure 2C and 2D) indicated significant difference in beta diversity between the two diet groups at the family level (P < 0.01), and the abundance variations of Bacteriodacea family were closely associated with the patterns of separation. Application of the Random Forests algorithm indicated that the diet types could be predicted with high accuracies based on the microbiome profiles of fecal samples (Figure 2E). MicrobiomeAnalyst also detected sex dimorphic changes in gut microbiota composition in response to WSD feeding. The dendrogram (Figure 2F) showed that the samples clustered more effectively according to diet as compared to sex. The heatmap (Figure 2G) showed two distinct abundance patterns for males and females when comparing the LFD to the WSD. Differential abundance analyses were performed using edgeR and DESeq2 at both OTU and family levels. Comparison of the result tables indicated that the significant features were largely consistent between the two methods.
DESIGN AND IMPLEMENTATION
MicrobiomeAnalyst is based on Java, R and JavaScript. In particular, the R package phyloseq (17) is used extensively for parsing different data formats, statistical analysis and visualization, with further optimizations for better computing efficiencies and visual effects. The Java Server Faces (JSF) technology is used as a high-performance web framework. The entire system is deployed on a Google Cloud server with 32GB of RAM and eight virtual CPUs with 2.6 GHz each. The performance of such implementation has been shown to be able to deal with ∼100s of users on a daily basis, based on the performance of our other tools deployed with the same configurations (36–38). MicrobiomeAnalyst has been tested with major modern browsers such as Google Chrome (5+), Mozilla Firefox (3+) and Microsoft Internet Explorer (9+).
COMPARISON WITH OTHER TOOLS
Several excellent web-based applications have been developed over the past decade to support microbiome data analysis (39–43). Most of these tools have been developed primarily for raw sequence processing, annotation and storage, with limited support for advanced statistical analysis and interactive visual exploration. MicrobiomeAnalyst complements these tools and data repositories by providing comprehensive support for statistical, visual and meta-analysis on the universal abundance tables and BIOM outputs. STAMP (44) and Shiny-phyloseq (45) are two options of locally installable applications equipped with graphical user interface. Based on the detailed comparisons among those web-based tools (Table 1), it is evident that MicrobiomeAnalyst offers a unique set of features and functions with regards to comprehensive statistical data analysis and visualization, metabolic network visual analysis, taxon set enrichment analysis and meta-analysis.
Table 1. Comparison of MicrobiomeAnalyst with other web-based tools. The URL for each tool is given below the table. Tools dedicated solely for sequence annotation are not included.
| Tools | Microbiome-Analyst | METAGEN-assist | EBI-Metagenomics | MG-RAST | VAMPS |
|---|---|---|---|---|---|
| Registration | No | No | Yes | Yes | Yes |
| Data Processing | |||||
| Input | Count tables; BIOM; mothur output | Count tables; BIOM; outputs from 4 tools | Sequences | Sequences | Sequences |
| Filtering | Abundance, variance, manual | Abundance, variance | – | Abundance | Abundance |
| Normalization | Scaling, transformation, rarefying | Scaling, transformation | – | Scaling, trans-formation | Scaling |
| Taxonomic Profiling | |||||
| Alpha-diversity | Multiple | – | – | Shannon | Multiple |
| Beta-diversity | PCoA & NMDS (2D & 3D) | PCA, PLS-DA | PCA | PCoA | PCoA & NMDS (2D only) |
| Functional profiling | |||||
| Functional prediction | PICRUSt & Tax4Fun | – | – | – | – |
| Functional annotation | COG & KEGG | – | GO | SEED, KEGG COG, eggNOG | – |
| Pathway visualization | Yes (JavaScript) | – | – | Yes (SVG) | – |
| Comparative analysis | |||||
| Differential analysis | Univariate methods, DESeq2, edgeR, metagenomeSeq | Univariate methods | – | – | – |
| Biomarker discovery & classification | LEfSe, Random Forests | SVM, Random Forests | – | – | – |
| Meta-analysis | |||||
| Taxon set enrichment analysis | 105 strain sets, 174 species sets, 42 others | – | – | – | – |
| Integration with public data | Visual analytics with 3D PCoA | – | – | – | – |
• MicrobiomeAnalyst: http://www.microbiomeanalyst.ca/
• METAGENassist: http://www.metagenassist.ca/
• EBI-Metagenomics: https://www.ebi.ac.uk/metagenomics/
• MG-RAST: http://metagenomics.anl.gov/
• VAMPS: https://vamps2.mbl.edu/
LIMITATIONS AND FUTURE DIRECTIONS
The data processing and statistical methods in MDP and SDP modules can be used for analysis and visualization of data for both human and environmental microbiome studies. However, the TSEA and PPD modules were developed based on resources primarily from human and mouse microbiome studies, and they are not suitable for analysis of data from other environments. Currently, MicrobiomeAnalyst does not support correlation or association analysis. Unlike other largely established analysis in which different algorithms usually produce results that agree with each other very well, current approaches for detecting correlations among taxa often gives very inconsistent results and could be misleading for inexperienced users. In addition, most of these methods require a large number samples and use computationally intensive re-sampling and permutation-based approaches to compute statistical significance, making it unsuitable for a real-time interactive web application. The current functions for meta-analysis focus on providing visual exploration against public data or supporting enrichment analysis against microbial signatures identified from other microbiome studies. In future releases, we intend to further enhance the support for more rigorous statistical meta-analysis (19,46).
CONCLUSIONS
As a new frontier in biomedical research, current microbiome studies and data analyses are mainly exploratory in nature. Despite the development of many new statistical algorithms in recent years, there is no single statistical method that performs universally well, as clearly shown by a recent large-scale benchmarking test (33). It is therefore critical to enable researchers in the microbiome field to easily explore their own datasets using a variety of algorithms, in real-time and through interactive visualization, to facilitate data understanding and hypothesis generation. MicrobiomeAnalyst fulfills these requirements by offering comprehensive support for diversity profiling, comparative analysis and metabolic network visual exploration. It also provides novel functions that allow users to interpret their findings with regard to curated taxonomic signatures or to compare their own data with public datasets. We believe MicrobiomeAnalyst fills a critical gap in current microbiome research. The microbiota is complex and dynamic, and to fully understand its behavior as a system and its interactions with the host, more than one type of omics data needs to be collected, analyzed and integrated. Indeed, multi-omics approaches are increasingly adopted for many microbiome studies (47). The future development of MicrobiomeAnalyst will focus on supporting these expanding trends, particularly in the integration of metabolomics data and systems biology (48–51).
FUNDING
This work was supported by the McGill Startup Fund and the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant. Funding for open access charge: NSERC.
Conflict of interest statement. None declared.
REFERENCES
- 1. Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012; 486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010; 464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huson D.H., Auch A.F., Qi J., Schuster S.C.. MEGAN analysis of metagenomic data. Genome Res. 2007; 17:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meyer F., Paarmann D., D'Souza M., Olson R., Glass E.M., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A. et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008; 9:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009; 75:7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010; 7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McDonald D., Clemente J.C., Kuczynski J., Rideout J.R., Stombaugh J., Wendel D., Wilke A., Huse S., Hufnagle J., Meyer F. et al. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. Gigascience. 2012; 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li H.Z. Microbiome, metagenomics, and high-dimensional compositional data analysis. Annu. Rev. Stat. Appl. 2015; 2:73–94. [Google Scholar]
- 9. White J.R., Nagarajan N., Pop M.. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009; 5:e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C.. Metagenomic biomarker discovery and explanation. Genome Biol. 2011; 12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Filzmoser P., Hron K., Reimann C.. Univariate statistical analysis of environmental (compositional) data: problems and possibilities. Sci. Total Environ. 2009; 407:6100–6108. [DOI] [PubMed] [Google Scholar]
- 12. Paulson J.N., Stine O.C., Bravo H.C., Pop M.. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods. 2013; 10:1200–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McMurdie P.J., Holmes S.. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014; 10:e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gloor G.B., Reid G.. Compositional analysis: a valid approach to analyze microbiome high-throughput sequencing data. Can. J. Microbiol. 2016; 62:692–703. [DOI] [PubMed] [Google Scholar]
- 17. McMurdie P.J., Holmes S.. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013; 8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koren O., Goodrich J.K., Cullender T.C., Spor A., Laitinen K., Backhed H.K., Gonzalez A., Werner J.J., Angenent L.T., Knight R. et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012; 150:470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sze M.A., Schloss P.D.. Looking for a signal in the noise: revisiting obesity and the microbiome. MBio. 2016; 7, doi:10.1128/mBio.01018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma W., Huang C., Zhou Y., Li J., Cui Q.. MicroPattern: a web-based tool for microbe set enrichment analysis and disease similarity calculation based on a list of microbes. Sci. Rep. 2017; 7:40200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weiss S., Xu Z.Z., Peddada S., Amir A., Bittinger K., Gonzalez A., Lozupone C., Zaneveld J.R., Vázquez-Baeza Y., Birmingham A. et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017; 5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dixon P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003; 14:927–930. [Google Scholar]
- 23. Langille M.G.I., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A., Clemente J.C., Burkepile D.E., Thurber R.L.V., Knight R. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013; 31:814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Asshauer K.P., Wemheuer B., Daniel R., Meinicke P.. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics. 2015; 31:2882–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Breiman L. Random forests. Mach. Learn. 2001; 45:5–32. [Google Scholar]
- 26. Knights D., Costello E.K., Knight R.. Supervised classification of human microbiota. FEMS Microbiol. Rev. 2011; 35:343–359. [DOI] [PubMed] [Google Scholar]
- 27. Schloss P.D., Iverson K.D., Petrosino J.F., Schloss S.J.. The dynamics of a family's gut microbiota reveal variations on a theme. Microbiome. 2014; 2:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Subramanian S., Huq S., Yatsunenko T., Haque R., Mahfuz M., Alam M.A., Benezra A., DeStefano J., Meier M.F., Muegge B.D. et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014; 510:417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reddy T.B., Thomas A.D., Stamatis D., Bertsch J., Isbandi M., Jansson J., Mallajosyula J., Pagani I., Lobos E.A., Kyrpides N.C.. The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta)genome project classification. Nucleic Acids Res. 2015; 43:D1099–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wattam A.R., Davis J.J., Assaf R., Boisvert S., Brettin T., Bun C., Conrad N., Dietrich E.M., Disz T., Gabbard J.L. et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017; 45:D535–D542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Merico D., Isserlin R., Stueker O., Emili A., Bader G.D.. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010; 5:e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li J., Jia H., Cai X., Zhong H., Feng Q., Sunagawa S., Arumugam M., Kultima J.R., Prifti E., Nielsen T. et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014; 32:834–841. [DOI] [PubMed] [Google Scholar]
- 33. Thorsen J., Brejnrod A., Mortensen M., Rasmussen M.A., Stokholm J., Al-Soud W.A., Sorensen S., Bisgaard H., Waage J.. Large-scale benchmarking reveals false discoveries and count transformation sensitivity in 16S rRNA gene amplicon data analysis methods used in microbiome studies. Microbiome. 2016; 4:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nishida K., Ono K., Kanaya S., Takahashi K.. KEGGscape: a Cytoscape app for pathway data integration. F1000Res. 2014; 3:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Turnbaugh P.J., Ridaura V.K., Faith J.J., Rey F.E., Knight R., Gordon J.I.. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009; 1:6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fan Y., Siklenka K., Arora S.K., Ribeiro P., Kimmins S., Xia J.. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016; 44:W135–W141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xia J., Gill E.E., Hancock R.E.. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015; 10:823–844. [DOI] [PubMed] [Google Scholar]
- 38. Xia J., Sinelnikov I.V., Han B., Wishart D.S.. MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Res. 2015; 43:W251–W257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wilke A., Bischof J., Gerlach W., Glass E., Harrison T., Keegan K.P., Paczian T., Trimble W.L., Bagchi S., Grama A. et al. The MG-RAST metagenomics database and portal in 2015. Nucleic Acids Res. 2016; 44:D590–D594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huse S.M., Mark Welch D.B., Voorhis A., Shipunova A., Morrison H.G., Eren A.M., Sogin M.L.. VAMPS: a website for visualization and analysis of microbial population structures. BMC Bioinformatics. 2014; 15:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mitchell A., Bucchini F., Cochrane G., Denise H., ten Hoopen P., Fraser M., Pesseat S., Potter S., Scheremetjew M., Sterk P. et al. EBI metagenomics in 2016–an expanding and evolving resource for the analysis and archiving of metagenomic data. Nucleic Acids Res. 2016; 44:D595–D603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen I.A., Markowitz V.M., Chu K., Palaniappan K., Szeto E., Pillay M., Ratner A., Huang J., Andersen E., Huntemann M. et al. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 2017; 45:D507–D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arndt D., Xia J., Liu Y., Zhou Y., Guo A.C., Cruz J.A., Sinelnikov I., Budwill K., Nesbo C.L., Wishart D.S.. METAGENassist: a comprehensive web server for comparative metagenomics. Nucleic Acids Res. 2012; 40:W88–W95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Parks D.H., Tyson G.W., Hugenholtz P., Beiko R.G.. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014; 30:3123–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McMurdie P.J., Holmes S.. Shiny-phyloseq: Web application for interactive microbiome analysis with provenance tracking. Bioinformatics. 2015; 31:282–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xia J., Fjell C.D., Mayer M.L., Pena O.M., Wishart D.S., Hancock R.E.. INMEX–a web-based tool for integrative meta-analysis of expression data. Nucleic Acids Res. 2013; 41:W63–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Quinn R.A., Navas-Molina J.A., Hyde E.R., Song S.J., Vazquez-Baeza Y., Humphrey G., Gaffney J., Minich J.J., Melnik A.V., Herschend J. et al. From sample to multi-omics conclusions in under 48 hours. mSystems. 2016; 1, doi:10.1128/mSystems.00038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wikoff W.R., Anfora A.T., Liu J., Schultz P.G., Lesley S.A., Peters E.C., Siuzdak G.. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McHardy I.H., Goudarzi M., Tong M., Ruegger P.M., Schwager E., Weger J.R., Graeber T.G., Sonnenburg J.L., Horvath S., Huttenhower C. et al. Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome. 2013; 1:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ursell L.K., Haiser H.J., Van Treuren W., Garg N., Reddivari L., Vanamala J., Dorrestein P.C., Turnbaugh P.J., Knight R.. The intestinal metabolome: an intersection between microbiota and host. Gastroenterology. 2014; 146:1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Magnusdottir S., Heinken A., Kutt L., Ravcheev D.A., Bauer E., Noronha A., Greenhalgh K., Jager C., Baginska J., Wilmes P. et al. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol. 2017; 35:81–89. [DOI] [PubMed] [Google Scholar]