Abstract
There is a continuous interest in increasing proteins stability to enhance their usability in numerous biomedical and biotechnological applications. A number of in silico tools for the prediction of the effect of mutations on protein stability have been developed recently. However, only single-point mutations with a small effect on protein stability are typically predicted with the existing tools and have to be followed by laborious protein expression, purification, and characterization. Here, we present FireProt, a web server for the automated design of multiple-point thermostable mutant proteins that combines structural and evolutionary information in its calculation core. FireProt utilizes sixteen tools and three protein engineering strategies for making reliable protein designs. The server is complemented with interactive, easy-to-use interface that allows users to directly analyze and optionally modify designed thermostable mutants. FireProt is freely available at http://loschmidt.chemi.muni.cz/fireprot.
INTRODUCTION
Proteins are widely used in numerous biomedical and biotechnological applications. However, naturally occurring proteins cannot usually withstand the harsh industrial environment, since they are mostly evolved to function at mild conditions (1). Protein engineering has revolutionized the utilization of naturally available proteins for different industrial applications by improving various protein features such as stability, activity or enantioselectivity to surpass their natural limitations. Protein stability is generally strongly correlated with its expression yield (2), half-life (3), serum survival time (4) and performance in the presence of denaturing agents (5). Thus, stability is one of the key determinants of proteins applicability in biotechnological processes.
In the ideal case, the saturation mutagenesis would be applied to evaluate every possible mutation on every position of the engineered protein (6). However, such a search space would be enormous and the experimental evaluation can delay the design of truly thermostable protein for months or even years. Therefore, there are demands for effective and precise predictive computation of protein stability. To satisfy this goal a number of in silico tools have been developed recently. Some of these tools such as EASE-MM (7), I-Mutant (8) or mCSM (9) are based on machine learning techniques. Others are using so-called energetic functions. These programs can be further categorized into two groups. The first group utilizes a physical effective energy function for simulating the fundamental forces between atoms and is represented by the programs like Rosetta (10) and Eris (11). The second group is based on statistical potentials for which the energies are derived from frequencies of residues or atom contacts reported in the datasets of experimentally characterized protein mutants, e.g. PopMuSiC (12) and FoldX (13). However, due to the potentially antagonistic effect of mutations, only single-point mutations are usually predicted in silico and have to be followed by laborious and costly protein expression, purification and characterization. Single-point mutations typically enhance the melting temperature of target proteins by units of degree (3,14). A much higher degree of stabilization can be achieved by constructing multiple-point mutants (15). We have recently developed the FireProt (16), combining energy- and evolution-based approaches for reliable design of stable multiple-point mutants. The protocol includes several preceding filters that accelerate the calculation by omitting potentially deleterious mutations. FireProt is currently available only in a stand-alone format and requires extensive experience in bioinformatics to carry out all necessary steps of the work flow. Currently, we are aware of only one server for design of stable multiple-point mutants - PROSS (17), utilizing Rosetta modeling and phylogenetic sequence information in its computation core.
Here, we present a web version of FireProt for the automated design of thermostable proteins. FireProt integrates sixteen computational tools and utilizes both sequence and structural information. FireProt web server provides users with thermostable proteins, constructed by three distinct strategies: (i) evolution-based approach, utilizing back-to-consensus analysis; (ii) energy-based approach, evaluating change in free energy upon mutation and (iii) combination of both evolution-based and energy-based approaches. In our view, it is very important to have this integrated approach, since phylogenetic analysis enables identification of the mutations stabilized by entropy, which cannot be predicted by force field calculations (Beerens et al., under review). The server allows users to include preferred mutations into the thermostable protein, to generate corresponding structures and sequences for gene syntheses. Compared to the previously published FireProt protocol (16), minimum effort and no bioinformatics knowledge is required from users to calculate and analyze the results. Furthermore, all input parameters and computational protocols were optimized to minimize otherwise highly time demanding procedure. The server was complemented with a graphical interface allowing users to directly analyze the protein of interest and design multiple-point mutants.
MATERIALS AND METHODS
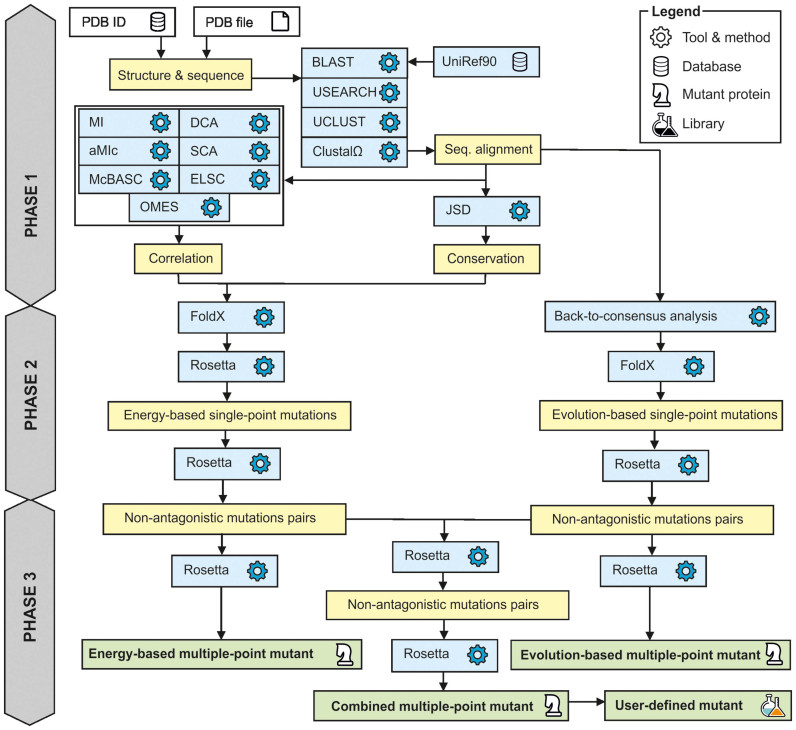
The basic workflow of FireProt strategy is outlined in Figure 1. In order to design a highly reliable thermostable multiple-point mutant, a protein defined by the user is annotated using several prediction tools and databases (Phase 1). With this knowledge in hand, energy- and evolution-based approach is applied to assemble a list of potentially stabilizing single-point mutations (Phase 2). Finally, three multiple-point mutants are generated in an additive manner, while removing potentially antagonistic effects of mutations (Phase 3).
Figure 1.
Workflow of FireProt strategy.
Phase 1: Annotation of the protein
Initially, the user is requested to specify the protein structure, either by providing its PDB ID or by uploading a user-defined PDB file. The biological assembly of the target protein is then automatically generated by the MakeMultimer tool (http://watcut.uwaterloo.ca/tools/makemultimer/). Sequence homologs are obtained by performing a BLAST search (18) against the UniRef90 database (19), using the target protein sequence as an input query. Identified homologs are then aligned with the query protein using USEARCH (20), while sequences whose identity with the query is below or above the user defined thresholds (default: 30 and 90%) are excluded from the list of homologs. The remaining sequences are clustered using UCLUST (20), with a 90% identity threshold to remove close homologs. The cluster representatives are sorted based on the BLAST query coverage and by default, the first 200 of them are used to create a multiple sequence alignment with Clustal Omega tool (21). The multiple sequence alignment is used to: (i) estimate the conservation coefficient of each residue position in the protein based on the Jensen–Shannon entropy (22); (ii) identify correlated positions employing a consensual decision of the OMES (23), MI (24), aMIc (25), DCA (26), SCA (27), ELSC (28), McBASC (29) and (iii) analyze amino acid frequencies at individual positions within the protein.
Phase 2: Prediction of single-point mutations
In accordance with the original FireProt protocol, potentially stabilizing single-point mutations are identified via two separate branches: one relying on the estimation of the change of free energy upon mutation and second utilizing back-to-consensus approach.
The first, energy-based approach is employing FoldX and Rosetta tools that performed best on our testing dataset. Preceding filters accelerate the calculation by omitting potentially deleterious mutations. Prior to the identification of the single-point mutations itself, the target protein structure is amended and minimized. FoldX protocol is utilized to fill in the missing atoms in the residues and patched structure is consequently minimized with Rosetta minimization module. Conserved and correlated positions are immediately excluded from further analysis. It was observed that functional and structural constraints in proteins generally lead to the conservation of amino acid residues (30–33). Similarly, correlated residues ordinarily help to maintain protein function, folding or stability (34–36). Mutations conducted on these positions are therefore considered unsafe by current FireProt strategy, even though there is certainly a space for more sophisticated treatment of correlated positions, which will be further developed in future versions of FireProt server.
The remaining positions are subjected to saturation mutagenesis by using FoldX tool. Mutations with predicted ddG over given threshold (default: –1 kcal/mol) are steered away and rest is forwarded to Rosetta calculations. Finally, the mutations predicted by Rosetta as strongly stabilizing (default cut-off: –1 kcal/mol) are tagged as potential candidates for the design of the multiple-point mutants.
A high time demands of Rosetta analysis were one of the most excruciating issues with the original FireProt protocol. Even with the application of filters over 100 mutations was usually left for precise, but slow, Rosetta calculations. For this reason, we have evaluated several force fields and Rosetta protocols with the newly assembled dataset containing 1573 mutations from ProTherm database (37) and HotMuSiC dataset (38). Based on the results of the evaluations, the best trade-off between the time requirements and precision was selected. With Rosetta protocol 3, we have achieved more than tenfold increase in calculation speed while preserving high prediction accuracy. Details on dataset construction and protocols evaluation can be found in the Supplement 1 (Supplementary Tables S1–S5).
The second approach is based on the information obtained from multiple sequence alignment. The most common amino acid in each position of protein sequence often provides a non-negligible effect on protein stability (39–42). Therefore, FireProt implements majority and frequency ratio approach to identify mutations at positions where the wild-type amino acid differs from the most prevalent one. By default, the single out mutations are located in the positions where the consensus residue is present in at least 50% of all analyzed sequences (majority method) or where consensus residue frequency is 40% and is at least five times more frequent than the wild-type amino acid (frequency ratio method). These thresholds were chosen in accordance to the previously published HotSpot Wizard method (43). Selected mutations are evaluated by FoldX and the stabilizing ones are listed as candidate mutations for the engineering of multiple-point mutant.
Phase 3: Design of thermostable protein
In total, three protein designs are provided by FireProt strategy. The first design includes only the mutations from energy-based approach, the second contains the mutations suggested by the evolution-based approach and the third is the combination of both. Naturally, because of potentially antagonistic effects between individual mutations, we cannot combine individual mutations blindly.
To avoid possible clashes, FireProt strategy is trying to minimize antagonistic effects by utilizing Rosetta. In the first step, all pairs of single-point mutations within the range of 10 Å are evaluated separately for energy- and evolution-based approach. Once change in free energy is obtained for all residue pairs, FireProt starts to introduce them into the multiple-point mutant in the order based on their predicted stability, excluding the mutations that are colliding with already included mutations. Algorithm stops once there are no mutations left or the stabilizing effect of analyzed pair drops below defined threshold.
Upon the completion of previous step, procedure is repeated this time considering only the pairs between the mutations chosen for the construction of energy- and evolution-based mutants. Finally, structures of all three mutants are modeled using the Rosetta protocol 16.
DESCRIPTION OF THE WEB SERVER
Input
The only required input to the web server is a tertiary structure of the protein of interest, provided either as a PDB ID or a user-defined PDB file. The user can then choose a predefined biological unit generated by the MakeMultimer tool or manually select chains for which the calculation should be performed. The calculations can be configured in either basic or advanced mode.
In the basic mode, user is allowed to change the setting of BLAST search and alignment construction. The advanced mode expands the list of modifiable parameters by the ones connected with: (i) the identification of consensus residues by majority and frequency ratio approach, (ii) the thresholds used by FoldX and Rosetta prediction tools and (iii) the decision threshold employed in the consensual analysis of correlated positions. Advanced mode allows expert users to fine-tune the parameters of calculation according to studied systems. However, the presented default values are optimized to provide reliable results for most of the systems and we therefore do not advice their change in the general scenarios.
Output
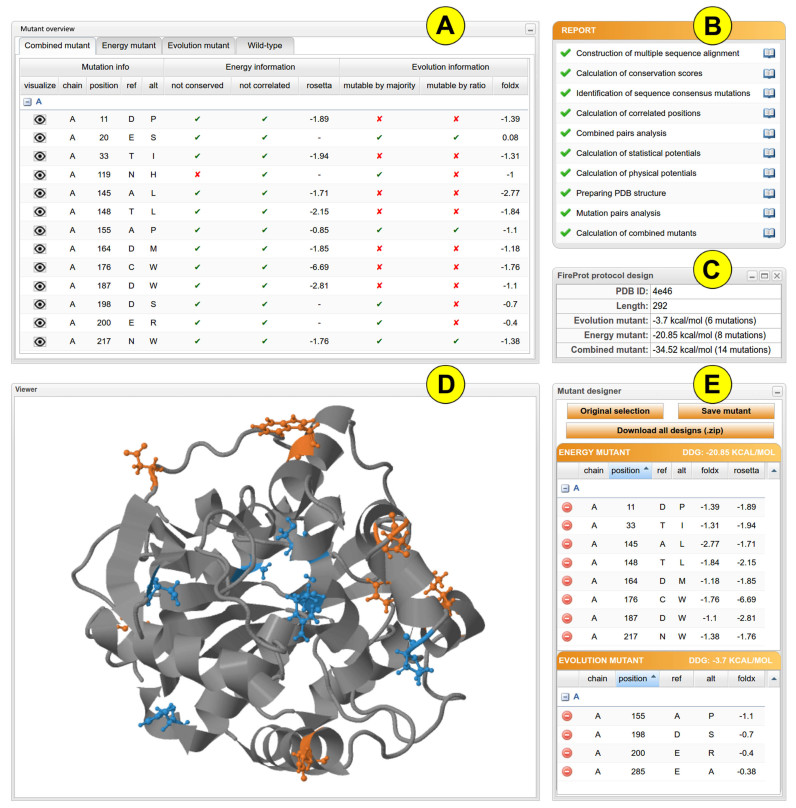
Upon submission, a unique identifier is assigned to each job to track the calculation and the ‘Results browser’ informs the user about the status of the individual steps in the FireProt workflow (Figure 2B). Once the job is finished, users can either directly download the results in the .zip archive or navigate themselves into the ‘Results page’ for further analysis. The ‘Results page’ is intuitively organized into several panels as described below.
Figure 2.
FireProt's graphical user interface showing the results obtained for the haloalkane dehalogenase DhaA (PDB ID: 4e46). (A) The ‘Mutant overview’ panel provides a list of mutations introduced into protein structure. (B) The ‘Report’ panel shows the status of calculation in the individual steps of the computational pipeline. (C) The ‘Protocol design’ panel provides general information about FireProt designs. (D) The JSmol ΄Viewer΄ allows interactive visualization of the protein. (E) The ‘Mutant designer’ panel enables manual adjustment of a new combined mutant.
Protein visualization
The wild-type and the mutant structure is interactively visualized in the web browser (Figure 2D) utilizing the Jsmol applet (http://wiki.jmol.org/index.php/JSmol). Users can switch between different protein visualization styles and also highlight selected amino acids in the protein structure. Residues that were included into energy-based mutant are colored in orange, evolution-based mutations are in blue and all other residues are in gray. User selected residues that were not part of any mutant are underlined in red.
Mutant overview
The ‘Mutant overview’ panel is organized into four tabs (Figure 2A). The first three tabs provide information about mutations included into combined, energy-based and evolution-based mutant. The checkbox, allowing users to visualize the chosen residues in Jsmol applet, can be found in each row together with all data relevant for a given computational approach. The last tab contains the list of all residues in the wild-type structure. While ‘wild-type’ tab is active, the wild-type structure is visualized in Jsmol applet instead of the mutated one and the user is allowed to introduce user-defined mutations into multiple-point mutant via the ‘plus’ icon in the last column.
General information
The ‘FireProt protocol design’ panel provides users with general information about the target protein and the designs constructed by FireProt strategy, such as a number of mutations and estimated change in free energy (Figure 2C).
Mutant designer
The ‘Mutant designer’ panel allows the user to design own multiple-point mutant by managing mutations divided into energy- and evolution-based subset. If all mutations in the subset have their predicted energy values assigned, a total change in Gibbs free energy is immediately estimated assuming simple additivity. Users can also generate an amino acid sequence from the designed multiple-point mutant that combines mutations included into energy- and evolution-based subsets. All prepared designs can be downloaded in one .zip archive (Figure 2E).
EXPERIMENTAL VALIDATION
The original FireProt strategy was experimentally verified with three proteins (haloalkane dehalogenase DhaA, PDB ID 4E46; γ-hexachlorocyclohexane dehydrochlorinase LinA, PDB ID 3A76; and fibroblast growth factor 2, PDB ID 4OEE) and provided respective stabilization of proteins ΔTm = 25, 21 and 15°C (Table 1). The original protocol was modified to enable fully automated calculation at the reasonable time, while maintaining high prediction accuracy (Supplementary Table S6). Prediction of eight multiple-point mutants using this modified protocol was validated using the data of FRESCO (44) and identified mutations were compared with another online protein stabilization tool PROSS (17). FireProt and PROSS showed similar predictive power, correctly identifying 29 and 20 potentially stabilizing positions, respectively (Supplementary Table S7).
Table 1. Experimental validation of FireProt strategy.
| Protein | Energy-based mutations | Evolution-based mutations | ΔTm [°C] |
|---|---|---|---|
| PDB ID | |||
| 4E46 | 8 | 3 | +25 |
| 3A76 | 4 | 3 | +21 |
| 4OEE | 4 | 2 | +15 |
CONCLUSIONS AND OUTLOOK
FireProt is a web server that provides users with a one-stop-shop solution for the design of thermostable multiple-point mutant proteins. In comparison with the standalone FireProt strategy (16), all default parameters and computational protocols were optimized to increase the calculation speed, while maintaining the prediction accuracy. The designs produced by the FireProt workflow were experimentally verified and thus users can obtain highly reliable thermostable proteins with minimal experimental effort. The server is complemented by an easy-to-use graphical interface that allows users to interactively analyze individual mutations selected as a part of energy- or evolution-based approach together with the ability to design their own multiple-point mutants on top of our robust strategy.
The automation of the whole procedure makes the process of the design of thermostable proteins accessible to users without any prior expertise in bioinformatics since it eliminates the need to select, install and evaluate tools, optimize their parameters, and interpret intermediate results. However, the energy-based approach of the FireProt strategy depends on the quality of provided protein structure and therefore the prediction accuracy might be compromised in the case of low-resolution structures or homology models.
In the future, we plan to implement new strategies such as a design based on the analysis of correlated positions that would contribute to the construction of the final combined mutant, elimination of highly flexible regions and introduction of disulfide bridges. Also, we plan to equip FireProt with several new filters, e.g. exclusion of the amino acids located in the close neighborhoods of the active sites or the ones participating in oligomerization.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the developers of PROSS Adi Goldenzweig and Dr Sarel Fleishman (Weizmann Institute of Science, Israel) for providing the data set for validation purposes. Computational resources were supplied by the Ministry of Education, Youth and Sports of the Czech Republic under the Projects CESNET (Project No. LM2015042) and CERIT-Scientific Cloud (Project No. LM2015085) provided within the program Projects of Large Research, Development and Innovations Infrastructures.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This research and open access charge was funded by Ministry of Education, Youth and Sports of the Czech Republic from the National Programme of Sustainability II [LQ1605, LO1214]; European Regional Development Fund [ELIXIR-CZ LM2015047]; Grant Academy of the Czech Republic [16-06096S]; Brno University Technology [FIT-S-17-3964].
Conflict of interest statement. None declared.
REFERENCES
- 1. Modarres H.P., Mofrad M.R., Sanati-Nezhad A.. Protein thermostability engineering. RSC Adv. 2016; 6:115252–115270. [Google Scholar]
- 2. Ferdjani S., Ionita M., Roy B., Dion M., Djeghaba Z., Rabiller C., Tellier C.. Correlation between thermostability and stability of glycosidases in ionic liquid. Biotechnol. Lett. 2011; 33:1215–1219. [DOI] [PubMed] [Google Scholar]
- 3. Wijma H.J., Floor R.J., Janssen D.B.. Structure- and sequence-analysis inspired engineering of proteins for enhanced thermostability. Curr. Opin. Struct. Biol. 2013; 23:588–594. [DOI] [PubMed] [Google Scholar]
- 4. Gao D., Narasimhan D.L., Macdonald J., Brim R., Ko M.C., Landry D.W., Woods J.H., Sunahara R.K., Zhan C.G.. Thermostable variants of cocaine esterase for long-time protection against cocaine toxicity. Mol. Pharmacol. 2009; 75:318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Polizzi K.M., Bommarius A.S., Broering J.M., Chaparro-Riggers J.F.. Stability of biocatalysts. Curr. Opin. Chem. Biol. 2007; 11:220–225. [DOI] [PubMed] [Google Scholar]
- 6. Gray K.A., Richardson T.H., Kretz K., Short J.M., Bartnek F., Knowles R., Kan L., Swanson P.E., Robertson D.E.. Rapid evolution of reversible denaturation and elevated melting temperature in a microbial haloalkane dehalogenase. Adv. Synth. Catal. 2001; 343:607–617. [Google Scholar]
- 7. Folkman L., Stantic B., Sattar A., Zhou Y.. EASE-MM: sequence-based prediction of mutation-induced stability changes with feature-based multiple models. J. Mol. Biol. 2016; 428:1394–1405. [DOI] [PubMed] [Google Scholar]
- 8. Capriotti E., Fariselli P., Casadio R.. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005; W306–W310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pires D.E., Ascher D.B., Blundell T.L.. mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics. 2014; 30:335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kellogg E.H., Leaver-Fay A., Baker D.. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins. 2011; 79:830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yin S., Ding F., Dokholyan N.V.. Modeling backbone flexibility improves protein stability estimation. Structure. 2007; 15:1567–1576. [DOI] [PubMed] [Google Scholar]
- 12. Dehouck Y., Grosfils A., Folch B., Gilis D., Bogaerts P., Rooman M.. Fast and accurate predictions of protein stability changes upon mutations using statistical potentials and neural networks: PoPMuSiC-2.0. Bioinformatics. 2009; 25:2537–2543. [DOI] [PubMed] [Google Scholar]
- 13. Guerois R., Nielsen J.E., Serrano L.. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 2002; 320:369–387. [DOI] [PubMed] [Google Scholar]
- 14. Gumulya Y., Reetz M.T.. Enhancing the thermal robustness of an enzyme by directed evolution: least favorable starting points and inferior mutants can map superior evolutionary pathways. ChemBioChem. 2011; 12:2502–2510. [DOI] [PubMed] [Google Scholar]
- 15. Bommarius A.S., Paye M.F.. Stabilizing biocatalysts. Chem. Soc. Rev. 2013; 42:6534–6565. [DOI] [PubMed] [Google Scholar]
- 16. Bednar D., Beerens K., Sebestova E., Bendl J., Khare S., Chaloupkova R., Prokop Z., Brezovsky J., Baker D., Damborsky J.. FireProt: energy- and evolution-based computational design of thermostable multiple-point mutants. PLoS Comput. Biol. 2015; 11:e1004556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goldenzweig A., Goldsmith M., Hill S.E., Gertman O., Laurino P., Ashani Y., Dym O., Unger T., Albeck S., Prilusky J. et al. Automated structure- and sequence-based design of proteins for high bacterial expression and stability. Mol. Cell. 2016; 63:337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Suzek B.E., Wang Y., Huang H., McGarvey P.B., Wu C.H.. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015; 31:926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010; 26:2460–2461. [DOI] [PubMed] [Google Scholar]
- 21. Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 2011; 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Capra J.A., Singh M.. Predicting functionally important residues from sequence conservation. Bioinformatics. 2007; 23:1875–1882. [DOI] [PubMed] [Google Scholar]
- 23. Kass I., Horovitz A.. Mapping pathways of allosteric communication in GroEL by analysis of correlated mutations. Proteins. 2002; 48:611–617. [DOI] [PubMed] [Google Scholar]
- 24. Korber B.T.M., Farber R.M., Wolpert D.H., Lapedes A.S.. Covariation of mutations in the V3 loop of human immunodeficiency virus type 1 envelope protein: an information theoretic analysis. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:7176–7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee B.C., Kim D.. A new method for revealing correlated mutations under the structural and functional constraints in proteins. Bioinformatics. 2009; 25:2506–2513. [DOI] [PubMed] [Google Scholar]
- 26. Weigt M., White R.A., Szurmant H., Hoch J.A., Hwa T.. Identification of direct residue contacts in protein–protein interaction by message passing. Proc. Natl. Acad. Sci. U.S.A. 2008; 106:67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lockless S.W., Ranganathan R.. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999; 286:295–299. [DOI] [PubMed] [Google Scholar]
- 28. Dekker J.P., Fodor A., Aldrich R.W., Yellen G.. A perturbation-based method for calculating explicit likelihood of evolutionary co-variance in multiple sequence alignments. Bioinformatics. 2004; 20:1565–1572. [DOI] [PubMed] [Google Scholar]
- 29. Valencia A. Multiple sequence alignments as tools for protein structure and function prediction. Compar. Funct. Genomics. 2003; 4:424–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Benner S.A., Gerloff D.. Patterns of divergence in homologous proteins as indicators of secondary and tertiary structure: a prediction of the structure of the catalytic domain of protein kinases. Adv. Enzyme Regul. 1991; 31:121–181. [DOI] [PubMed] [Google Scholar]
- 31. Brenner S. The molecular evolution of genes and proteins: a tale of two serines. Nature. 1988; 334:528–530. [DOI] [PubMed] [Google Scholar]
- 32. Cooperman B.S., Baykov A.A., Lahti R.. Evolutionary conservation of the active site of soluble inorganic pyrophosphatase. Trends Biochem. Sci. 1992; 17:262–266. [DOI] [PubMed] [Google Scholar]
- 33. Howell N. Evolutionary conservation of protein regions in the protonmotive cytochrome b and their possible roles in redox catalysis. J. Mol. Evol. 1989; 29:157–169. [DOI] [PubMed] [Google Scholar]
- 34. Gobel U., Sander C., Schneider R., Valencia A.. Correlated mutations and residue contacts in proteins. Proteins. 1994; 18:309–317. [DOI] [PubMed] [Google Scholar]
- 35. Neher E. How frequent are correlated changes in families of protein sequences. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taylor W.R., Hatrick K.. Compensating changes in protein multiple sequence alignments. Protein Eng. 1994; 7:341–348. [DOI] [PubMed] [Google Scholar]
- 37. Bava K.A., Gromiha M.M., Uedaira H., Kitajima K., Sarai A.. ProTherm, version 4.0: thermodynamic database for proteins and mutants. Nucleic Acids Res. 2004; 32:D120–D121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pucci F., Bourgeas R., Rooman M.. Predicting protein thermal stability changes upon point mutations using statistical potentials: introducing HoTMuSiC. Scientific Rep. 2016; 6:23257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Amin N., Liu A.D., Ramer S., Aehle W., Meijer D., Metin M., Wong S., Gualfetti P., Schellenberger V.. Construction of stabilized proteins by combinatorial consensus mutagenesis. Protein Eng. Des. Select. 2004; 17:787–793. [DOI] [PubMed] [Google Scholar]
- 40. Lehmann M., Loch C., Middendorf A., Studer D., Lassen S.F., Pasamontes L., van Loon A.P., Wyss M.. The consensus concept for thermostability engineering of proteins: further proof of concept. Protein Eng. 2002; 15:403–411. [DOI] [PubMed] [Google Scholar]
- 41. Pey A.L., Rodriguez-Larrea D., Bomke S., Dammers S., Godoy-Ruiz R., Garcia-Mira M.M., Sanchez-Ruiz J.M.. Engineering proteins with tunable thermodynamic and kinetic stabilities. Proteins. 2008; 71:165–174. [DOI] [PubMed] [Google Scholar]
- 42. Sullivan B.J., Nguyen T., Durani V., Mathur D., Rojas S., Thomas M., Syu T., Magliery T.J.. Stabilizing proteins from sequence statistics: the interplay of conservation and correlation in triosephosphate isomerase stability. J. Mol. Biol. 2012; 420:384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bendl J., Stourac J., Sebestova E., Vavra O., Musil M., Brezovsky J., Damborsky J.. HotSpot wizard 2.0: automated design of site-specific mutations and smart libraries in protein engineering. Nucleic Acids Res. 2016; 44:W479–W487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Floor R.J.1, Wijma H.J., Colpa D.I., Ramos-Silva A., Jekel P.A., Szymański W., Feringa B.L., Marrink S.J., Janssen D.B.. Computational library design for increasing haloalkane dehalogenase stability. ChemBioChem. 2014; 15:1660–1672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.