Abstract
Background.
In previous clinical trials, antiangiogenic therapies such as bevacizumab did not show efficacy in patients with newly diagnosed glioblastoma (GBM). This may be a result of the heterogeneity of GBM, which has a variety of imaging-based phenotypes and gene expression patterns. In this study, we sought to identify a phenotypic subtype of GBM patients who have distinct tumor-image features and molecular activities and who may benefit from antiangiogenic therapies.
Methods.
Quantitative image features characterizing subregions of tumors and the whole tumor were extracted from preoperative and pretherapy perfusion magnetic resonance (MR) images of 117 GBM patients in 2 independent cohorts. Unsupervised consensus clustering was performed to identify robust clusters of GBM in each cohort. Cox survival and gene set enrichment analyses were conducted to characterize the clinical significance and molecular pathway activities of the clusters. The differential treatment efficacy of antiangiogenic therapy between the clusters was evaluated.
Results.
A subgroup of patients with elevated perfusion features was identified and was significantly associated with poor patient survival after accounting for other clinical covariates (P values <.01; hazard ratios > 3) consistently found in both cohorts. Angiogenesis and hypoxia pathways were enriched in this subgroup of patients, suggesting the potential efficacy of antiangiogenic therapy. Patients of the angiogenic subgroups pooled from both cohorts, who had chemotherapy information available, had significantly longer survival when treated with antiangiogenic therapy (log-rank P=.022).
Conclusions.
Our findings suggest that an angiogenic subtype of GBM patients may benefit from antiangiogenic therapy with improved overall survival.
Keywords: angiogenesis, antiangiogenic therapy, patient stratification, quantitative perfusion-weighted imaging, radiogenomic analysis
Importance of the study
Our study shows that antiangiogenic therapies significantly prolonged the overall survival of a subtype of glioblastoma (GBM) patients. Two recent large randomized clinical trials published in New England Journal of Medicine reported that bevacizumab did not show efficacy in improving overall survival in newly diagnosed GBM patients. This result is consistent with our analysis of the pooled patient population. However, by leveraging magnetic resonance (MR) perfusion imaging features, we found that a subtype of GBM patients were enriched for angiogenesis pathways and elevated intratumor perfusion features. In this image-based GBM subtype, patients treated with antiangiogenic therapies had significantly longer survival than those who were not treated (log-rank P=.022), with a median survival difference of more than 1 year. In contrast, antiangiogenic therapy did not show such efficacy in patients who were not in this subtype. Thus, the perfusion feature may potentially be used as an imaging biomarker to facilitate clinical decision-making for personalized treatment.
Glioblastoma (GBM, World Health Organization [WHO] grade IV) is the most common and most aggressive brain cancer in adults. Despite multimodal therapy of surgical resection, radiation, and chemotherapy, the median survival of GBM in the last several decades has remained poor at less than 2 years.1 Studies show that GBM is a heterogeneous disease reflected by mixed genetic patterns, varied radiographic phenotypes, and disparate clinical outcomes. Thus, defining characteristic phenotypes of GBM that distinguish clinically relevant subgroups could enable tailoring treatment to these subgroups.
Therapeutic drugs targeting tumor biological processes are being developed and evaluated for their efficacy in improving patient clinical outcomes.2 Recent advances in cancer immunotherapy in mouse models show promising results for potentially identifying peptides arising from tumor-specific mutations that may trigger a therapeutic immune response.3 Angiogenesis is a prominent pathophysiological process in GBM that is defined by the formation of new blood vessels to supply nutrients and oxygen to rapidly proliferating tumor cells via upregulation of vascular endothelial growth factor A (VEGF-A).4 The antiangiogenic therapy bevacizumab, a humanized monoclonal antibody against VEGF-A to block angiogenesis, was approved for recurrent GBM patients.5,6 A subsequent clinical trial evaluating bevacizumab in newly diagnosed GBM patients found no survival advantage for the treatment.7,8 These patients were assessed as a uniform group with the same clinical diagnosis; however, the fact that GBM is a heterogeneous disease suggests the potential for stratifying patients into subgroups and assessing subgroup-specific responses to antiangiogenic therapy.
Recent large-scale studies using The Cancer Genome Atlas9 (TCGA) database have provided a comprehensive genomic, epigenetic, transcriptional, and protein-level characterization of GBM,9,10 with the ultimate goal of translating this molecular understanding to inform clinical decisions. The integrated analysis of imaging and genomics data is establishing bridges that link our understanding of tissue-level features to molecular counterparts that may help characterize new aspects of disease.11 A recent study has identified molecular signatures associated with prognostic clusters based on tumor morphological features.12 Another study found that tumor location associated with poor survival has a distinct molecular profile.13
Magnetic resonance imaging (MRI) is used as the primary modality for the clinical diagnosis of GBM. Prominent imaging features of GBM include heterogeneous enhancement with central necrotic regions on contrast-enhanced T1-weighted images.1 Dynamic susceptibility-weighted contrast-enhanced (DSC) perfusion MRI is an advanced MR technique that has increasingly become an integral part of the diagnostic workup for GBM.14 Whereas T1-weighted imaging shows morphological phenotypes of GBM, perfusion-weighted imaging (PWI) detects functional and physiological phenotypes of tumor vascular characteristics of cancers noninvasively, allowing indirect assessment of angiogenesis.14,15 Relative cerebral blood volume (rCBV) quantified from PWI enables voxel-based measurement across the contrast-enhancing lesion (CEL), thus showing regional microvascular variation that can characterize GBM lesions.16,17
In this study, we sought to identify novel patient subgroups with newly diagnosed GBM using quantitative perfusion MRI to define their molecular profiles and assess the treatment response to antiangiogenic therapy in these patient subgroups.
Materials and Methods
Patient Cohorts
Health Insurance Portability and Accountability Act (HIPAA) compliant institutional review board approval was obtained with informed consent from all patients. Patients aged 18 years or older with de novo GBM who underwent 3-dimensional presurgical gadolinium-based contrast-enhanced T1-weighted and DSC T2*-weighted perfusion MRI exams were identified retrospectively from 2 independent patient cohorts. The first cohort comprised 68 cases in The Cancer Imaging Archive (TCIA) collected from 2 institutions. Patient-matched microarray gene expression data, gene expression-based subtypes previously defined by TCGA, clinical chemotherapy drug information, and overall survival were downloaded from The Cancer Genome Atlas (TCGA).9 A total of 20 cases were removed from the TCGA cohort due to several data quality issues (Supplementary Methods). The second cohort comprised 79 patients from the Stanford University Medical Center Medical Center (SUMC). A total of 10 patients were excluded from the SUMC cohort (Supplementary Methods). Thus, 48 patients in the TCGA cohort and 69 patients in the SUMC cohort were used in subsequent analyses.
Antiangiogenic chemotherapy as part of the therapeutic regimen—regardless of being adjuvant or in progression—was annotated for both cohorts. Chemotherapy information was available for 25 and 30 patients in the TCGA and SUMC cohorts, respectively. For the TCGA cohort, antiangiogenic treatments included Angiocept, Avastin/bevacizumab, cilengitide, enzastaurin, sorafenib, thalidomide, and vandetanib.18 Among the 9 patients given antiangiogenic therapies in the TCGA cohort, 3 were treated both in their initial treatment and at tumor progression, and the other 6 at progression or recurrence. In contrast, except for 1 patient treated with enzastaurin, Avastin was the only antiangiogenic therapy given to patients in the SUMC cohort (Supplementary Methods). Among the 27 patients whose antiangiogenic treatment dates were available in the SUMC cohort, 2 patients were administered adjuvant antiangiogenic treatment concurrent with temozolomide as the first-line therapy, and 25 received Avastin at tumor recurrence.
DSC MR PWI Data Acquisition Protocol
The images data of the TCGA cohort were collected from 2 institutions and downloaded from the Cancer Imaging Archive (http://cancerimagingarchive.net/).19 The PWI from both institutions in TCGA were obtained with T2*-weighted gradient-echo echo-planar imaging (EPI). The perfusion images from institution 1 (N=35) were acquired with a 1.5T or 3T MR machine (TE: 40ms; TR: 1550ms or 1900ms; flip angle: 90°) with a section thickness of 5 or 6mm. The perfusion images from institution 2 (N=13) were collected with a 1.5T MR machine (TE: 54ms; TR: 1250ms or 2000ms; flip angle: 30°) with section thicknesses ranging from 3 to 5mm. Perfusion images were acquired during passage of 0.1mmol/kg gadopentetate dimeglumine (Magnevist; Bayer healthCare, Berlin) administered at a rate of 5mL/second for patients in both institutions in TCGA.20 Contrast bolus preload was not employed.
The T2*-weighted gradient-echo EPI perfusion images in the SUMC cohort (N=69) were acquired with a 1.5T MR machine (TE: 40ms; TR: 1800ms or 1113ms; flip angle: 60° or 90°) with a section thickness of 5mm during passage of 0.1mmol/kg of gadopentetate dimeglumine (Magnevist; Bayer healthCare, Berlin) or gadobenate dimglumine (MultiHance, Bracco Imaging, Milan) administered at a rate of 4mL/second. Acquisition time was 2 minutes. Contrast bolus preload was not employed.
Image Processing Pipeline for Computation of PWI Features
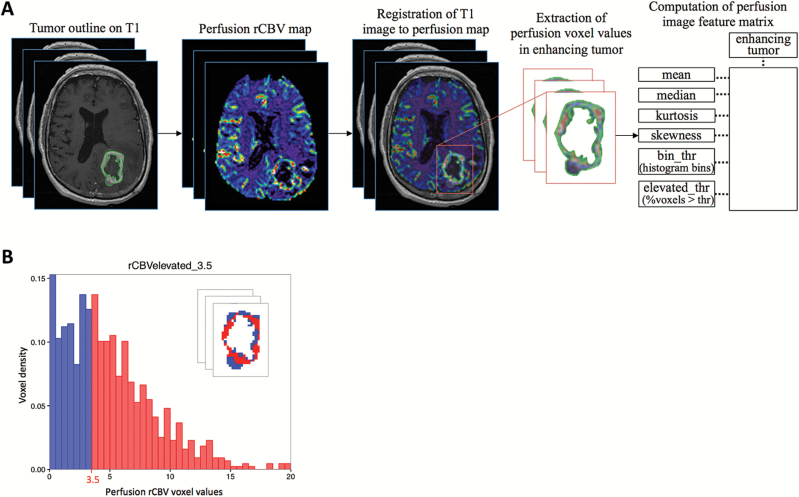
We developed and applied an image analysis pipeline to generate quantitative voxel-based PWI features from the enhancing regions of the GBM tumors (Fig. 1), similar to that previously described (Supplementary Methods).13 We generated relative cerebral blood volume (rCBV) maps using FDA-approved IB Neuro perfusion analysis software (v1.1; Imaging Biometrics, LLC, Elm Grove, WI, USA), a plugin integrated into the OsiriX platform. The perfusion values generated by IB Neuro were normalized to the normal-appearing white matter in the hemisphere contralateral to that of the tumor. The volumes of the transformed tumor ROI and the rCBV map were superimposed to extract voxel-based rCBV values in the enhancing region of the GBM tumor (implemented in a script in Matlab; www.mathworks.com/products/matlab/).
Fig. 1.
Perfusion image-processing pipeline for generating quantitative perfusion-weighted imaging (PWI) features. (A) The enhancing tumor region (excluding central necrosis) was segmented on T1 images. Relative cerebral blood volume (rCBV) maps were derived from perfusion images. The T1 images and segmented tumor masks were registered to the perfusion images. Perfusion voxel values in the enhancing tumor region were extracted and then used to compute quantitative PWI features. (B) An illustration of computation of an imaging feature, rCBVelevated_3.5 that measures the percentage of the tumor with voxel rCBV values >3.5. The red histogram bins >3.5 correspond to the tumor voxels colored in red in the inset.
Quantile Normalization of Pooled PWI Tumor Voxel Values Between 2 Cohorts
Due to variation arising from different scanners/vendors and different institutions in imaging data acquisition, there may be “batch effects” in perfusion voxel values between the 2 cohorts (Fig. S1). Batch effects are also commonly observed in molecular data, such as multiple batches of microarray experiments. Quantile normalization is widely used to correct for batch effects in molecular data.21 Here we quantile-normalized the PWI tumor voxel values pooled from all patients between the 2 cohorts. The voxel values of the TCGA cohort were used to quantile-normalize those of the SUMC cohort using the normalize.quantile.use.target function in the “preprocessCore” bioconductor R package.21
Quantification of PWI Features
We sought to extract features that capture perfusion image phenotypes of both the whole tumor and tumor heterogeneity. After the quantitative image analysis pipeline, we quantified a total of 46 nonparametric voxel-based PWI features in the CEL of each GBM tumor (Fig. 1A), including 6 summary statistics describing the bulk tumor characteristics and 40 histogram-based features quantifying regional variation and intratumoral heterogeneity of PWI voxel values. The 6 summary statistics included mean, median, variance, maximum, skewness, and kurtosis.22 The histogram-based features consisted of 20 histogram bins (rCBVbin) at an interval of 0.5 ranging from 0.5 to 10, and 20 features that measured elevated perfusion tumor burden (the fraction of the tumor with rCBV voxel values greater than a threshold [rCBVelevated]), where the same thresholds were used for generating histogram bins (Fig. 1B).
Discovery of PWI-based Subtypes
We performed hierarchical consensus clustering with agglomerative average linkage to discover PWI-based clusters in GBM patients23 (Supplementary Methods). The PWI features were normalized by mean-centering each feature. The resulting clusters were represented and visualized using t-distributed stochastic neighbor embedding (T-SNE) implemented in R, with a pairwise distance metric of (1-r), where r is the Pearson’s correlation coefficient.24 The maximum number of iterations was set to 2,000 to keep the cost (error) below 0.5.
Identification of Important PWI Features Associated With Each Cluster
To validate the reproducibility of patient clusters, we built a random forest model25 using the TCGA cohort to predict cluster assignment of the SUMC cohort, which was compared to the clusters identified from unsupervised consensus clustering above. Similarly, we predicted the cluster assignment of the TCGA cohort using the SUMC cohort and reported the prediction accuracy. The importance of the PWI features was evaluated using the Gini index.25 Feature selection of a subset of PWI features that achieved the highest 10-fold cross validation accuracy was identified using a recursive feature elimination (RFE) algorithm implemented in an R package caret.26
Survival Analysis
We performed Kaplan-Meier survival analysis with the log-rank test on categorical clinical variables, including age > 60 years, sex, solitary or multicentric tumor phenotype, gene expression-based subtypes, and the discovered PWI-based groups. These variables were also used to construct a multivariate Cox proportional hazards survival regression model to assess the clinical significance of PWI-based groups in association with overall survival after accounting for other clinical prognostic covariates.
We ran Kaplan-Meier survival analysis to assess the prognostic value of antiangiogenic treatment in cluster II patients, who were predicted to respond to antiangiogenic therapy. The overall survival of patients stratified by PWI-based group and gene expression-based subtype was visualized using a boxplot. All statistical analyses were performed using R (version 3.3).
Molecular Pathway Analysis
Gene set enrichment analysis (GSEA, http://www.broad.mit.edu/gsea) was performed to identify upregulated gene sets and pathways in the PWI-based clusters (Supplementary Methods).
Results
Characterization of Patient Cohorts
The median age in the TCGA and SUMC cohorts was 61 years (range, 30y– 84y) and 60.5 years (range, 21y–91y), respectively. Table 1 shows survival analysis of clinical variables, where known prognostic variables such as KPS and multicentric tumor phenotype are significantly associated with survival in both cohorts, consistent with previous reports.9,10
Table 1.
Clinical variables and the perfusion-weighted imaging (PWI)-based subgroup as covariates in the survival analysis of glioblastoma (GBM) patients. Univariate and multivariate Cox proportional hazard models show that PWI-based subgroups are significantly associated with survival after accounting for the other clinical variables in both The Cancer Genome Atlas (TCGA) and the Stanford University Medical Center (SUMC) cohorts. Contrast enhancing lesion (CEL) tumor volume was dichotomized by the median. KPS is available for N=34 patients in TCGA. Statistically significant values are shown in bold.
| Clinical variable | TCGA | SUMC cohort | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate Cox | Multivariate Cox | Univariate Cox | Multivariate Cox | |||||
| HR (95% CI) |
P value | HR (95% CI) |
P value | HR (95% CI) |
P value | HR (95% CI) |
P value | |
| Age at initial diagnosis > 60 y | 1.2 [0.7, 2.3] |
.48 | 1.5 [0.6, 3.8] |
.35 |
2.7
[1.4, 5.3] |
.0044 |
3.4
[1.6, 7.4] |
.0016 |
| Sex=male | 0.7 [0.4, 1.4] |
.39 | - | - | 1.9 [0.9, 3.8] |
.074 | - | - |
| Large CEL volume (cm3) | 1.3 [0.7, 2.5] |
.35 | 1.4 [0.6, 3.3] |
.47 | 1.2 [0.6, 2.3] |
.64 | 1.3 [0.7, 2.6] |
.39 |
| Multicentric tumor phenotype |
3.0
[1.2, 7.5] |
.019 | 0.5 [0.07, 3.3] |
.45 |
2.1
[1.0, 4.4] |
.048 | 1.9 [0.8, 4.3] |
.12 |
| KPS < 80 |
3.1
[1.4, 6.8] |
.0043 |
3.9
[1.4, 10.7] |
.0078 |
2.8
[1.5, 5.4] |
.0017 |
3.0
[1.5, 6.2] |
.0026 |
| PWI-based subgroup=2 |
2.3
[1.2, 4.4] |
.0092 |
4.4
[1.6, 11.8] |
.0033 |
2.6
[1.3, 5.1] |
.0041 |
3.5
[1.7, 7.4] |
.0010 |
Abbreviations: CEL, contrast-enhancing lesion; CI, confidence interval; HR, hazard ratio; SUMC, Stanford University Medical Center; PWI, perfusion-weighted imaging; TCGA, The Cancer Genome Atlas
Unsupervised Clustering Using PWI Features Identifies 2 Prognostic Patient Subgroups
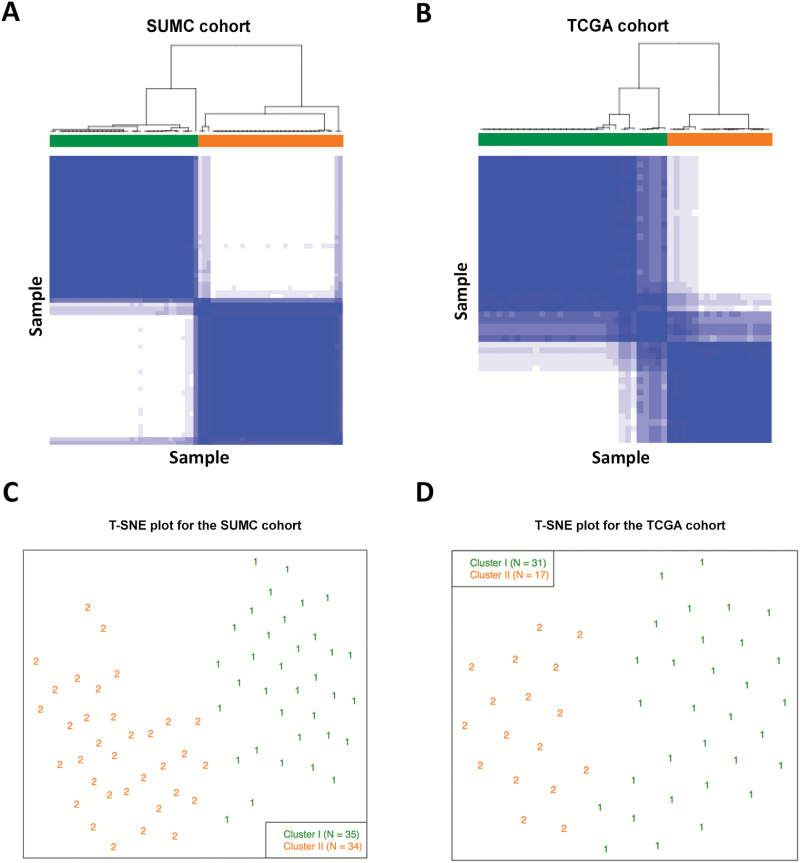
Unsupervised consensus clustering using the 46 PWI features produced 2 clusters in both the TCGA and the SUMC cohorts (Fig. 2A, B). We then computed the overall average silhouette width27 for the 2 clusters to evaluate the validity of the number of clusters. The average silhouette widths for the 2 cohorts were 0.59 and 0.66, providing supportive evidence that the 2 clusters are robust (Figs. S2, S3). Cluster II formed a distinct cluster from cluster I, as visualized by the T-SNE plots of both cohorts (Fig. 2C, D).
Fig. 2.
Unsupervised clustering in both cohorts. Consensus clustering of patients based on perfusion-weighted imaging (PWI) features in (A) the Stanford University Medical Center (SUMC) and (B) The Cancer Genome Atlas (TCGA) cohorts consistently identified 2 clusters that were well separated, as shown by the t-distributed stochastic neighbor embedding (T-SNE) plots of (C) the SUMC and (D) TCGA cohorts. In the consensus matrices in (A) and (B), solid blue indicates the 2 samples that always cluster together in one group, whereas white indicates those that never cluster together.
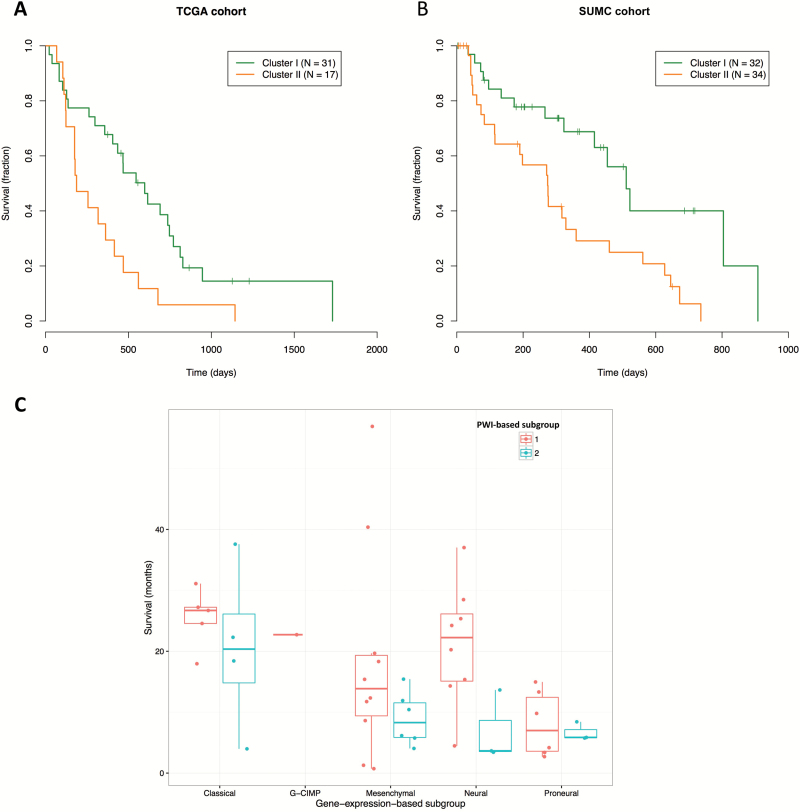
Kaplan-Meier survival analysis showed that cluster II patients had significantly worse survival than cluster I patients in both the TCGA (log-rank P=.0092; HR=2.30) (Fig. 3A) and SUMC (log-rank P=.0041; HR=2.58) cohorts (Fig. 3B). Multivariate Cox analysis showed that this survival difference for cluster II in TCGA remained significant (log-rank P=.0033; HR=4.39) after accounting for other clinical variables, including age > 60 years, CEL volume, multicentric tumor phenotype, and KPS (Table 1). Similarly, the SUMC cohort confirmed that group II patients had significantly worse survival (log-rank P=.0010; HR=3.49), independent of other clinical covariates (Table 1). These results suggested that leveraging a comprehensive set of PWI features characterizing both the bulk tumor and intratumoral heterogeneity enabled us to identify robust clinically relevant subgroups.
Fig. 3.
Kaplan-Meier curves of patients dichotomized into clusters (clusters I and II) in both cohorts revealed that patients in cluster II had significantly worse survival than those in cluster I. (A) Kaplan-Meier curve for the 2 clusters in the The Cancer Genome Atlas (TCGA) cohort (log-rank P=.0092; HR=2.30). (B) Kaplan-Meier curve for the 2 clusters in the Stanford University Medical Center (SUMC) cohort (log-rank P=.0041; HR=2.58). Three patients in cluster I were removed due to missing overall survival information. (C) Box plot of patients’ overall survival stratified by gene expression-based subgroup and perfusion-weighted imaging (PWI)-based subtype. Right-censored patients were included in the subtype visualization because the overall survival of each right-censored patient was above the median survival of its corresponding subtype.
Corroborating with the result obtained from all patients, the PWI-based cluster II was associated with worse survival than cluster I consistently across different gene expression-based subtypes, most prominently in the neural, classical, and mesenchymal subtypes (Fig. 3C). Also, the non-G-CIMP, proneural subtype (log-rank P=.0053, HR=4.6) was significantly correlated with worse survival than the other subtypes (Table S3). The multivariate Cox analysis showed that both the PWI-based cluster II and the gene expression-based non-G-CIMP proneural subtype were significant indicators of poor prognosis (Tables S3, S4).
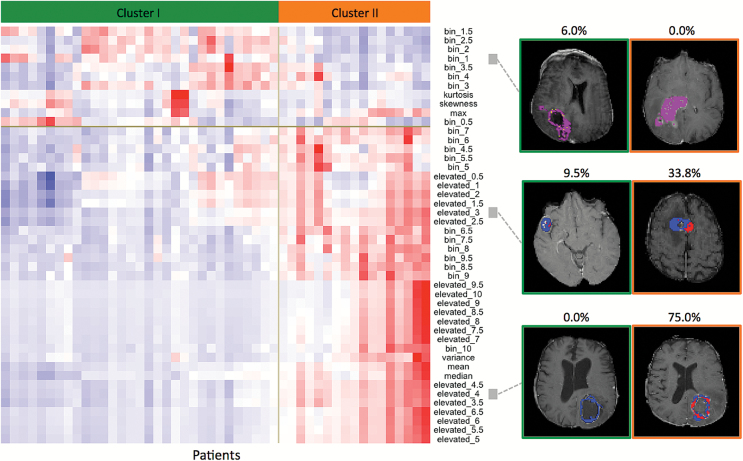
Cluster II Patients Are Associated With High Intratumoral PWI Features
Extracted from the whole enhancing tumor, the 6 summary PWI features alone were not consistently associated with the discovered clusters in the 2 cohorts, confirming previous reports20 (Supplementary Results). In contrast, intratumoral PWI features were more informative for detecting patient subgroups than the summary features. Heatmaps of the PWI features revealed the difference between the 2 clusters of patients, with most histogram-based regional PWI features in cluster II being larger than those in cluster I (Fig. 4). As shown in the example images of 3 PWI features in the 2 clusters (Fig. 4), cluster II in TCGA was positively associated with a larger number of voxels at a low-to-medium cutoff (eg, such as rCBVelevated_3 and rCBVelevated_4) corresponding to a large fraction of voxels with values greater than the cutoff (colored in red for visualization) (Fig. 4). A random forest trained on the TCGA cohort confirmed that rCBVelevated features at low to medium cutoffs were predictive of the 2 clusters (Fig. S8A). These PWI feature patterns, which are characteristics of the 2 clusters, were similarly observed in the SUMC cohort (Fig. S8B). Since many of these PWI features are highly correlated (redundant) (Fig. S9), the recursive feature elimination algorithm selected a handful of features that are predictive the 2 clusters, including rCBVelevated_2.5 and rCBVelevated_3 for the TCGA cohort and rCBVelevated_2.5 and rCBVmedian for the SUMC cohort (Fig. S8).
Fig. 4.
Two clusters of glioblastoma (GBM) patients with distinct perfusion-weighted imaging (PWI) features as illustrated by 3 example features observed on representative image slices (the analysis was performed in 3D). Left: matrix of patients (columns) and the quantitative image features of glioblastoma (GBM) contrast-enhancing lesion (CEL) regions (rows). Right: Colored perfusion maps superimposed on the aligned anatomical T1 images show example images of 3 linked PWI features in the 2 clusters with their actual values specified on the top. In the 2 example images for relative cerebral blood volume (rCBV)bin_1, yellow indicates the percentage of voxel with values ≥0.5 and <1, and purple indicates voxel values ≥1 or <0.5. In the example images for rCBVelevated_3 and rCBVelevated_4, red represents voxels above the threshold, and those below are colored in blue. Thus, the rCBVelevated feature is the proportion of the red area of the whole tumor.
To validate the generalizability of the significant PWI features associated with the clusters to unseen cases, we used the random forest classifier constructed using the SUMC cohort to classify patients of the TCGA cohort into 2 groups. When comparing the classifier-based stratification with the unsupervised clustering approach above, the accuracy of predicting the TCGA cohort using a model trained on all PWI features in the SUMC cohort was 95.8% (46/48), and the model trained on the selected subset of features was 97.9% (47/48). The classifier-based stratification trained on the SUMC cohort remained significantly associated with survival in TCGA (log-rank P=.030; HR=1.98). Similarly, the classification accuracy was 92.8% (64/69) for training on all features in TCGA and predicting on the SUMC cohort, and was 94.2% (65/69) for training on the selected subset of features in TCGA. The classifier-based stratification of the SUMC cohort trained on TCGA was significant in correlating with survival (log-rank P=.012; HR=2.26).
PWI-based Cluster II Patients Are Enriched for Angiogenesis
We ran gene set enrichment analysis (GSEA)28 to identify molecular activities that are different between the 2 clusters. A total of 13 gene sets, including angiogenesis-signaling pathway, vasculature development, and response to hypoxia, were found to be significantly enriched in cluster II compared with cluster I (FDR P<.05) (Table S1). Shared genes contributing to the core enrichment of both the hypoxia signaling and the angiogenesis pathways consisted of angiogenin (ANG), VEGF-A, and transforming growth factor beta 2 (TGFB2, also called glioblastoma-derived T-cell suppressor factor). Upregulation of angiogenesis pathways found in cluster II suggests the potential for treatment efficacy using antiangiogenic therapy in this subgroup of patients.
PWI-based Cluster II Patients Given Antiangiogenic Treatment Have Better Survival
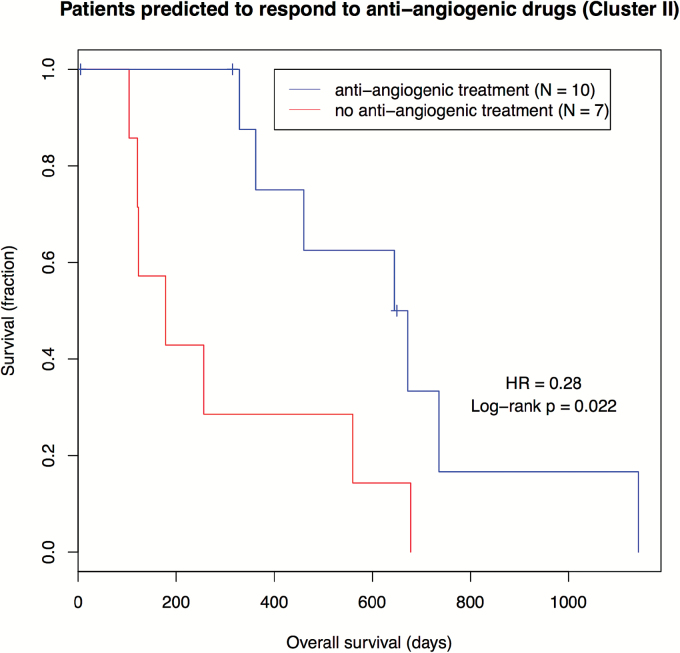
We next evaluated whether the PWI-based quantitative imaging features can be used as biomarkers for predicting treatment response to antiangiogenic therapy in GBM patients based on identifying the cluster in which they belong. Because chemotherapy treatment information was only available for a subset of patients in both of our cohorts (Fig. S10), we combined patients with chemotherapy information from both cohorts to increase statistical power. Antiangiogenic treatment did not prolong overall survival in all patients as a single group (log-rank P=.15; HR=0.59), consistent with results reported in a recent large-scale clinical trial.7 In the cluster II patients who were predicted to respond to antiangiogenic treatment from both cohorts, those treated with antiangiogenic therapies (median survival: 552.5 d) had significantly longer survival than those who were not given the antiangiogenic therapy (median survival: 178 d) (log-rank P=.022; HR=0.28) (Fig. 5), with a median survival difference >1 year (374.5 d). In contrast, antiangiogenic treatment (N=26/37) did not confer survival advantage in the cluster I patients (log-rank P=.77; HR=0.86), as might be predicted from the differential PWI feature and molecular analyses. More specifically, the median survival for patients treated with and without antiangiogenic therapy in cluster I was 439 and 546 days, respectively.
Fig. 5.
Antiangiogenic treatment significantly improves overall survival of patients in cluster II. In the subgroup of patients who were predicted to respond to antiangiogenic treatment based on perfusion-weighted imaging (PWI) features (cluster II), patients treated with antiangiogenic therapies were significantly associated with longer survival than those who were not given the antiangiogenic therapy (log-rank P=.022).
Discussion
We identified an angiogenic subtype of patients with newly diagnosed GBM who could potentially benefit from antiangiogenic therapy as part of their treatment regimen, with improved clinical outcome (survival). This subtype, which is distinguished by elevated PWI features, is characterized by worse overall survival, enrichment of angiogenesis and hypoxia pathways, and significantly better survival in patients treated with antiangiogenic therapy than those who were not treated. The results were consistently reproduced in 2 independent cohorts.
Recent large phase 3 randomized clinical trials of bevacizumab therapy in newly diagnosed GBM reported that bevacizumab therapy did not show efficacy in improving overall survival,7,8,29 consistent with our analysis of the pooled patient population that is not subdivided by subtype (log-rank P=.15). Poor treatment efficacy across all patients may result from heterogeneous patterns in genetic profile, imaging phenotype, and clinical outcome across GBM patients.18 To improve the current one-treatment-fits-all approach, there is a need to identify biomarkers that stratify patients into clinically relevant subgroups. In this study, our results showed that antiangiogenic therapy significantly improved overall survival in a distinct subgroup of GBM patients identified by quantitative PWI features (log-rank P=.022; HR=0.28) (Fig. 5) that may be useful as clinically actionable biomarkers for making treatment decisions. In corroboration, our molecular analysis provided molecular insight showing the subgroup enriched with proangiogenic factor VEGF-A, which is the target for antiangiogenic drugs such as bevacizumab. Thus, perfusion imaging may provide a promising noninvasive biomarker predictive of response to antiangiogenic therapy.
While we sought to overcome disease heterogeneity by clustering patients into subgroups, intratumoral heterogeneity—which arises from multiple tumor subclones and leads to treatment failure and drug resistance—also needs to be addressed to ultimately enable personalized treatment.30,31 Quantitative voxel-based perfusion MR image analysis provides a volumetric characterization of the tumor that allows us to examine spatial variation in tumor rCBV. In addition to statistics summarizing characteristics of the whole tumor, we comprehensively captured intratumoral heterogeneity by quantifying histogram bins and elevated features at 20 uniform thresholds from 0.5 to 10 (Fig. 1). Our analysis revealed that poor prognosis was associated with a large fraction of high rCBV voxels measured by rCBVelevated, suggesting that the extent of highly vascular subregions in the tumor may be correlated with the level of aggressiveness in the heterogeneous tumor. Previous perfusion studies using summary features were limited by the small number of features and only dichotomized patients by the feature median, which were often unable to uncover novel patient subtypes when the underlying subgroups were not balanced.20 Here we leveraged PWI features quantifying intratumoral heterogeneity to identify subtypes associated with overall survival in presurgical, newly diagnosed GBM patients. Except for manual segmentation of the tumor region, our computational approach for identifying patients who would benefit from antiangiogenic treatment is fully automated and could be incorporated into the clinical workflow.
The patient clusters associated with survival were consistently reproducible across the 2 cohorts (Figs. 2, 3). We also validated the generalization of our clustering analysis by training on one cohort and testing on the other cohort. We trained a random forest classifier using the SUMC cohort to predict 2 subgroups in TCGA. The predicted subgroups remained significantly associated with survival (log-rank P=.030), and 95.8% (46/48) of patients in TCGA were assigned to the same clusters as those by the unsupervised consensus clustering approach. When swapping the training and testing cohorts, the predicted SUMC subgroups were significant in correlating with survival (log-rank P=.012), and the predicted cluster assignments of 92.8% (64/69) of patients were consistent with those from the unsupervised clustering approach. After feature selection, the prediction accuracies improved further (97.9% [47/48] and 94.2% [65/69]) when predicting TCGA using selected features of the SUMC cohort, and vice versa. This indicated that the selected features are predictive of the clusters and are robust when generalizing to unseen data.
There are several limitations in our study. First, our analysis was limited by the small number of patients with complete treatment information because many patients in the SUMC cohort had surgery at the Stanford Medical Center and were then transferred to local hospitals for follow-up treatments. Thus, these patients without complete clinical neuro-oncology notes had to be removed in the treatment analysis. A second limitation is potential “batch effects” between the 2 cohorts due to variability in perfusion imaging technique. We corrected the batch effects by quantile normalization of the SUMC cohort based on the TCGA cohort, and the prediction accuracies improved substantially (Supplementary results). There could possibly be remaining residual “batch effects,” and thus future studies with larger cohorts of patients undergoing the same perfusion imaging protocol could be helpful for effectively removing this variability.
A third limitation of our study was the variability in antiangiogenic treatments administered. Because this is a retrospective study, the administration of antiangiogenic treatment was heterogeneous across patients in both cohorts depending on the clinical presentation of disease recurrence and various other factors (Supplementary methods). There may be potential confounding variables in determining which patients received antiangiogenic treatment. For example, if there were clinical presentations correlated with the PWI-based cluster II that led to the decision of the antiangiogenic treatment administration (eg, neovascularization on subsequent MR images), the difference in survival between the 2 PWI-based clusters would be reduced by the effect of salvage therapies. The fact that we nonetheless observed the differential survival between the 2 PWI-based clusters (Fig. 3) suggests that this difference is substantial. We did not observe a significant association between antiangiogenic treatment administration and PWI-based cluster II (Fig. 10S), indicating that any potential variable correlated with the perfusion-based stratification has not previously been used systematically in the decision-making of administering antiangiogenic therapies in clinics. Moreover, the heterogeneous timing of treatments may affect the overall survival. However, this heterogeneity is present in both PWI-based clusters; factors such as the timing and duration of antiangiogenic therapies did not vary between the 2 PWI-based clusters. The key finding of our study is that antiangiogenic therapies showed efficacy in PWI-based cluster II but not in cluster I. Within the patients of PWI-based cluster II, those treated with antiangiogenic therapies had significantly longer overall survival than those who were not treated. One could argue that if the antiangiogenic therapies were given earlier, these patients may have had even longer overall survival. In contrast, this survival benefit from antiangiogenic therapy was not observed in patients of the PWI-based cluster I. In fact, the median overall survival of patients who were treated with antiangiogenic therapy (439 days) was slightly worse than that of patients who were not treated (546 days) in cluster I.
Our study provides a first approach leveraging PWI features as potential imaging biomarkers to stratify patients for personalized antiangiogenic treatment. Future large-cohort prospective studies evaluating treatment effectiveness of antiangiogenic treatment in the subtype of patients having elevated PWI features would be helpful to confirm our results.
In conclusion, we determined that an angiogenic subgroup of patients with worse overall outcome and high intratumoral quantitative PWI features has better response to antiangiogenic therapy than patients who lack these imaging features. Our analysis approach using quantitative PWI features as a discriminative, predictive phenotype could potentially be used in future clinical practice to evaluate GBM tumors and assist with therapeutic decision-making for targeted, personalized treatment.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
National Cancer Institute, National Institutes of Health (U01CA142555, U01CA190214 to D.L.R); National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health (R01EB020527 to O.G.).
Conflict of interest statement. none.
Supplementary Material
Acknowledgement
We would like to thank Dr Jarrett Rosenberg for helpful discussions on statistical analysis. We also want to acknowledge Drs. Geoffrey Appelboom, Joshua J. Loya, and Erick M. Westbroek for their help with treatment data collection and image annotations.
References
- 1. Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310(17):1842–1850. [DOI] [PubMed] [Google Scholar]
- 2. Thomas AA, Brennan CW, DeAngelis LM, Omuro AM. Emerging therapies for glioblastoma. JAMA Neurol. 2014;71(11):1437–1444. [DOI] [PubMed] [Google Scholar]
- 3. Yadav M, Jhunjhunwala S, Phung QT, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515(7528):572–576. [DOI] [PubMed] [Google Scholar]
- 4. Zhang X, Pagel MD, Baker AF, Gillies RJ. Reproducibility of magnetic resonance perfusion imaging. PLoS One. 2014;9(2):e89797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kreisl TN, Kim L, Moore K, et al. Phase II trial of single-agent bevacizumab followed by bevacizumab plus irinotecan at tumor progression in recurrent glioblastoma. J Clin Oncol. 2009;27(5):740–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Friedman HS, Prados MD, Wen PY, et al. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J Clin Oncol. 2009;27(28):4733–4740. [DOI] [PubMed] [Google Scholar]
- 7. Gilbert MR, Dignam JJ, Armstrong TS, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chinot OL, Wick W, Mason W, et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):709–722. [DOI] [PubMed] [Google Scholar]
- 9. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gevaert O, Mitchell LA, Achrol AS, et al. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology. 2014;273(1):168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Itakura H, Achrol AS, Mitchell LA, et al. Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Sci Transl Med. 2015;7(303):303ra138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu TT, Achrol AS, Mitchell LA, et al. Computational identification of tumor anatomic location associated with survival in 2 large cohorts of human primary glioblastomas. AJNR Am J Neuroradiol. 2016;37(4):621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barajas RF, Jr, Cha S. Benefits of dynamic susceptibility-weighted contrast-enhanced perfusion MRI for glioma diagnosis and therapy. CNS Oncol. 2014;3(6):407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hakyemez B, Erdogan C, Bolca N, Yildirim N, Gokalp G, Parlak M. Evaluation of different cerebral mass lesions by perfusion-weighted MR imaging. J Magn Reson Imaging. 2006;24(4):817–824. [DOI] [PubMed] [Google Scholar]
- 16. Hu LS, Eschbacher JM, Heiserman JE, et al. Reevaluating the imaging definition of tumor progression: perfusion MRI quantifies recurrent glioblastoma tumor fraction, pseudoprogression, and radiation necrosis to predict survival. Neuro Oncol. 2012;14(7):919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barajas RF, Jr, Phillips JJ, Parvataneni R, et al. Regional variation in histopathologic features of tumor specimens from treatment-naive glioblastoma correlates with anatomic and physiologic MR Imaging. Neuro Oncol. 2012;14(7):942–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu-Emerson C, Duda DG, Emblem KE, et al. Lessons from anti-vascular endothelial growth factor and anti-vascular endothelial growth factor receptor trials in patients with glioblastoma. J Clin Oncol. 2015;33(10):1197–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clark K, Vendt B, Smith K, et al. The cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jain R, Poisson L, Narang J, et al. Genomic mapping and survival prediction in glioblastoma: molecular subclassification strengthened by hemodynamic imaging biomarkers. Radiology. 2013;267(1):212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. [DOI] [PubMed] [Google Scholar]
- 22. Davnall F, Yip CS, Ljungqvist G, et al. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging. 2012;3(6):573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Monti S, Tamayo P, Mesirov J, Golub T. Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Mach Learn. 2003;52(1–2):91–118. [Google Scholar]
- 24. Maaten Lvd, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 25. Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2(3):18–22. [Google Scholar]
- 26. Kuhn M. Building predictive models in R using the caret package. J Stat Software. 2008;28(5):1-26. [Google Scholar]
- 27. Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:53–65. [Google Scholar]
- 28. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weller M, Yung WK. Angiogenesis inhibition for glioblastoma at the edge: beyond AVAGlio and RTOG 0825. Neuro Oncol. 2013;15(8):971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.