Abstract
G-protein coupled receptors (GPCRs) are key players in signal transduction and therefore a large proportion of pharmaceutical drugs target these receptors. Structural data of GPCRs are sparse yet important for elucidating the molecular basis of GPCR-related diseases and for performing structure-based drug design. To ameliorate this problem, GPCR-SSFE 2.0 (http://www.ssfa-7tmr.de/ssfe2/), an intuitive web server dedicated to providing three-dimensional Class A GPCR homology models has been developed. The updated web server includes 27 inactive template structures and incorporates various new functionalities. Uniquely, it uses a fingerprint correlation scoring strategy for identifying the optimal templates, which we demonstrate captures structural features that sequence similarity alone is unable to do. Template selection is carried out separately for each helix, allowing both single-template models and fragment-based models to be built. Additionally, GPCR-SSFE 2.0 stores a comprehensive set of pre-calculated and downloadable homology models and also incorporates interactive loop modeling using the tool SL2, allowing knowledge-based input by the user to guide the selection process. For visual analysis, the NGL viewer is embedded into the result pages. Finally, blind-testing using two recently published structures shows that GPCR-SSFE 2.0 performs comparably or better than other state-of-the art GPCR modeling web servers.
INTRODUCTION
G-protein coupled receptors (GPCRs) are the largest family of integral membrane receptors consisting of more than 800 members in humans and clustering into five main groups based on phylogenetic criteria (1,2). They transduce a wide variety of extracellular signals to within the cell including ions, hormones, neurotransmitters and sensory stimuli. Due to their fundamental role in signal transduction, a large proportion of medical drugs target these receptors (3). GPCR structural data are important for both understanding the molecular mechanisms underlying diseases caused by mutations in these receptors as well as for performing structure-based drug design. Despite recent advances in stabilizing and crystallizing GPCRs, it is still difficult to obtain experimental structures of them (4). Currently, structural data of Class A GPCRs are restricted to around 30 members. All of these GPCR structures share a common molecular architecture of seven transmembrane helices (TMHs). Hence, the deficit in experimental GPCR structure data can be resolved by building molecular models of GPCRs of unknown structure using homology modeling techniques. Many researchers working on GPCRs are not experienced homology modelers and are therefore unable to benefit from the information that can be gleaned from such three-dimensional (3D) models. Methods that provide high-quality homology models of GPCRs are therefore highly useful to such researchers. As a consequence, various methodologies have been developed for modeling GPCRs and provided for usage as web servers: GPCRM (5), GoMoDo (6), GPCR-ModSim (7) and GPCR-SSFE (8) employ homology modeling techniques whereas GPCR-I-TASSER (9) is a threading assembly method. GPCRM employs a profile–profile comparison and multiple structural alignments for averaging the structure by Modeller. The loops are refined by Modeller and Rosetta. GoMoDo includes Modeller-driven single template homology modeling and offers small molecule docking blindly or by using experimental information. GPCR-ModSim is designed to combine automated modeling (Modeller based) and molecular dynamics equilibration of GPCRs in different conformational states. As a default the protocol uses a single template approach, however multiple template based homology modeling is possible by user selection for each topological section of the GPCR. In contrast to these Modeller based approaches, GPCR-I-TASSER relies on a hybrid protocol to construct the GPCR models by integrating experimental constraints from the GPCR-RD database and uses either a threading or an ab initio TM helix assembly.
The first version of GPCR-SSFE was published in 2011, based at that time on five crystal structures, providing an automatic pipeline for Class A GPCR homology modeling and a comprehensive set of pre-calculated 7TMH homology models (5025) accessible via a web server (8). Template selection is based on the structure–sequence relationship and uses our published workflow whereby the template for each TMH is selected individually thus allowing either a single template or a fragment-based approach to be used for model building (10). GPCR-SSFE's models have been frequently cited and used, for example, to help generate new hypotheses on the enteroendocrine fat sensor GPR119 (11), rationalize the design of potent CB1 antagonists (12) and the server is linked as a partner tool on the GPCRdb website (13).
Many new structural templates have been released since the publication of the first version (including diverse GPCR subtypes) necessitating an update to GPCR-SSFE. These structures provide important insight into general and distinctive structural features (which were previously unknown), shedding light on the variability of GPCR 7TMH architecture. Moreover, the accuracy of homology models is important for their successful usage e.g. in drug design and consequently the increased number of templates results in greater precision of homology model features, including helix or loop properties. These improved models enable better predictions and more accurate dockings of allosteric or endogenous ligands into so far unsolved GPCR structures.
Here, we present an updated and extended version of the web server (http://www.ssfa-7tmr.de/ssfe2). In comparison to our previous version, GPCR-SSFE 2.0 benefits from a considerably enlarged pool of templates (27 inactive crystal structures in contrast to the five templates used in the original version), provides models of the entire serpentine domain by using an evaluated automated version of SuperLooper2 (SL2) (14) for loop modeling and TMH template selection now follows a fingerprint correlation scoring strategy by using a sequence fingerprint database corresponding to distinct structural features. This latter feature allows the optimal template to be selected for each TMH using a weighted structure-feature-matrix based on the presence or absence of fingerprint features. For receptor and fragment visualization, we use NGL (15,16), which adopts capabilities of modern web browsers, such as WebGL for molecular graphics. GPCR-SSFE 2.0 performs comparably or better than other published web servers for modeling Class A GPCRs. Being a fragment-based approach, it has the advantage that it is able to capture structural features that are not identified using single template sequence similarity alone. GPCR-SSFE 2.0 stores a comprehensive set of pre-calculated models comprising 1002 human, mouse and rat GPCR sequences, providing Class A GPCR 3D structural data to the wider GPCR research community.
MATERIALS AND METHODS
Technical features of the GPCR-SSFE 2.0 web server
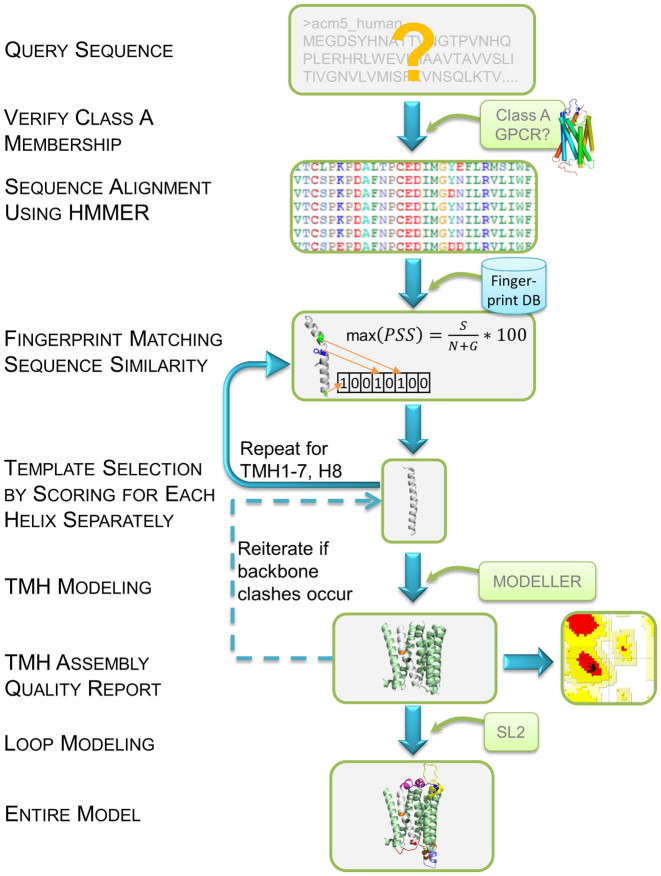
An overview of the workflow used by GPCR-SSFE 2.0 is shown in Figure 1. GPCR-SSFE 2.0 was built by combining an Apache web server (http://www.apache.org), PHP Hypertext Pre-processor scripts (PHP5) and a relational database management system (MySQL 5.5). User-submitted job requests are subject to both client- and server-side validation. Upon validation, the request is saved to the MySQL database and a job with the ID of the corresponding database record started. Each job utilizes a series of Python scripts as well as the following free bioinformatics software and tools: Biopython (17) (sequence parsing), HMMER3 (18) (sequence alignment), MODELLER 9.14 (19) (homology modeling), SL2 (loop modeling) (14) and PROCHECK (20) (quality assessment). The web interface is based on HTML 5, JavaScript and Cascading Style Sheets (CSS) and is cross-browser responsive. The homology model structures are displayed using the NGL viewer, which allows interactive display of even large molecular complexes and is unaffected by the retirement of third-party plug-ins such as Flash or Java-Applets (15). This viewer can easily access and utilize available structural data without any further installations. Loop modeling is provided by a customized SL2 web service, which is queried directly and automatically by SSFE 2.0 using a Python script.
Figure 1.
An overview of the workflow used by GPCR-SSFE 2.0 for template selection and homology modeling. A query protein sequence is first verified for Class A GPCR membership. Upon passing this check, sequence alignment is performed using a HMMER3 derived hidden markov model (HMM) profile. Alignment of the transmembrane helices (TMHs) is used to identify matching fingerprint motifs from our database (see Figure 2). Template selection is performed separately for each helix using the fingerprint scoring strategy and sequence similarity score. Once template selection has been carried out for the 7TMHs and helix 8 (H8) modeling is performed using Modeller. Where backbone clashes occur between two helices, the second-best scoring template is chosen instead and Modeller is re-run. PROCHECK is used to carry out quality checks with a Ramachandran plot produced. SL2 is used to perform loop modeling with selection of the loops being done interactively by the user. Both the TMH models and entire models are available for download by the user.
Fingerprint identification for improved template selection
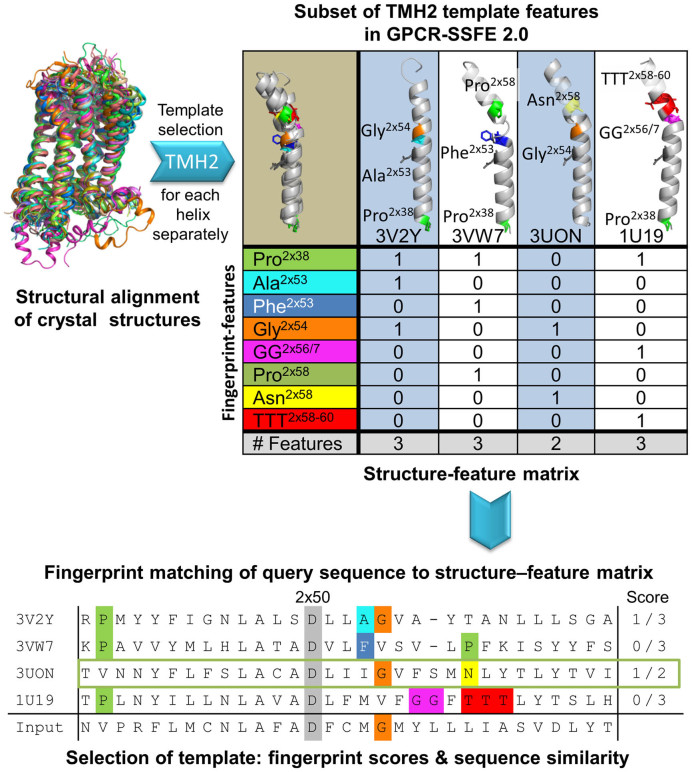
The highest resolution structure of all Class A GPCRs having an inactive crystal structure (totaling 27 at the time) were superimposed using the highly conserved residues found within the 7TMHs (as defined by the Ballesteros–Weinstein nomenclature (21)) and manually inspected for structural features that deviate from a regular α-helical backbone (kinks, bulges, etc) or which are not conserved in all members (Figure 2). These features must correlate with a corresponding sequence pattern (fingerprint) such as proline distortions (22,23), serine/threonine residues modifying proline-induced α-helical kinks by forming side chain hydrogen bonds with the helix backbone (24,25), glycine patterns, conserved motifs, lack of conserved residues etc. More than 150 fingerprint motifs are stored in our relational database. To ensure proper assembling of the TMH fragment-based models, the superimposed Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) (26) co-ordinates were used for homology modeling.
Figure 2.
Fingerprint identification and scoring strategy used by GPCR-SSFE 2.0 for template selection. The set of available crystal structures (templates) were superimposed using the most conserved amino acids in each of the 7TMHs. The structures were manually examined for structural features that deviate from a regular α-helical backbone (kinks, bulges, etc.) or which are not conserved in all members. The fingerprint features are stored in a relational database. Here we show a subset of TMH2 template features in GPCR-SSFE 2.0–Sphingosine 1-phosphate receptor 1 (PDB ID: 3V2Y) has Pro 2×39 and Ala 2×53; Proteinase-activated receptor 1 (PDB ID: 3VW7) has Pro 2×39, Phe 2×53 and Pro 2×58; Muscarinic acetylcholine receptor M2 (PDB ID: 3UON) has Gly 2×54 and Asn 2×58 and bovine Rhodopsin (PDB ID: 1U19) has Pro 2×39, GG 2×56/7 and TTT 2×58–60. These fingerprint features give rise to different helical conformations (straight and different degrees of kink). The aligned input query sequence is then checked against our database of fingerprint features for any matches; in this case, it matches Gly 2×54 and Asn 2×58. The scores of the different templates are then calculated based on the number of matching fingerprint features divided by the total number of fingerprint features that the template has in that helix. The highest scoring template is then chosen for homology modeling (in this instance 3UON for TMH2).
Sequence alignment and scoring strategy
The user uploads a Class A GPCR protein sequence in FASTA format (see workflow in Figure 1) and on passing the Class A GPCR sequence verification check, the server creates a multiple sequence alignment (MSA) using a profile hidden markov model (HMM). HMMER3 was used to generate the HMM from a MSA of 27 template structures plus 51 other class A GPCRs (18). The 51 GPCRs were selected so as to maximize the coverage of the phylogenetic tree for Class A GPCRS (1). After alignment, template selection is carried out for each TMH based on an updated version of our previously published workflow; extending on the structural feature workflows utilized in the original version of GPCR-SSFE (10), we have now implemented a new scoring strategy which uses a weighted structure-feature matrix to calculate the best template per helix based on the presence or absence of fingerprint features (e.g. proline distortions, conserved motifs and TMH extensions) (Figure 2). Where more than one template structure has the highest fingerprint score, the structure with the highest sequence similarity to the query protein is then chosen as the template. Where no fingerprint features are identified in the query protein, the template with the highest sequence similarity score out of the pool of 27 is selected for homology modeling. Template selection for helix 8 (H8) follows a different strategy. The template structures were clustered into groups based on the position of the junction between TMH7 and H8 when superimposed (see Supplementary Data, Figure S1). The template for H8 is selected from the cluster containing the template(s) chosen for TMH7, with sequence similarity used as the basis for selection. This ensures that clashes or large distances are not introduced between TMH7 and H8.
Modeling procedures
Initial modeling is performed for the 7TMHs and H8 using Modeller 9.14. Three models are produced for each GPCR but only the one with the lowest DOPE score (i.e. the most energetically favorable model) is returned (27). If Modeller reports backbone clashes between helices, the second-best scoring template of one of these helices is selected instead and the modeling repeated. Where multiple templates score most highly for a particular helix, a homology model is built using each set of templates. PROCHECK is used to perform stereochemical quality checking of the models with a Ramachandran plot of each model being generated.
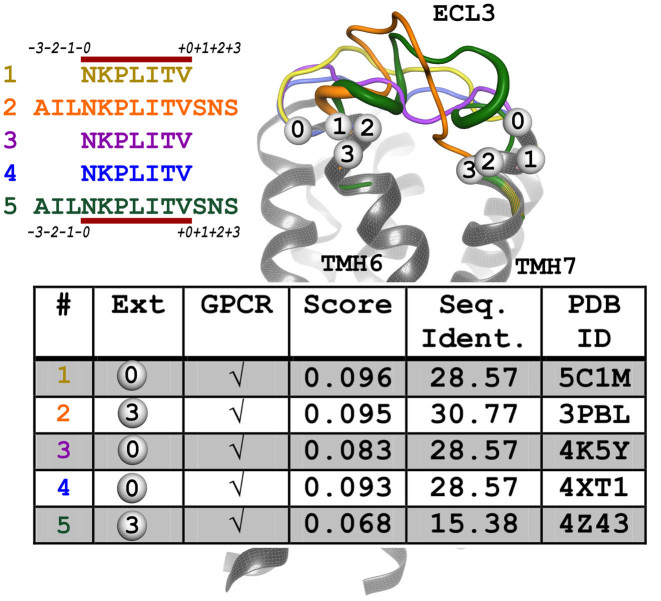
Loop regions are modeled using the SL2 interactive web service (15). Loop candidates are selected from a pre-calculated database currently containing more than 900 million protein fragments with a residue length of 3–35 derived from all entries of the RCSB PDB. Selection of the loops is primarily based on target–template sequence similarity and geometrical fit of stem atoms of the template loop to the receptor (28). To facilitate selection of loops by the user, a customized workflow was constructed for GPCR-SSFE 2.0. The customized SL2 utilizes two loop fragment databases, one containing only fragments from GPCR structures and the other containing fragments derived from all membrane protein (LIMP) entries deposited in the PDB, including GPCRs. The semi-automated approach of SL2 was upgraded allowing fully automated prediction of conformations of intra- and extracellular loops (ECLs). For loops modeled by the SL2 workflow, 100 loop candidates have been calculated from each of the GPCR and LIMP databases ranging in length from the original length of the loop up to symmetrical N- and C- terminal extensions of length 1–3 (see Figure 3). Thus, up to 800 fragment candidates per loop are calculated. GPCR loop templates are given five times higher scores. A list containing all candidates is sorted according to the score and re-ranked using a sliding window of size 2 to favor a candidate with less clashes if two results have a score differing <25%. In general, only candidates with <10 clashes are allowed. Afterward, the top five candidates for each loop are provided to the user for visual inspection. After selecting a set of loops a complete receptor model comprising the entire serpentine domain can be downloaded by the user.
Figure 3.
Output of customized SL2 method for extracellular loop (ECL) 3 of thyroid-stimulating hormone receptor (TSHR). On the right, the TSHR serpentine domain model is shown as cartoon representation in gray with the modeled ECL3 loops as colored tubes (thicker lines indicate structurally defined conformations like helices). N- and C-terminal extensions of the loop sequence, which enlarge the gap into the transmembrane domain of the GPCR are indicated by a numbered bubble. On the left, the modeled loop sequences are listed with their color code and the N- and C-terminal extensions are annotated. The minimal loop length is indicated by the red line. The table is extracted from the results page, providing information about the extension, the origin of the fragment (GPCR and PDB ID:), the fitting score and sequence identity. The user can select the most suitable loop by visual inspection. For visualization purposes only, the loops are elongated by 3 amino acids. Therefore, the loops reach further into the transmembrane region, giving an impression of the concatenated/complete structure.
GPCR-SSFE 2.0 WEB SERVER
Database of pre-calculated models
Using the modeling strategy outlined above, GPCR-SSFE 2.0 stores pre-calculated models for 1002 Class A GPCRs: 252 human (all non-olfactory), 432 mouse and 318 rat. The sequences were downloaded from the GPCRdb in June 2016 (13). Results can be retrieved by either browsing or searching the results. By navigating to the ‘BROWSE’ menu option, the user is presented with a list of 10 Class A GPCR subgroups: Aminergic, Peptide, Protein, Lipid, Melatonin, Nucleotide, Steroid, Alicarboxylic acid, Sensory and Orphan receptors. These subgroups correspond to the endogenous ligand type (as used by the GPCRdb). Each subgroup can be expanded by clicking on it, revealing further subgroupings (receptor family and subtype) or (if the final node is reached) a list of GPCRs within the subtype corresponding to models from different species. Clicking on the UniProt entry name (29) of a GPCR will take the user to the results page for that particular receptor. Alternatively, users may retrieve results by entering the UniProt entry name onto the ‘SEARCH’ webpage.
GPCR-SSFE 2.0 results page
The results returned by GPCR-SSFE 2.0 include: (i) the templates used for analysis; (ii) the MSA of the 27 templates and the query sequence for each individual TMH and H8; (iii) the MSA of the profile HMM GPCR sequences and the query sequence, which spans the entire serpentine domain; (iv) the HMMER2 e-value assigned to the full-length MSA; (v) the template suggestions (and reasons) for the seven TMHs and H8; (vi) the sequence similarity score between the suggested template(s) and the query sequence; (vii) the rationale for the selected templates such as matched fingerprint motifs; (viii) an embedded NGL viewer displaying the TMH homology model(s) of the query GPCR based on the template suggestions; (ix) a link to the SL2 predictions for the intra- and extracellular loops (see below for more details); (x) a Ramachandran plot of each generated TMH model and (xi) links to UniProt and the GPCRdb. Files containing the sequence alignments, modeled PDB structure(s), Ramachandran plot(s) and loop sequences are made available for download.
SuperLooper2 results page
The NGL viewer is embedded in this page allowing the visual inspection of the loop predictions by SL2. For each of the intracellular loops (ICLs) and ECLs, the five top-scoring predictions are listed with the top hit for all loops automatically being loaded into the gap of the protein model. Alternative loop conformations can be selected from the drop-down menu. For each candidate, the score, the RCSB PDB entry-code and sequence of the template protein, the number of clashes and the sequence identity between target and template are listed in a table. If no suitable loop candidate was found, the gap remains open. The completed structure (initial serpentine domain model plus selected loops) can be downloaded by clicking the download button. Side chains of the loops are included but require further optimization to select the most appropriate rotamers. Clashes between sidechains in the loops and the TMHs can be eliminated by carrying out energy minimization on the entire structure using, for example, the ModRefiner web server (30). Likewise, where the conserved cysteines in TMH3 and ECL2 are present in the sequence but not in the model, minimization may shift their side chain orientations to allow the conserved disulphide bridge to form (functionality to deal specifically with this issue will be implemented in the next web server update).
Submitting a modeling job to GPCR-SSFE 2.0
Where GPCR-SSFE 2.0 does not store modeling results for a particular Class A GPCR, for example species not stored in the database or newly identified orphan GPCRs, users can submit their GPCR sequence to GPCR-SSFE 2.0 for analysis and homology model building. For such cases, users should navigate to the ‘RUN’ webpage and enter their GPCR sequence (by either uploading a file or by copying and pasting it) and email address (optional). Upon completion of homology modeling (taking ∼5 min for the TMHs and a further 5–10 min for the loop modeling) the results page is displayed (and where an email address was provided, a web-link is emailed to the user). Results are stored on the server for 7 days. The results page looks exactly like those retrieved when searching the database of pre-calculated models.
Performance
Since completion of the large-scale modeling and extension of GPCR-SSFE 2.0, several new non-active Class A GPCR structures have been published recently such as: human endothelin type B (ETB) receptor; PDB ID: 5GLI (31) and human CC Chemokine receptor 2 (CCR2); PDB ID: 5T1A (32). These receptor structures therefore provide us with an ideal means of assessing the performance of the models produced by GPCR-SSFE 2.0 as they were not included in the pool of structural templates used by GPCR-SSFE 2.0 and we can calculate the root-mean-square deviation (RMSD) of our predicted models to the crystal structures.
Two Chemokine structures have already been published and are included in our pool of templates (CCR5; PDB ID: 4MBS and CXCR4; PDB ID: 4ODU), therefore template selection and homology modeling of CCR2 should be relatively straightforward. Indeed, for CCR2 we find that GPCR-SSFE 2.0, GPCR-ModSim and GPCR-I-TASSER all select a chemokine crystal structure as the best template for homology modeling, resulting in comparable model accuracies for both the transmembrane and full length models (Table 1).
Table 1. The RMSD of ccr2_human and ednrb_human homology models compared to their crystal structures.
| Method of template selection and modeling | Accuracy (RMSD) | |||
|---|---|---|---|---|
| CCR2_HUMAN1 | EDNRB_HUMAN2 | |||
| GPCR-SSFE 2.0 | 0.72 Å | (0.89 Å) | 1.30 Å | (2.33 Å) |
| GPCRM (Rosetta optimized) | 0.72 Å | (0.82 Å) | 1.60 Å | (2.36 Å) |
| Max %sequence similarity per helix + Modeller | 0.72 Å | 1.62 Å | ||
| GPCR-I-TASSER HGmod (2014) | 0.73 Å | (0.83 Å) | 1.64 Å | (1.91 Å) |
| GoMoDo | 1.53 Å | (1.72 Å) | 1.78 Å | (2.01 Å) |
| GPCR-ModSim | 0.78 Å | (0.92 Å) | 1.97 Å | (2.36 Å) |
| GPCR-SSFE 1.0 | 1.87 Å | 2.01 Å | ||
1RMSD between 5T1A and ccr2_human model in TMH region (left) and full length (brackets).
2RMSD between 5GLI and ednrb_human model in TMH region (left) and full length (brackets).
The ETB receptor's closest homolog is the κ-type opioid receptor (PDB ID: 4DJH) with 75% sequence similarity. The multiple fragment-based approach utilized by GPCR-SSFE 2.0 produced the most accurate model for the TMHs compared to other state-of-the-art modeling servers tested using default settings, all of which select a single template for homology modeling (Table 1). Models produced using the highest sequence similarity template per helix have similar resolutions to the second best scoring server, GPCRM. However, when loops are included in the RMSD calculations for the ETB receptor, GPCR-I-TASSER and GoMoDo scored better (Table 1). For assessing the performance of GPCR-SSFE 2.0 in building full length models, we used the first suggested loop for each of the ICLs and ECLs for calculating the RMSD (excluding loops from the actual crystal structure of the tested protein). Nevertheless, a particular strength of the loop modeling procedure implemented in GPCR-SSFE 2.0 is that it is the first multi-template based service which allows loops to be selected interactively. Thus, utilizing their expert knowledge, experimental data or similarity to crystallized GPCR structures, the RMSD values for models built by GPCR-SSFE 2.0 may be improved depending on the loops selected by the user.
An advantage of our fingerprint-based method is that sequence differences causing slight backbone changes such as bulges or kinks are considered in more detail, which will be discussed further in the following section.
Example
A case study that demonstrates the utility of our fragment-based approach to homology modeling involves the thyroid-stimulating hormone receptor (TSHR). A distinct feature of TSHR compared to other Class A GPCRs is that in TMH5 it lacks the highly conserved proline in position 5×50 (modified Ballesteros–Weinstein nomenclature that considers the structural alignment of bulges (33)) instead having an alanine (Ala593) at this position. GPCR crystal structures that have this conserved proline have a bulged TMH5 conformation causing a kink and twist toward the extracellular end of the helix whereas those having a different amino acid at this position have a regular α-helical TMH5 e.g. the Sphingosine 1-phosphate receptor 1 (alanine in position 5×50; PDB ID: 3V2W (34)), the P2Y12 receptor (asparagine in position 5×50; PDB ID: 4NTJ (35)) and the Lysophosphatidic-Acid-Receptor 1 (threonine in position 5×50, LPAR1, PDB ID: 4Z34 (36)). Mutagenesis studies have suggested that the alanine at position 5×50 in TSHR most likely also causes a regular α-helix conformation in this receptor (37,38).
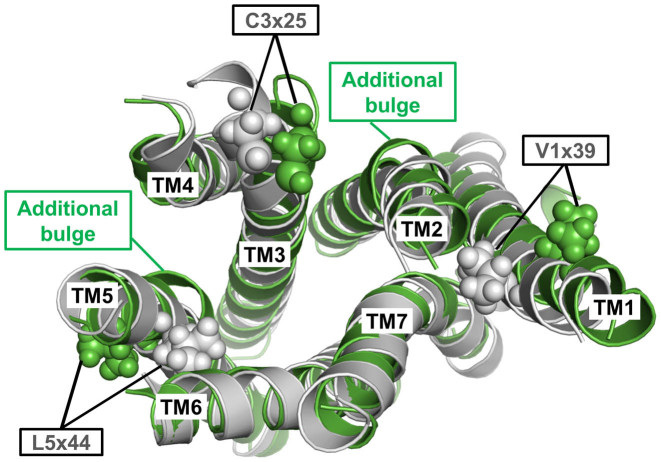
The fragment-based inactive TMH model of TSHR built by GPCR-SSFE 2.0 uses 6 of the 27 different template structures for model building (Supplementary Data, Table S1). Figure 4 shows a comparison between this multiple-template fragment model (shown in gray) with the best matching single template TSHR model (shown in green) based on the β2 adrenergic receptor ADRB2 (PDB ID: 2RH1 (39)) with an overall sequence identity of 24%. Comparison of the two model structures reveals that the single template model has additional bulges in TMH2 and TMH5 and clearly shows different side chain orientations of the highly conserved cysteine in TMH3 as well as Val421 (position 1×39) and Leu587 (position 5×44) (Figure 4). Conservative hydrophobic substitution to isoleucine and valine at these respective positions lead to constitutive activation of TSHR (40) inferring that these side chains are unlikely to be orientated toward the membrane as is the case with the single template model. Thus, the activating roles of these mutations are better rationalized by the structural data when these side chains point toward neighboring helices as seen in the fragment-based multiple template TSHR model (Figure 4). This clearly demonstrates the advantage of using a multiple-template fragment-based approach in achieving an improved accuracy over single template based methods or those based on sequence similarity alone.
Figure 4.
Structural superposition of the multiple-template fragment-based model (gray) with the best matching single template TSHR serpentine domain model based on the β2 adrenergic receptor (PDB ID: 2RH1) (green). The latter model has additional bulges in TMH2 and 5 and not only has shifted locations of the highly conserved cysteine in TMH3 but clearly has different orientations of the side chains V421 (position 1×39) and L587 (position 5×44). Constitutively activating mutations by slight hydrophobic alterations (V421I; l587V) (40) of these two positions are better explained when these side chains point toward neighboring helices as in the fragment-based model (gray) but are incompatible with them being orientated toward the membrane as observed in the single template TSHR model (green).
Documentation
To aid usability of the web server, there is both a facts and questions page as well as a tutorials page, linking to various tutorial videos that are available on our YouTube channel.
CONCLUSIONS
GPCR-SSFE 2.0 is a web server dedicated to template selection and homology modeling of GPCRs, which has been updated to include the latest structural data and extended with new components. It is user-friendly and allows non-expert users to access 3D structural data that might otherwise be unattainable; GPCR-SSFE 2.0 stores and makes freely available, a comprehensive and up-to-date set of pre-calculated homology models of human, mouse and rat GPCRs. Template selection is done individually for each of the 7TMHs and H8, flexibly allowing for both single template models and fragment-models to be built, depending on the similarity of a query sequence to the available templates. It uses a database of sequence fingerprint features correlating with observed structural features in the templates to guide template selection. This allows sequence differences causing slight backbone changes such as bulges or differently oriented kinks to be considered in more detail. Loop modeling results are now provided using SL2, with the advantage of user-guided loop selection allowing their knowledge-based input to drive the selection process, a limitation of automatic methods of loop modeling. Up-to-date visualization is provided by the NGL viewer. We demonstrate that the models produced by GPCR-SSFE 2.0 are as good or better than other published methods with the benefit of being able to capture structural features that sequence similarity alone is unable to do. In summary, compared to other approaches the fingerprint-driven fragment approach used by GPCR-SSFE 2.0 can achieve an improved accuracy in the predicted TMH regions, which is essential for in silico ligand docking and virtual screening.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank members of the GLISTEN Cost action (CM1207) for helpful discussions.
Footnotes
Present address: Franziska Kreuchwig, Cardiovascular and Metabolic Sciences, Max Delbrück Center of Molecular Medicine, 13125 Berlin, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) [KR 1273/4-2 to G.K., DFG HI 1502/1-2, BI 893/8 to P.W.H]. The publication of this article was funded by the Open Access Fund of the Leibniz Association.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bjarnadóttir T.K., Gloriam D.E., Hellstrand S.H., Kristiansson H., Fredriksson R., Schiöth H.B.. Comprehensive repertoire and phylogenetic analysis of the G protein-coupled receptors in human and mouse. Genomics. 2006; 88:263–273. [DOI] [PubMed] [Google Scholar]
- 2. Fredriksson R., Lagerström M.C., Lundin L.-G., Schiöth H.B.. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003; 63:1256–1272. [DOI] [PubMed] [Google Scholar]
- 3. Overington J.P., Al-Lazikani B., Hopkins A.L.. How many drug targets are there. Nat. Rev. Drug Discov. 2006; 5:993–996. [DOI] [PubMed] [Google Scholar]
- 4. Venkatakrishnan A.J., Deupi X., Lebon G., Tate C.G., Schertler G.F., Babu M.M.. Molecular signatures of G-protein-coupled receptors. Nature. 2013; 494:185–194. [DOI] [PubMed] [Google Scholar]
- 5. Latek D., Bajda M., Filipek S.. A hybrid approach to structure and function modeling of G protein-coupled receptors. J. Chem. Inf. Model. 2016; 56:630–641. [DOI] [PubMed] [Google Scholar]
- 6. Sandal M., Duy T.P., Cona M., Zung H., Carloni P., Musiani F., Giorgetti A., Overington J., Al-Lazikani B., Hopkins A. et al. GOMoDo: a GPCRs online modeling and docking webserver. PLoS One. 2013; 8:e74092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Esguerra M., Siretskiy A., Bello X., Sallander J., Gutiérrez-de-Terán H.. GPCR-ModSim: a comprehensive web based solution for modeling G-protein coupled receptors. Nucleic Acids Res. 2016; 44:W455–W462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Worth C.L., Kreuchwig A., Kleinau G., Krause G.. GPCR-SSFE: a comprehensive database of G-protein-coupled receptor template predictions and homology models. BMC Bioinform. 2011; 12:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang J., Yang J., Jang R., Zhang Y.. GPCR-I-TASSER: a hybrid approach to G protein-coupled receptor structure modeling and the application to the human genome. Structure. 2015; 23:1538–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Worth C.L., Kleinau G., Krause G.. Comparative sequence and structural analyses of G-protein-coupled receptor crystal structures and implications for molecular models. PLoS One. 2009; 4:e7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Engelstoft M.S., Norn C., Hauge M., Holliday N.D., Elster L., Lehmann J., Jones R.M., Frimurer T.M., Schwartz T.W.. Structural basis for constitutive activity and agonist-induced activation of the enteroendocrine fat sensor GPR119. Br. J. Pharmacol. 2014; 171:5774–5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Advani P., Joseph B., Ambre P., Pissurlenkar R., Khedkar V., Iyer K., Gabhe S., Iyer R.P., Coutinho E.. In silico optimization of pharmacokinetic properties and receptor binding affinity simultaneously: a ‘parallel progression approach to drug design’ applied to β-blockers. J. Biomol. Struct. Dyn. 2016; 34:384–398. [DOI] [PubMed] [Google Scholar]
- 13. Isberg V., Mordalski S., Munk C., Rataj K., Harpsøe K., Hauser A.S., Vroling B., Bojarski A.J., Vriend G., Gloriam D.E.. GPCRdb: an information system for G protein-coupled receptors. Nucleic Acids Res. 2016; 44:D356–D364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ismer J., Rose A.S., Tiemann J.K.S., Goede A., Preissner R., Hildebrand P.W.. SL2: an interactive webtool for modeling of missing segments in proteins. Nucleic Acids Res. 2016; 44:W390–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rose A.S., Hildebrand P.W.. NGL Viewer: a web application for molecular visualization. Nucleic Acids Res. 2015; 43:W576–W579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rose A.S., Bradley A.R., Valasatava Y., Duarte J.M., Prlić A., Rose P.W.. Web-based molecular graphics for large complexes. Proceedings of the 21st International Conference on Web3D Technology - Web3D’ 16. 2016; NY: ACM Press; 185–186. [Google Scholar]
- 17. Cock P.J.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009; 25:1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eddy S.R., Altschul S., Gish W., Miller W., Myers E., Lipman D., Altschul S., Madden T., Schaffer A., Zhang J. et al. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011; 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Šali A., Blundell T.L.. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993; 234:779–815. [DOI] [PubMed] [Google Scholar]
- 20. Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M.. and IUCr (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291. [Google Scholar]
- 21. Ballesteros J.A., Weinstein H.. [19]Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995; 25:366–428. [Google Scholar]
- 22. Deupi X. Quantification of structural distortions in the transmembrane helices of GPCRs. Methods Mol. Biol. 2012; 914:219–235. [DOI] [PubMed] [Google Scholar]
- 23. Devillé J., Rey J., Chabbert M.. An indel in transmembrane helix 2 helps to trace the molecular evolution of Class A G-protein-coupled receptors. J. Mol. Evol. 2009; 68:475–489. [DOI] [PubMed] [Google Scholar]
- 24. Deupi X., Olivella M., Govaerts C., Ballesteros J.A., Campillo M., Pardo L.. Ser and Thr residues modulate the conformation of pro-kinked transmembrane a-helices. Biophys. J. 2004; 86:105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deupi X., Olivella M., Sanz A., Dölker N., Campillo M., Pardo L.. Influence of the gÀ conformation of Ser and Thr on the structure of transmembrane helices. J. Struct. Biol. 2009; 169:116–123. [DOI] [PubMed] [Google Scholar]
- 26. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shen M., Sali A.. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006; 15:2507–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hildebrand P.W., Goede A., Bauer R.A., Gruening B., Ismer J., Michalsky E., Preissner R.. SuperLooper–a prediction server for the modeling of loops in globular and membrane proteins. Nucleic Acids Res. 2009; 37:W571–W574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. UniProt: a hub for protein information Nucleic Acids Res. 2015; 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu D., Zhang Y.. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys. J. 2011; 101:2525–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shihoya W., Nishizawa T., Okuta A., Tani K., Dohmae N., Fujiyoshi Y., Nureki O., Doi T.. Activation mechanism of endothelin ETB receptor by endothelin-1. Nature. 2016; 537:363–368. [DOI] [PubMed] [Google Scholar]
- 32. Zheng Y., Qin L., Zacarías N.V.O., de Vries H., Han G.W., Gustavsson M., Dabros M., Zhao C., Cherney R.J., Carter P. et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature. 2016; 540:458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Isberg V., de Graaf C., Bortolato A., Cherezov V., Katritch V., Marshall F.H., Mordalski S., Pin J.-P., Stevens R.C., Vriend G. et al. Generic GPCR residue numbers - aligning topology maps while minding the gaps. Trends Pharmacol. Sci. 2015; 36:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hanson M.A., Roth C.B., Jo E., Griffith M.T., Scott F.L., Reinhart G., Desale H., Clemons B., Cahalan S.M., Schuerer S.C. et al. Crystal structure of a lipid G protein-coupled receptor. Science. 2012; 335:851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang K., Zhang J., Gao Z.-G., Zhang D., Zhu L., Han G.W., Moss S.M., Paoletta S., Kiselev E., Lu W. et al. Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature. 2014; 509:115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chrencik J.E., Roth C.B., Terakado M., Kurata H., Omi R., Kihara Y., Warshaviak D., Nakade S., Asmar-Rovira G., Mileni M. et al. Crystal structure of antagonist bound human lysophosphatidic acid receptor 1. Cell. 2015; 161:1633–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kleinau G., Hoyer I., Kreuchwig A., Haas A.-K., Rutz C., Furkert J., Worth C.L., Krause G., Schülein R.. From molecular details of the interplay between transmembrane helices of the thyrotropin receptor to general aspects of signal transduction in family a G-protein-coupled receptors (GPCRs). J. Biol. Chem. 2011; 286:25859–25871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chantreau V., Taddese B., Munier M., Gourdin L., Henrion D., Rodien P., Chabbert M., Vassart G., Dumont J., Vassart G. et al. Molecular insights into the transmembrane domain of the thyrotropin receptor. PLoS One. 2015; 10:e0142250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rasmussen S.G.F., Choi H.-J., Rosenbaum D.M., Kobilka T.S., Thian F.S., Edwards P.C., Burghammer M., Ratnala V.R.P., Sanishvili R., Fischetti R.F. et al. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature. 2007; 450:383–387. [DOI] [PubMed] [Google Scholar]
- 40. Kleinau G., Haas A.-K., Neumann S., Worth C.L., Hoyer I., Furkert J., Rutz C., Gershengorn M.C., Schulein R., Krause G.. Signaling-sensitive amino acids surround the allosteric ligand binding site of the thyrotropin receptor. FASEB J. 2010; 24:2347–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.