Abstract
With currently more than 126 000 publicly available structures and an increasing growth rate, the Protein Data Bank constitutes a rich data source for structure-driven research in fields like drug discovery, crop science and biotechnology in general. Typical workflows in these areas involve manifold computational tools for the analysis and prediction of molecular functions. Here, we present the ProteinsPlus web server that offers a unified easy-to-use interface to a broad range of tools for the early phase of structure-based molecular modeling. This includes solutions for commonly required pre-processing tasks like structure quality assessment (EDIA), hydrogen placement (Protoss) and the search for alternative conformations (SIENA). Beyond that, it also addresses frequent problems as the generation of 2D-interaction diagrams (PoseView), protein–protein interface classification (HyPPI) as well as automatic pocket detection and druggablity assessment (DoGSiteScorer). The unified ProteinsPlus interface covering all featured approaches provides various facilities for intuitive input and result visualization, case-specific parameterization and download options for further processing. Moreover, its generalized workflow allows the user a quick familiarization with the different tools. ProteinsPlus also stores the calculated results temporarily for future request and thus facilitates convenient result communication and re-access. The server is freely available at http://proteins.plus.
INTRODUCTION
Three-dimensional (3D) structures of macromolecules are often the starting point for achieving an in-depth understanding of protein function. Their use has a long tradition in early-phase drug design applying tools like homology modeling, molecular docking and molecular dynamics simulation. Before any of these methods can be applied, the structure must be pre-processed and usually further analyzed. The preparation of a macromolecular model often includes the addition of hydrogen atoms, the identification of potential binding sites and the assembly of alternative conformations. While there have been substantial efforts of the worldwide Protein Data Bank (PDB) (1) to include information on the quality of deposited structures (2–5), additional validation of the atomic position reliability can be required for highly specific and more demanding applications. Visualization approaches are generally required for the analysis and interpretation of structural data and can further assist communication tasks like the illustration of molecular interactions. Other examples for advanced structure-based applications are the assessment of binding site druggability or the analysis of protein–protein interactions (PPI).
A wide range of tools has been developed to address these issues. However, the usability of these tools is occasionally restricted by platform dependencies, installation obstacles or non-trivial user interfaces. Especially command line tools might be challenging for non-expert users. Therefore, it is desirable to circumvent these issues by providing web services offering platform-independent usage and easy-to-use interfaces. For two of our own approaches, we already provided a web server (6,7). Both had their own interface fitting the specific requirements of the underlying methods. Thus, adding new functionalities or tools requires parallel refactoring or the development of a new web service. This does not only lead to a lack of interoperability but might also constitute a barrier for the users who need to familiarize themselves with different interfaces. In order to address these issues, we developed ProteinsPlus which currently integrates the two former and four new state-of-the-art approaches. It also offers a unified, easy-to-use interface via a single web server. The integrated services cover a broad range of elementary tasks frequently occurring in structure-related life sciences.
THE ProteinsPlus SERVER
The main objective during the development of ProteinsPlus was to create a general workflow to access and preprocess structural data for all kinds of life science research. The resulting workflow starts with the selection of a PDB ID or the upload of a custom PDB file and optionally a ligand file in SD format as input. ProteinsPlus gives an immediate visual impression of the overall protein structure and contained ligand molecules. Afterward, the user can choose an application service of interest (see below), set additional tool configurations and start the calculation. The results will automatically be displayed after the calculation is finished. To provide the best possible user experience, ProteinsPlus uses a caching system to store calculation results. With this system users can access results at a later time and share them with colleagues.
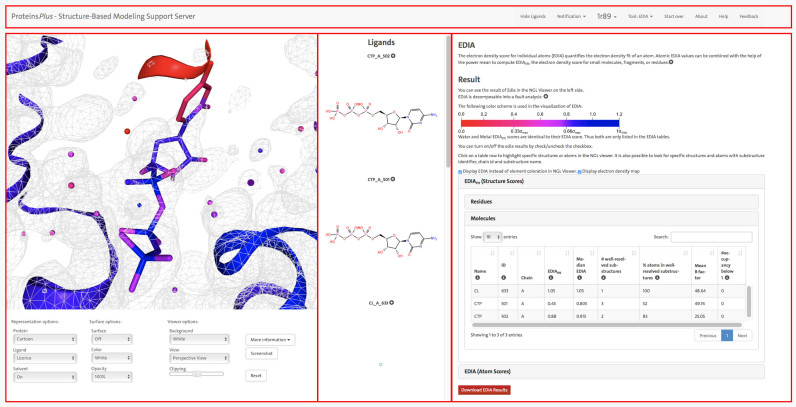
In order to allow for processing various kinds of structure-based tasks, a unified interface is needed that facilitates the integration of different services and meets high usability standards. The single main interface (cf. Figure 1) is divided into three panels and has a menu bar at the top to display additional target related information and to control the panels. The first panel visualizes 3D structural information with the NGL web viewer (8). Below is a control panel that allows to switch between different graphical representations, change the background color, display a molecular surface, clip the scene in z-direction and take a screenshot of the visualized data. If the given PDB file contains ligand molecules, these are additionally depicted as standard structure diagrams in the second panel and are further annotated with their PDB identifier and a unique SMILES string (9) (which is hidden per default). A click on a specific structural diagram highlights the ligand in the NGL viewer panel and also selects the ligand for the tool configuration. The third panel displays all tool related information and offers the ability to set options and trigger the calculations. After a calculation is finished, the result page will also be displayed in this panel. Depending on the applied tool, the result page contains various opportunities to manipulate the structure representation in the NGL viewer panel. This includes the visualization of calculated structural elements, the coloring of the depicted elements and the possibility to automatically focus on certain substructures. Linking the individual results with a commonly used 3D visualization supports the general understanding of different structural properties and simplifies the result interpretation.
Figure 1.
EDIA analysis for the crystal structure of an archaeal class I CCA-adding enzyme in complex with cytidine-5΄-triphosphate (CTP) (PDB ID: 1R89 (10)). This figure demonstrates how the ProteinsPlus web server can be used to assess the quality of a protein structure and analyze potential uncertainties in the structure. The panel on the right side shows the results from the EDIA calculation along with a short description of the quality measure. The detailed results for the EDIAm (structure score) for molecular substructures are displayed. For CTP 501 A, the EDIAm score is very low, indicating possible uncertainties in the structure. The binding site of this CTP molecule is shown in the left panel in the NGL web viewer, allowing a detailed visual inspection. All atoms in the structure are colored according to their individual EDIA score (as explained in the right panel). Additionally, the electron density map (2fo-fc) at 1 σ is displayed. It is clearly recognizable that most atoms in the cytosine moiety receive very low EDIA scores. This is consistent with the observation that around these atoms no electron density is observed at 1σ. The figure also highlights the menu bar at the top and all three panels with red rectangles, the NGL viewer with the control panel on the left, the ligand panel with structure diagrams in the middle and the tool panel with the result page of EDIA at the right.
Currently, the ProteinsPlus server comprises six services addressing the most important tasks at the beginning of structure analysis. The following sections introduce the main aspects of these approaches.
Protoss—hydrogen prediction
A common barrier to the application of three-dimensional structures is the incomplete representation of the respective macromolecules in many available data sources. This is primarily reasoned in shortcomings of the respective structure elucidation methods. For example, in the case of X-ray crystallography, insufficient resolution leads almost generally to the lack of hydrogen atom positions and frequently also impedes a differentiation of similar chemical elements which, in turn, increases the risk of erroneous side-chain orientations. Besides that, another common problem is the lack of additional information on bond orders and atom hybridization in many publicly distributed structural data sources. This is especially relevant for the interpretation of complexed ligands and atypical residues. However, a multitude of structure-based applications rely on a detailed representation of the considered molecules. For example, an accurate assessment of molecular interactions normally requires the knowledge of all atom positions, especially for the investigation of strongly directed interactions like hydrogen bonds. Therefore, several approaches have been developed for completing a structural model by missing elements such as hydrogen atoms and bond types and additionally improving unlikely side-chain orientations. (11–20)
The ProteinsPlus server allows to tackle these tasks by applying our hydrogen prediction software Protoss (21,22). Starting with a macromolecular structure, Protoss first identifies unknown bond types on the basis of atom distance analysis. Following this, possible alternative states of polar moieties are detected and mutual energetic influences of these states are analyzed resulting in an interaction network. Finally, Protoss selects an optimal state for each group on the basis of a network optimization algorithm. The selected states eventually define the presence and position of polar hydrogen atoms as well as the orientation of ambiguous side chains. It is noteworthy that Protoss is able to consider alternative states of arbitrary chemical moieties (cf. Figure 2 for an example), while the vast majority of competitive tools focuses on the treatment of groups occurring in proteinogenic amino acids. Our large scale evaluation studies demonstrated that Protoss, in comparison to alternative approaches, benefits from this more elaborate modeling of chemical variability in terms of improved optimization capabilities for molecular interaction networks of protein–ligand interfaces. In the ProteinsPlus web interface, the completed structures are visualized in the NGL viewer panel and provided for download in PDB format. Processed ligand molecules and atypical residues can additionally be downloaded in SD format. Due to its low computation times, the results of a Protoss calculation can mostly be provided within a few seconds.
Figure 2.
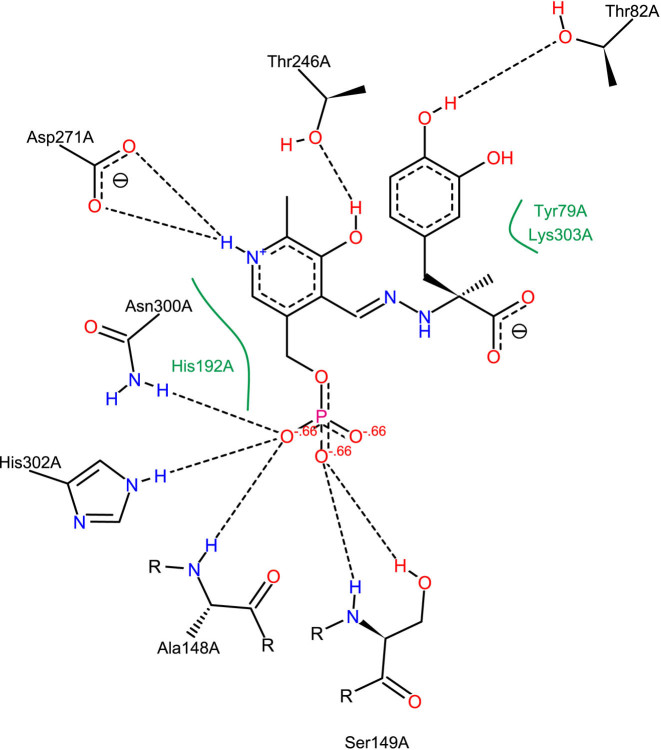
PoseView interaction diagram of dopa decarboxylase in complex with the inhibitor carbidopa (PDB ID: 1JS3 (23)). The automatically generated depiction clearly illustrates the molecular interactions described in the primary publication, e.g. the ‘salt bridge between the carboxylate group of ASP 271 and the protonated pyridine nitrogen’ (23). The PoseView interaction analysis is based on hydrogen orientations and protonation states calculated with Protoss (22).
PoseView—2D interaction diagrams
The increasing amount of protein–ligand complex structures—both from experimental sources and computational predictions—makes the availability of efficient visual inspection tools mandatory. The classic approach of inspecting such structure collections is looking at each of them in a 3D representation. This requires the user to rotate and translate the view until all features are visible. It can neither be used for the comparative visualization of many complexes nor for print and share. In text books and scientific publications, 2D representations which illustrate the key interactions between protein and ligand are frequently applied in this case. Various tools exist to condense the information about participating amino acids and relevant interactions into a 2D structure diagram. MOE (24) and LeView (25) create diagrams that depict the ligand in atomic detail while residues of the pocket are shown as circles. LigPlot+ (26) and PoseView (27) show all interacting structural elements in atomic detail. Unlike LigPlot+, which generates 2D coordinates by flattening out the input 3D structure, PoseView generates structure diagrams from scratch focussing only on the best layout. Thus, it is able to draw about 80% of the Ligand Expo PDB subset without overlaps (28). Furthermore, PoseView aims at depicting all structure diagrams following the IUPAC drawing conventions. It is also integrated into the RCSB PDB website itself. An example of a PoseView diagram is given in Figure 2.
The ProteinsPlus server facilitates to create PoseView interaction diagrams for ligands from PDB structures or additionally provided custom molecules in a fully automated fashion. Before identifying the involved amino acids, Protoss (see preceding section) is used for pre-processing the active site to define the protonation as well as tautomeric form of the protein and ligand. The resulting interaction diagram can be viewed directly in the browser and can be downloaded in various file formats (PDF, SVG and PNG).
EDIA—structural quality elucidation
Like any other experimental technique, structure elucidation has its limitations related to resolution and precision. Therefore, the examination of structural uncertainty is an advisable initial step for all applications based on macromolecular models. For structures determined with X-ray crystallography, a number of measures exist that objectively quantify the electron density fit, e.g. the real-space correlation coefficient (29) or the real-space difference density Z-score (30). Recently, we developed the electron density score for individual atoms (EDIA) (31) as a measure for estimating how well each atom position in a certain structure is supported by the experimental electron density. For all life scientists basing their research on individual structural features of a protein or a nucleic acid, it is essential to know this degree of experimental support for each atom, functional group or ligand molecule.
Based on a 2fo-fc map, EDIA applies a grid-based approach to analyze the electron density distribution in a sphere around a certain atom considering both, density shape and intensity. It avoids the use of annotated B-factors by using a statistically determined resolution dependent B-factor. Therefore, EDIA overcomes known weaknesses of existing approaches like strong shape dependency (4) and tolerating overly flexible atoms that cause weak, stretched out electron density. The EDIA formula can be decomposed to allow an automatic analysis explaining the reasons for a low EDIA score. Furthermore, EDIA scores can be combined using the power mean to score molecular fragments (EDIAm) and thus facilitate the identification of well resolved substructures. EDIAm is also a very valuable addition to calculating RMSD values for investigation of re-docking capability, since the EDIAm truthfully reports the displacement from the experimental data while the RMSD reports the displacement from the interpreted coordinates.
Within ProteinsPlus, EDIA and EDIAm scores are presented in an interactive table and the structure in the NGL viewer panel is recolored based on the EDIA coloring scheme. This allows an instantaneous differentiation of well resolved (blue) and weakly supported (red) substructures (see Figure 1). For comparison, the electron density can be displayed at a level of 1σ. Additionally, the result tables and the 3D visualization contain mutual links that allow to focus on a certain substructures in the NGL viewer panel by selecting an element from the result tables or filtering the entries of the result tables by clicking a certain residue in the viewer area. The download package consists of all EDIA and EDIAm scores in combination with the structure in a PDB file containing EDIA values in the B-factor column and the error analysis in the occupancy column. All EDIA scores of an average-sized structure can be computed in ∼4 min.
SIENA—structure ensemble assembly
When working with experimental structures of macromolecules, another highly relevant limitation is the inherent incapability of a single structure to properly represent the molecule's flexibility or other variations like its mutation sensitivity. As a straightforward approach to circumvent this drawback, multiple structures of the same target can be employed, often even without major adaption of the applied tools. Ideally, such ensembles can also be compiled from experimental data. While this remains difficult for nucleic acids, for which so far only a limited amount of refined structures exist, for many proteins there is already a sufficient number of structural alternatives available. The required ensemble generation process involves the challenge of selecting an appropriate set of structures. This includes the differentiation of desired and undesired variations as well as the identification of structural artefacts and inconsistencies in data annotation. Furthermore, typical preprocessing steps like a residue-wise alignment, superposition and hydrogen prediction (cf. Protoss) can support the direct applicability of the ensemble. In order to support all these tasks, we have developed an adaptive ensemble assembly approach called SIENA (32) that allows a case-specific generation and preprocessing of structure ensembles. Due to the high relevance of molecular interactions for protein functions, SIENA has a specific focus on the treatment of user-defined substructures like protein binding sites. SIENA achieves a quick access to alternative structures by a combination of an indexed database and an alignment technique (33) that is specifically geared to the processing of alternative binding site conformations. Additionally, it provides a set of various filters that allow a use-case specific adaption of the ensemble compilation. Among others, this includes functionalities for the assertion of structural consistency and an interaction-driven approach for ensemble reduction leading to a small but diverse set of representative structures. Various evaluation experiments highlight that SIENA allows for accurate and efficient ensemble preprocessing for sequence identities over than 70%.
Within the ProteinsPlus server, SIENA can be triggered with a user-defined binding site query in combination with various filtering conditions to eliminate unwanted structures. Typical application scenarios like flexibility analysis, virtual screening and ligand pose comparison are supported by a one-click selection opportunity of predefined parameterization settings. The superimposed structures of the resulting binding site ensembles, which are usually provided within a few seconds, can be visualized in the NGL visualization area individually. Furthermore, the ProteinsPlus server allows to download the generated ensemble in form of an archive file that contains all superimposed structures, a sequence alignment of the binding site residues and a statistical overview of certain ensemble measures like binding site RMSD or the number of mutated amino acids.
DoGSiteScorer—binding site detection
Target assessment is one of the major challenges in early drug discovery. Besides aspects such as medical rationale and commercial attractiveness, knowledge about the ability of a target to bind a drug like molecule, i.e. called druggability, is of utmost importance (35). The binding site of a protein is the key to its function. Given a protein structure, the first step is, thus, the identification of potential cavities and a precise description of them. If a ligand-bound structure is available, this ligand defines the binding site. Nevertheless, additional allosteric or novel sites in ligand-free structures are of interest in prospective analyzes. In such cases, automatic methods to predict and rank cavities are investigated, e.g. FPocket (36), SiteMap (37) or DoGSiteScorer (38). Binding site detection methods rely solely on the 3D structure of the protein and use geometric and/or energetic information to detect cavities. Furthermore, these methods are able to estimate the druggable potential of a pocket using linear combinations (37), exponential functions (36) or machine learning models (38) derived from selected pocket descriptors, such as volume, enclosure or hydrophobicity.
DoGSiteScorer, is a grid-based pocket detection (39) and druggability prediction (38) method. The (sub)pocket detection step (39) has been evaluated on several benchmark dataset (Weisel dataset (40), PDBbind (41), sc-PDB (42)) and showed superior results. For druggability prediction (38), DoGSiteScorer uses a small set of physico-chemical and geometric descriptors combined with a support vector machine (SVM) trained and evaluated on the freely available druggability dataset (DD) (43). Validation on the complete DD yielded 88% correct predictions. DoGSiteScorer has been applied in several studies (>180 citations of references (7,38,39)) and was listed within the selected online resources supporting drug discovery in 2013 (44).
DoGSiteScorer is part of the ProteinsPlus server and can be used to detect binding sites on a target of interest (see Figure 3). It discloses information about the properties of the detected pockets as well as their druggability. This knowledge can be used to prioritize targets for drug discovery or structures/binding sites for docking; or to compare pockets. As input, only a protein structure is required (PDB format or PDB ID). After pocket calculation, a sortable table appears that lists all pockets, together with the values for pocket surface, volume and druggability score. Additional descriptors can be displayed upon request. Per default, the largest pocket is shown in mesh representation in the NGL visualization (color corresponds to the table). Additional pockets can interactively be en-/disabled. All data, the pocket volumes (CCP4 format), the pocket residues (PDB format) as well as the full descriptor table (text format), is available for download.
Figure 3.
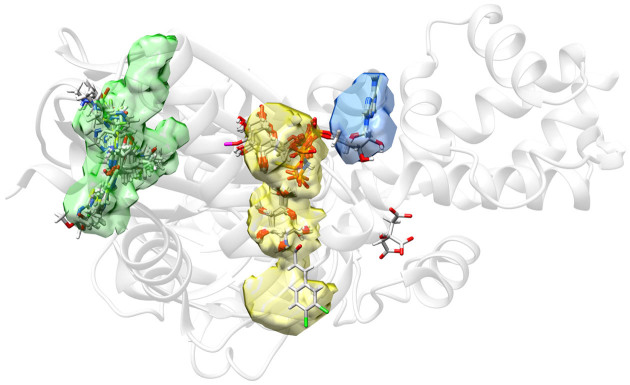
Predicted pockets using DoGSiteScorer for Hexokinase IV in complex with α-D-glucose only (PDB ID: 3QIC (34)). An ensemble was generated with SIENA using 3QIC as query structure and α-D-glucose as reference ligand. The figure includes all ligand molecules from this ensemble with more than six heavy atoms and within a distance of 5 Å from any protein atom in the 3QIC structure. As indicated by the superposition of ligands and DoGSiteScorer pocket predictions, the two best-ranked pockets correspond very well to the allosteric binding site (green) and the substrate binding site (yellow). Interestingly, the allosteric binding site is identified as the most druggable pocket (Drug-Score calculated by DoGSiteScorer: 0.81), which is in good agreement with the distribution of activating ligands found by SIENA. The ATP binding site, which is relatively solvent exposed, is not detected as one pocket but still well covered when considering the union of the two neighboring pockets depicted in yellow and blue.
HyPPI—protein–protein interactions classification
PPIs play key roles in biological regulatory pathways. Therefore, they are of central importance for the understanding of biological processes. Furthermore, they are of special interest for the development of small molecule modulators and lately received more attention in drug discovery (45–47). The PDB contains a substantial amount of structural data related to protein–protein complexes. However, the asymmetric unit (the smallest structure that cannot be recreated using symmetry operations) deposited in the PDB file is not necessarily composed of a biological-relevant protein–protein complex. The protein–protein complex might only be due to crystallization conditions (crystal artefact) or the biological-relevant complex must be generated by applying symmetry operations first. Since experimental methods for the determination of the oligomeric state of a complex are costly and time-consuming, it is of interest to develop an automated discrimination of biological complexes (permanent or transient) and crystal artefacts. Diverse methods exist which try to predict PPIs based on the computation of free energies or classification models based on physico-chemical and geometrical descriptors, e.g. PQS (48), NOXclass (49), EPIC (50), PISA (51), DiMoVo (52), CRK (53), OringPV (54), IPAC (55) or IChemPIC (56). Most of those methods achieve high accuracies of 85–97%. However, they use a large amount of descriptors to discriminate those complexes (22–213 descriptors).
The prediction tool HyPPI underlying ProteinsPlus discriminates biological complexes and crystal artefacts. The most promising descriptors we found to characterize the different PPIs are the hydrophobic binding energy and the proportion of the interface ratios (IFquotient). The hydrophobic binding energy is calculated according to the desolvation term of the HYDE scoring function (57). The IFquotient measures the proportion of the subunits’ relative interface area with respect to the molecular surface of the unbound subunit. Thus, it represents the symmetry of the PPI. Using only these two descriptors for the discrimination of biological complexes and crystal artefacts, we achieve a state-of-the-art accuracy of 92.5% on our training set of 254 complexes (49) and 77.9% on an independent test set (152 complexes from different sources (58–62)) which is comparable to the performance of the aforementioned tools. Within the ProteinsPlus server, the discrimination of a PPI can be triggered with HyPPI by selecting the respective subunits. As a result, the probability for each class—biological (permanent or transient) versus crystal artefact—is given. This way, the user directly gets an indication of the reliability of the classification.
SUMMARY AND OUTLOOK
ProteinsPlus presents a unified interface for various structure-based modeling tools. It makes the installation of large modeling software packages for an initial inspection of protein structural data dispensable. Therefore, the server is of special interest to life scientists with an occasional need to work with protein structures. The integrated NGL web viewer gives a first impression of the input structure and the calculated results. Thanks to the caching system, users can also share the results or check them later without any further calculation. With currently six tools, the unified easy-to-use interface and the generalized workflow, the ProteinsPlus web server is a valuable resource for structure-based life science research. For the future, we plan to extend its functionality by additional modeling techniques and further improve its usability, e.g. by predefined use case parameterizations and by a pipeline functionality which allows to use previously calculated results as input for other integrated tools.
ACKNOWLEDGEMENTS
We would like to thank Alexander Rose for his support in integrating the NGL viewer on ProteinsPlus. Figure 3 was generated with the UCSF Chimera package (63).
FUNDING
Developement of ProteinsPlus by de.NBI (in part); German Federal Ministry of Education and Research [031L0105]. Funding for open access charge: German Federal Ministry of Education and Research (BMBF) [031L0105].
Conflict of interest statement. Protoss is part of the BioSolveIT product SeeSAR; M.R. is a co-founder and stakeholder of BioSolveIT.
REFERENCES
- 1. Berman H., Henrick K., Nakamura H.. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003; 10:980–980. [DOI] [PubMed] [Google Scholar]
- 2. Adams P.D., Aertgeerts K., Bauer C., Bell J.A., Berman H.M., Bhat T.N., Blaney J.M., Bolton E., Bricogne G., Brown D. et al. . Outcome of the first wwPDB/CCDC/D3R ligand validation workshop. Structure. 2016; 24:502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Montelione G.T., Nilges M., Bax A., Güntert P., Herrmann T., Richardson J.S., Schwieters C.D., Vranken W.F., Vuister G.W., Wishart D.S. et al. . Recommendations of the wwPDB {NMR} validation task force. Structure. 2013; 21:1563–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Read R.J., Adams P.D., Arendall W.B., Brunger A.T., Emsley P., Joosten R.P., Kleywegt G.J., Krissinel E.B., Lütteke T., Otwinowski Z. et al. . A new generation of crystallographic validation tools for the Protein Data Bank. Structure. 2011; 19:1395–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gore S., Velankar S., Kleywegt G.J.. Implementing an X-ray validation pipeline for the Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 2012; 68:478–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stierand K., Maaß P.C., Rarey M.. Molecular complexes at a glance: automated generation of two-dimensional complex diagrams. Bioinformatics. 2006; 22:1710–1716. [DOI] [PubMed] [Google Scholar]
- 7. Volkamer A., Kuhn D., Rippmann F., Rarey M.. DoGSiteScorer: a web-server for automatic binding site prediction, analysis, and druggability assessment. Bioinformatics. 2012; 28:2074–2075. [DOI] [PubMed] [Google Scholar]
- 8. Rose A.S., Hildebrand P.W.. NGL Viewer: a web application for molecular visualization. Nucleic Acids Res. 2015; 43:W576–W579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weininger D. SMILES, a chemical language and information system. 1. introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988; 28:31–36. [Google Scholar]
- 10. Xiong Y., Li F., Wang J., Weiner A.M., Steitz T.A.. Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes. Mol. Cell. 2003; 12:1165–1172. [DOI] [PubMed] [Google Scholar]
- 11. Brünger A.T., Karplus M.. Polar hydrogen positions in proteins: empirical energy placement and neutron diffraction comparison. Proteins. 1988; 4:148–156. [DOI] [PubMed] [Google Scholar]
- 12. Bass M.B., Hopkins D.F., Jaquysh W. A.N., Ornstein R.L.. A method for determining the positions of polar hydrogens added to a protein structure that maximizes protein hydrogen bonding. Proteins. 1992; 12:266–277. [DOI] [PubMed] [Google Scholar]
- 13. McDonald I.K., Thornton J.M.. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994; 238:777–793. [DOI] [PubMed] [Google Scholar]
- 14. McDonald I.K., Thornton J.M.. The application of hydrogen bonding analysis in X-ray crystallography to help orientate asparagine, glutamine and histidine side chains. Protein Eng. 1995; 8:217–224. [DOI] [PubMed] [Google Scholar]
- 15. Hooft R.W., Sander C., Vriend G.. Positioning hydrogen atoms by optimizing hydrogen-bond networks in protein structures. Proteins. 1996; 26:363–376. [DOI] [PubMed] [Google Scholar]
- 16. Word J.M., Lovell S.C., Richardson J.S., Richardson D.C.. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 1999; 285:1735–1747. [DOI] [PubMed] [Google Scholar]
- 17. Li X., Jacobson M.P., Zhu K., Zhao S., Friesner R.A.. Assignment of polar states for protein amino acid residues using an interaction cluster decomposition algorithm and its application to high resolution protein structure modeling. Proteins. 2007; 66:824–837. [DOI] [PubMed] [Google Scholar]
- 18. Bayden A.S., Fornabaio M., Scarsdale J.N., Kellogg G.E.. Web application for studying the free energy of binding and protonation states of protein-ligand complexes based on HINT. J. Comput. Aided Mol. Des. 2009; 23:621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Labute P. Protonate3D: assignment of ionization states and hydrogen coordinates to macromolecular structures. Proteins. 2009; 75:187–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krieger E., Dunbrack R.L. Jr, Hooft R.W., Krieger B.. Baron R. Assignment of protonation states in proteins and ligands: combining pKa prediction with hydrogen bonding network optimization. Computational Drug Discovery and Design. 2012; NY: Springer; 405–421. [DOI] [PubMed] [Google Scholar]
- 21. Lippert T., Rarey M.. Fast automated placement of polar hydrogen atoms in protein-ligand complexes. J. Cheminf. 2009; 1:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bietz S., Urbaczek S., Schulz B., Rarey M.. Protoss: a holistic approach to predict tautomers and protonation states in protein-ligand complexes. J. Cheminf. 2014; 6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burkhard P., Dominici P., Borri-Voltattorni C., Jansonius J.N., Malashkevich V.N.. Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase. Nat. Struct. Biol. 2001; 8:963–967. [DOI] [PubMed] [Google Scholar]
- 24. Clark A.M., Labute P.. 2D depiction of protein-ligand complexes. J. Chem. Inf. Model. 2007; 47:1933–1944. [DOI] [PubMed] [Google Scholar]
- 25. Caboche S. LeView: automatic and interactive generation of 2D diagrams for biomacromolecule/ligand interactions. J. Cheminform. 2013; 5:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laskowski R.A., Swindells M.B.. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011; 51:2778–2786. [DOI] [PubMed] [Google Scholar]
- 27. Stierand K., Rarey M.. From modeling to medicinal chemistry: automatic generation of two-dimensional complex diagrams. Chemmedchem. 2007; 2:853–860. [DOI] [PubMed] [Google Scholar]
- 28. Stierand K., Rarey M.. Drawing the PDB: protein-ligand complexes in two dimensions. ACS Med. Chem. Lett. 2010; 1:540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jones T., Kjeldgaard M.. [10] Electron density map interpretation. Methods Enzymol. 1997; 277:173–208. [DOI] [PubMed] [Google Scholar]
- 30. Tickle I.J. Statistical quality indicators for electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 2012; 68:454–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nittinger E., Schneider N., Lange G., Rarey M.. Evidence of water molecules—a statistical evaluation of water molecules based on electron density. J. Chem. Inf. Model. 2015; 55:771–783. [DOI] [PubMed] [Google Scholar]
- 32. Bietz S., Rarey M.. SIENA: efficient compilation of selective protein binding site ensembles. J. Chem. Inf. Model. 2016; 56:248–259. [DOI] [PubMed] [Google Scholar]
- 33. Bietz S., Rarey M.. ASCONA: rapid detection and alignment of protein binding site conformations. J. Chem. Inf. Model. 2015; 55:1747–1756. [DOI] [PubMed] [Google Scholar]
- 34. Liu Q., Shen Y., Liu S., Weng J., Liu J.. Crystal structure of E339K mutated human glucokinase reveals changes in the ATP binding site. FEBS Lett. 2011; 585:1175–1179. [DOI] [PubMed] [Google Scholar]
- 35. Volkamer A., Rarey M.. Exploiting structural information for drug-target assessment. Future Med. Chem. 2014; 6:319–331. [DOI] [PubMed] [Google Scholar]
- 36. Le Guilloux V., Schmidtke P., Tuffery P.. Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics. 2009; 10:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Halgren T.A. Identifying and characterizing binding sites and assessing druggability. J. Chem. Inf. Model. 2009; 49:377–389. [DOI] [PubMed] [Google Scholar]
- 38. Volkamer A., Kuhn D., Grombacher T., Rippmann F., Rarey M.. Combining global and local measures for structure-based druggability predictions. J. Chem. Inf. Model. 2012; 52:360–372. [DOI] [PubMed] [Google Scholar]
- 39. Volkamer A., Griewel A., Grombacher T., Rarey M.. Analyzing the topology of active sites: on the prediction of pockets and subpockets. J. Chem. Inf. Model. 2010; 50:2041–2052. [DOI] [PubMed] [Google Scholar]
- 40. Weisel M., Proschak E., Schneider G.. PocketPicker: analysis of ligand binding-sites with shape descriptors. Chem. Cent. J. 2007; 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang R., Fang X., Lu Y., Wang S.. The PDBbind database: collection of binding affinities for protein-ligand complexes with known three-dimensional structures. J. Med. Chem. 2004; 47:2977–2980. [DOI] [PubMed] [Google Scholar]
- 42. Kellenberger E., Muller P., Schalon C., Bret G., Foata N., Rognan D.. sc-PDB: an annotated database of druggable binding sites from the Protein Data Bank. J. Chem. Inf. Model. 2006; 46:717–727. [DOI] [PubMed] [Google Scholar]
- 43. Schmidtke P., Barril X.. Understanding and predicting druggability. A high-throughput method for detection of drug binding sites. J. Med. Chem. 2010; 53:5858–5867. [DOI] [PubMed] [Google Scholar]
- 44. Villoutreix B.O., Lagorce D., Labbé C.M., Sperandio O., Miteva M.A.. One hundred thousand mouse clicks down the road: selected online resources supporting drug discovery collected over a decade. Drug Discov. Today. 2013; 18:1081–1089. [DOI] [PubMed] [Google Scholar]
- 45. Wells J.A., McClendon C.L.. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007; 450:1001–1009. [DOI] [PubMed] [Google Scholar]
- 46. Ivanov A.A., Khuri F.R., Fu H.. Targeting protein-protein interactions as an anticancer strategy. Trends Pharmacol. Sci. 2013; 34:393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Villoutreix B.O., Kuenemann M.A., Poyet J.L., Bruzzoni-Giovanelli H., Labbé C., Lagorce D., Sperandio O., Miteva M.A.. Drug-like protein-protein interaction modulators: challenges and opportunities for drug discovery and chemical biology. Mol. Inform. 2014; 33:414–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Henrick K., Thornton J.M.. PQS: a protein quaternary structure file server. Trends Biochem. Sci. 1998; 23:358–361. [DOI] [PubMed] [Google Scholar]
- 49. Zhu H., Domingues F.S., Sommer I., Lengauer T.. NOXclass: prediction of protein-protein interaction types. BMC Bioinformatics. 2006; 7:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Block P., Paern J., Hüllermeier E., Sanschagrin P., Sotriffer C.A., Klebe G.. Physicochemical descriptors to discriminate protein-protein interactions in permanent and transient complexes selected by means of machine learning algorithms. Proteins. 2006; 65:607–622. [DOI] [PubMed] [Google Scholar]
- 51. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 52. Bernauer J., Bahadur R.P., Rodier F., Janin J., Poupon A.. DiMoVo: a voronoi tessellation-based method for discriminating crystallographic and biological protein-protein interactions. Bioinformatics. 2008; 24:652–658. [DOI] [PubMed] [Google Scholar]
- 53. Schärer M.A., Grütter M.G., Capitani G.. CRK: an evolutionary approach for distinguishing biologically relevant interfaces from crystal contacts. Proteins. 2010; 78:2707–2713. [DOI] [PubMed] [Google Scholar]
- 54. Liu Q., Li J.. Propensity vectors of low-ASA residue pairs in the distinction of protein interactions. Proteins. 2010; 78:589–602. [DOI] [PubMed] [Google Scholar]
- 55. Mitra P., Pal D.. Combining Bayes classification and point group symmetry under Boolean framework for enhanced protein quaternary structure inference. Structure. 2011; 19:304–312. [DOI] [PubMed] [Google Scholar]
- 56. Da Silva F., Desaphy J., Bret G., Rognan D.. IChemPIC: a random forest classifier of biological and crystallographic protein-protein interfaces. J. Chem. Inf. Model. 2015; 55:2005–2014. [DOI] [PubMed] [Google Scholar]
- 57. Schneider N., Lange G., Hindle S., Klein R., Rarey M.. A consistent description of HYdrogen bond and DEhydration energies in protein-ligand complexes: methods behind the HYDE scoring function. J. Comput. Aided Mol. Des. 2013; 27:15–29. [DOI] [PubMed] [Google Scholar]
- 58. Nooren I. M.A., Thornton J.M.. Structural characterisation and functional significance of transient protein-protein interactions. J. Mol. Biol. 2003; 325:991–1018. [DOI] [PubMed] [Google Scholar]
- 59. Bahadur R.P., Chakrabarti P., Rodier F., Janin J.. A dissection of specific and non-specific protein-protein interfaces. J. Mol. Biol. 2004; 336:943–955. [DOI] [PubMed] [Google Scholar]
- 60. De S., Krishnadev O., Srinivasan N., Rekha N.. Interaction preferences across protein-protein interfaces of obligatory and non-obligatory components are different. BMC Struct. Biol. 2005; 5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen Y.C., Lim C.. Common physical basis of macromolecule-binding sites in proteins. Nucleic Acids Res. 2008; 36:7078–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Madaoui H., Guerois R.. Coevolution at protein complex interfaces can be detected by the complementarity trace with important impact for predictive docking. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:7708–7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]