Abstract
The IntaRNA algorithm enables fast and accurate prediction of RNA–RNA hybrids by incorporating seed constraints and interaction site accessibility. Here, we introduce IntaRNAv2, which enables enhanced parameterization as well as fully customizable control over the prediction modes and output formats. Based on up to date benchmark data, the enhanced predictive quality is shown and further improvements due to more restrictive seed constraints are highlighted. The extended web interface provides visualizations of the new minimal energy profiles for RNA–RNA interactions. These allow a detailed investigation of interaction alternatives and can reveal potential interaction site multiplicity. IntaRNAv2 is freely available (source and binary), and distributed via the conda package manager. Furthermore, it has been included into the Galaxy workflow framework and its already established web interface enables ad hoc usage.
INTRODUCTION
The interaction of RNA molecules is an important factor for regulatory processes in all organisms. The in silico modeling and prediction of such RNA–RNA interactions is thus a central aspect in current research projects. In the last decades, various approaches to solve this problem have been proposed (see reviews (1,2)). To enable highly accurate predictions, state-of-the-art tools not only take the stability (energy) of possible RNA–RNA interactions into account but also consider the accessibility of the interacting subsequences (3). The latter is often incorporated by adding a pseudo-energy penalty proportional to the subsequences’ probability to be involved in intramolecular structure formation (4). Prediction quality can be further improved by addressing additional features known from experimentally validated interactions. For instance, bacterial small RNAs (sRNA) and eukaryotic microRNAs form highly stable subinteractions, which are considered to seed the formation of extended RNA–RNA duplexes (5,6).
IntaRNA, first introduced in (7), is one of the most widely used state-of-the-art RNA–RNA interaction prediction approaches (2,3). It is employed in prokaryotic (8–10) and eukaryotic (11–13) systems, and its web server (14,15) received ∼16 500 external jobs in 2016. Its fast heuristic prediction mode enables genome wide target prediction while its high prediction accuracy is based on the incorporation of interaction site accessibility and seed constraints.
However, IntaRNA has become technically outdated for the following reasons. Firstly, IntaRNA was fixed to an aged Vienna RNA library (v1.*) (16) and its energy parameters Turner99 (17). Secondly, it featured a strongly restricted user interface and was hard to maintain, update or extend.
Here, we introduce IntaRNAv2, a complete open-source reimplementation of the previous approach (from now on referred to as IntaRNAv1) with a strong focus on efficiency, modularity and flexibility. The implementation closely follows the software development guidelines suggested in (18) and enables enhanced and customizable RNA–RNA interaction prediction.
IntaRNAv2 enforces the seed interaction to be energetically favorable. Furthermore, a slightly changed treatment of dangling end energy contributions, compared to IntaRNAv1 and other approaches, has been incorporated. This, in concert with the usage of recent RNA energy parameters (from the Vienna RNA package v2.* (19)), enables an improvement of prediction quality. The newly performed benchmark uses a data set of 160 experimentally verified enterobacterial sRNA–mRNA interactions and evaluates genome wide screens for each interaction pair.
The extended IntaRNAv2 web interface within the Freiburg RNA tools [Freiburg RNA tools: http://rna.informatik.uni-freiburg.de/IntaRNA.] v4.4.2 (14,15) enables full control over energy parameters, seed constraints and accessibility computation. The user has to provide either lists of interacting RNAs (all versus all predictions) or can perform a genome wide target screen (providing the according NCBI accession number) for a given input RNA. Using an efficient dynamic programming scheme, the minimum free energy interactions (or suboptimals if requested) are listed and additional details can be retrieved for each predicted interaction pair. Furthermore, the new version provides minimal energy profiles for interacting RNA pairs that enable a detailed investigation of interaction alternatives.
METHODS
IntaRNAv2 reimplements and extends the original IntaRNA approach (7) and closely follows established rules for software development in systems biology, e.g. see (18).
Besides the heuristic prediction mode of IntaRNAv1 that allows fast, genome-wide predictions (7), IntaRNAv2 also features exact interaction prediction by reimplementing and extending the RNAup approach (4). Since seed constraints are optional for all prediction modes, IntaRNAv2 can emulate RNAup predictions (by disabling the seed requirement). Furthermore, the user interface enables full control of accessibility usage within RNA–RNA interaction prediction. Per default, unpaired probabilities are computed using the Vienna RNA package v2.* routines. Here, both global (RNAup-like) or local unpaired probabilities (as computed by RNAplfold (20)) can be selected. IntaRNAv2 also supports the loading of pre-computed accessibility data. This can significantly reduce the run time since the computation of the accessibility is highly time consuming (7). This feature is especially useful when screening a large target sequence (21). By disabling the integration of accessibility information, IntaRNAv2 can be used to emulate predictions from TargetScan (22) or RNAhybrid (23).
Seed stability constraints are a new feature of IntaRNAv2, which enable direct control over the stability of the seed interaction. To this end, an upper energy bound can be provided. This defaults to 0 kcal/mol and thus requires all seed interactions to be energetically favorable. Furthermore, the user can optionally provide a lower bound on the unpaired probability of the subsequences forming the seed to define whether highly accessible regions are to be favored.
Dangling end contributions incorporate a stabilizing effect due to the stacking of unpaired nucleotides directly neighboring base pairs (17). Since the interaction model of IntaRNA incorporates the accessibility, i.e. ‘unpairedness’, of the involved subsequences, an explicit dangling end case treatment was implemented in IntaRNAv1, i.e. dangling end contributions were only incorporated if the according nucleotide was also part of the accessible subsequence considered for interaction formation (see Supplementary material for further details).
IntaRNAv2 simplifies the dangling end treatment for interactions. Here, the conditional probability of the dangling end to be free when the interaction is formed is used to weight the according dangling end contributions (see Supplementary material for details). These conditional probabilities can be directly computed from the respective accessibility terms already available for the interaction scoring.
Minimal energy profiles, either position wise for each RNA (4) or intermolecular index pairwise for an RNA pair, can provide deeper insights into RNA–RNA interaction alternatives, since as for single RNA structure prediction, the optimal interaction is not necessarily the only or biologically correct option. With IntaRNAv2 both types of profiles can be generated by linking their generation with the respective dynamic programming computations. That is, whenever an interaction site (a cell in the according table) is computed, the respective minimal energy profiles are updated. This feature comes with a linear/quadratic overhead in computation time for single/pairwise profiles, respectively, since all indices within the according subsequences have to be updated. For instance, the run time of the heuristic prediction mode of O(n2) is increased to O(n3) for single sequence profiles and O(n4) for pairwise minimal energy profiles. While computationally demanding, the investigation of these profiles provides meaningful additional information about possible interaction patterns, as discussed in the Results section.
Suboptimal interaction enumeration also enables the investigation of interaction alternatives. In IntaRNAv1, only query interactions were allowed to overlap, since interaction alternatives in the target were of interest. IntaRNAv2 now enables explicit control where suboptimal interactions are allowed to overlap. Besides the (so far limited) possibility to constrain overlaps to one sequence, this enables the enumeration of variants of the minimum free energy interaction (overlap in both) as well as the suppression of any overlap in both.
Customizable output in CSV format complements the normal or detailed ASCII chart output of the interaction details. The CSV output provides for the first time a machine-readable output format of all interaction details. To this end, the user can specify all details of interest (columns of the CSV table) that will be reported for each predicted interaction.
For further options of IntaRNAv2 to constrain the interaction, seed site, accessibility computation, energy model, multi-threading, output formats, installation, docker/conda/galaxy packages, etc. we refer to its documentation [IntaRNA at github: https://github.com/BackofenLab/IntaRNA/.].
RESULTS AND DISCUSSION
In order to evaluate the impact of recent energy parameters and the altered energy computation (see Methods), we performed a benchmark on genome-wide interaction predictions. In recent comprehensive benchmarks (2,24,25), IntaRNA and RNAupwere shown to be the best performing tools that are not based on conservation. Thus, we limited our comparison to IntaRNAv1 and IntaRNAv2. To assess performance, we followed the procedure previously applied in (25). The dataset of verified RNA–RNA interactions was extended by several candidates that have been reported in recent literature (26–36). Together, this is a dataset of 160 verified sRNA–target interactions (see Supplementary Table S1).
For interaction prediction, we extracted 200 nucleotides upstream and 100 nucleotides downstream of the annotated start codon for each protein coding gene in Escherichia coli (NC_000913) and Salmonella (NC_003197). This gives rise to 4319 putative targets in E. coli and 4553 putative targets in Salmonella all of length 300 nucleotides with the start codon located at positions 201, 202 and 203. Target predictions were performed for sRNAs (see Supplementary FASTA) from both species individually with IntaRNAv1 and IntaRNAv2 (parameters see Supplementary material). The resulting predictions were ranked by energy score with the lowest energy ranking on position 1.
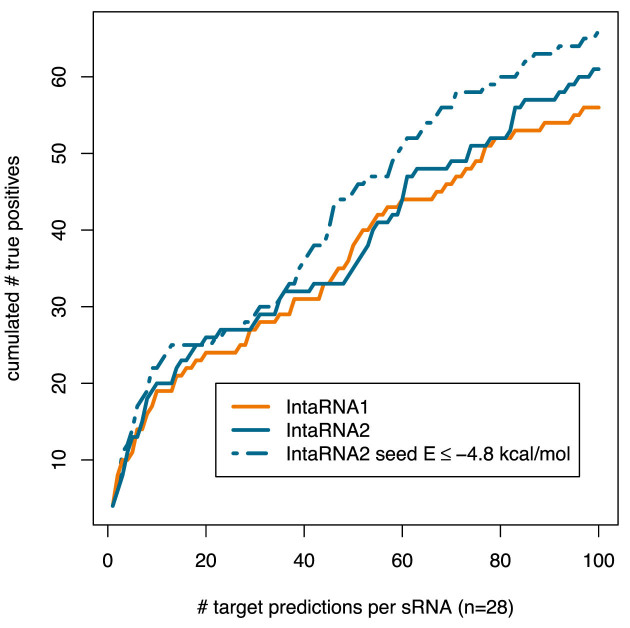
A comparison is provided in Figure 1, which shows a slightly increased performance of IntaRNAv2 compared to IntaRNAv1 using comparable parameterization for both tools with equivalent run time (data not shown). The memory requirement of IntaRNAv2 was drastically reduced by a factor of 6 (from an average of 79MB to 13MB).
Figure 1.
The figure shows the whole genome target prediction performance of IntaRNAv1 (orange) and IntaRNAv2 (blue) on a benchmark set of 160 experimentally verified enterobacterial (E. coli, Salmonella) RNA–RNA interactions including 28 different sRNAs (see Supplementary Table S1 and Supplementary FASTA). Furthermore, the performance of IntaRNAv2 was assessed with the same data set under enforcement of a seed energy E ≤ –4.8 kcal/mol (dashed blue). The x-axis represents the amount of predictions per sRNA at a given rank and the y-axis shows the cumulated number of true positives for all sRNA whole genome target predictions.
Seed evaluation
To investigate the impact of more restrictive seed constraints, we first evaluated the energy distribution of seed interactions (without ED and dangling end contributions). To this end, we exhaustively generated all seeds (without bulges) for 5–8 bp, which revealed the seed energies to be normally distributed (see Table 1 for details). We observe that the mean value follows the formula (bp − 4)* − 1.6, where –1.6 is the average contribution of an additional stacking base pair for the Turner04 energy parameters.
Table 1. Distribution of seed energies E for different seed lengths (= number of base pairs; no bulges allowed) with μ|σ = mean|standard deviation. The highlighted row corresponds to the seed constraints of the benchmark.
| Seed length | min(E) | max(E) | μ(E) | σ(E) |
|---|---|---|---|---|
| 5 | −9.2 | 8.3 | −1.6 | 2.5 |
| 6 | −12.5 | 9.6 | −3.2 | 2.8 |
| 7 | −15.8 | 9.9 | −4.8 | 3.1 |
| 8 | −19.1 | 11.2 | −6.4 | 3.4 |
For testing more tight constraints, we enforced the seed's stability to be above average (P-value: 0.5) and reran the benchmark. Since the number of seed base pairs was set to 7, only interactions with a seed that shows an energy below −4.8 kcal/mol are reported. The results are shown in Figure 1 and reveal that a noticeable performance increase can be obtained. Furthermore, the run time is heavily reduced (halved) since less than half the number of sequence pairs that showed a stable interaction without constraint enable such a stable seed (only 55 227 of 121 988).
While this looks promising, such boundaries have to be chosen carefully. For instance, the prediction for the verified interaction of DsrA with the rpoS mRNA (37) is completely lost (no seed with E ≤ −4.8 kcal/mol) while it is top ranked (rank 1) for both IntaRNAv1 as well as IntaRNAv2 without seed constraint (see Supplementary Table S1). Thus, further research is needed on whether or not general suggestions for upper seed energy bounds can be provided or if such bounds are to be chosen in a sequence and organism dependent manner.
Minimal energy profiles
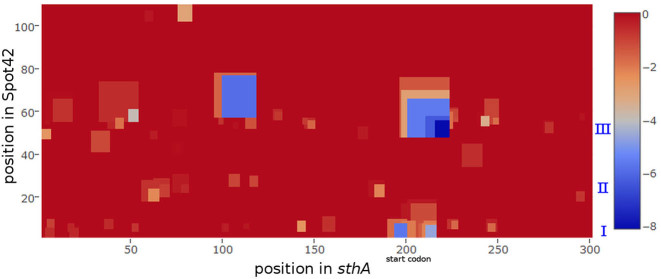
The extended web interface of IntaRNAv2 visualizes minimal energy profiles for interaction predictions that enable a sophisticated overview of interaction alternatives and their relative positioning. Figure 2 shows an example for the interaction of Spot42 with a region around the translation start site of the sthA (b3962) mRNA, which encodes a pyridine nucleotide transhydrogenase. Spot42 is known to interact with its targets via three accessible regions (I, II and III) (38), and the profile highlights sites I and III near the start codon. In fact, it has been shown that both regions I and III are important for regulation of the sthA mRNA (39), and this can also be deduced by investigating the profiles for the according Spot42 mutants (see Supplementary material).
Figure 2.
Minimal energy profile for all intermolecular index pairs covered by any predicted interaction of Spot42 with the sthA mRNA (with E < 0). Conserved accessible regions I, II and III of Spot42 known to interact are tagged on the right.
Interestingly, while region II does not seem to be centrally involved in the interaction, its mutation still slightly reduces regulation by Spot42 (39). This might stem from a minor stability reduction of both sites I and III (predicted minimal energy increase of about 0.5 kcal/mol) when region II is mutated.
CONCLUSIONS
The reimplementation of IntaRNA enables state-of-the-art energy parameters as well as seed constraint and accessibility incorporation for fast and accurate RNA–RNA interaction prediction. Increased prediction quality for genome-wide target predictions are also beneficial for comparative interaction prediction approaches like CopraRNA (25,40).
Based on an extended benchmark set, we show the enhanced prediction quality of IntaRNAv2 and highlight the possibility for further improvements, e.g. using more restrictive seed constraints. While promising, such constraints need careful tuning since they can lead to false negative predictions.
One of the new features of the IntaRNA web server is the visualization of minimal energy profiles for interacting RNAs. This enables the detailed study of alternative RNA–RNA interactions. This way, it is possible to see whether multiple interaction sites are likely and if these can occur in conjunction or only exclusively. Furthermore, mutational effects can be analyzed and thus guide wet-lab experiments, which attempt in depth validation of predicted target sites (33,39,41).
The new flexible framework is the foundation for further upcoming extensions of IntaRNA. One direction is to enable multi-site RNA–RNA interaction predictions. This way, hypotheses from the minimal energy profile investigations can be extended and further consolidated for successive experimental validation. Another direction is to enable further established input and output formats for an even more general embedding of IntaRNA into high-throughput workflow systems like Galaxy (42).
In summary, the reimplementation lays the groundwork for IntaRNA to remain among the state-of-the-art RNA–RNA interaction prediction tools in future. Thus, it will continue to be useful and available for both bioinformaticians and wet-lab experimentalists.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A.S. Richter, M. Uhl and S. Will for various ideas how to improve IntaRNA and Z. Wanli for its integration into the Galaxy tool shed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
German Research Foundation (DFG) [BA 2168/14-1 and BA 2168/16-1]. Funding for open access charge: Budget resources.
Conflict of interest statement. None declared.
REFERENCES
- 1. Backofen R., Hess W.R.. Computational prediction of sRNAs and their targets in bacteria. RNA Bio.l. 2010; 7:33–42. [DOI] [PubMed] [Google Scholar]
- 2. Lai D., Meyer I.M.. A comprehensive comparison of general RNA–RNA interaction prediction methods. Nucleic Acids Res. 2016; 44:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Umu S.U., Gardner P.P.. A comprehensive benchmark of RNA–RNA interaction prediction tools for all domains of life. Bioinformatics. 2016; btw728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mückstein U., Tafer H., Hackermüller J., Bernhart S.H., Stadler P.F., Hofacker I.L.. Thermodynamics of RNA–RNA binding. Bioinformatics. 2006; 22:1177–1182. [DOI] [PubMed] [Google Scholar]
- 5. Wagner E. G.H., Romby P.. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 2015; 90:133–208. [DOI] [PubMed] [Google Scholar]
- 6. Fabian M.R., Sonenberg N., Filipowicz W.. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010; 79:351–379. [DOI] [PubMed] [Google Scholar]
- 7. Busch A., Richter A.S., Backofen R.. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics. 2008; 24:2849–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Overloeper A., Kraus A., Gurski R., Wright P.R., Georg J., Hess W.R., Narberhaus F.. Two separate modules of the conserved regulatory RNA AbcR1 address multiple target mRNAs in and outside of the translation initiation region. RNA Biol. 2014; 11:624–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klahn S., Schaal C., Georg J., Baumgartner D., Knippen G., Hagemann M., Muro-Pastor A.M., Hess W.R.. The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E6243–E6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tobias N.J., Heinrich A.K., Eresmann H., Wright P.R., Neubacher N., Backofen R., Bode H.B.. Photorhabdus-nematode symbiosis is dependent on hfq-mediated regulation of secondary metabolites. Environ. Microbiol. 2016; doi:10.1111/1462-2920.13502. [DOI] [PubMed] [Google Scholar]
- 11. Starczynowski D.T., Morin R., McPherson A., Lam J., Chari R., Wegrzyn J., Kuchenbauer F., Hirst M., Tohyama K., Humphries R.K. et al. Genome-wide identification of human microRNAs located in leukemia-associated genomic alterations. Blood. 2011; 117:595–607. [DOI] [PubMed] [Google Scholar]
- 12. Villamizar O., Chambers C.B., Riberdy J.M., Persons D.A., Wilber A.. Long noncoding RNA Saf and splicing factor 45 increase soluble Fas and resistance to apoptosis. Oncotarget. 2016; 7:13810–13826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang F., Song W., Zhao H., Ma Y., Li Y., Zhai D., Pi J., Si Y., Xu J., Dong L. et al. The RNA-binding protein QKI5 regulates primary miR-124-1 processing via a distal RNA motif during erythropoiesis. Cell Res. 2017; 27:416–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith C., Heyne S., Richter A.S., Will S., Backofen R.. Freiburg RNA Tools: a web server integrating IntaRNA, ExpaRNA and LocARNA. Nucleic Acids Res. 2010; 38(Suppl):W373–W377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wright P.R., Georg J., Mann M., Sorescu D.A., Richter A.S., Lott S., Kleinkauf R., Hess W.R., Backofen R.. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 2014; 42:W119–W123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hofacker I.L., Fontana W., Stadler P.F., Bonhoeffer S., Tacker M., Schuster P.. Fast Folding and Comparison of RNA Secondary Structures. Monatsh. Chem. 1994; 125:167–188. [Google Scholar]
- 17. Turner D.H., Mathews D.H.. NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res. 2010; 38:D280–D282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. List M., Ebert P., Albrecht F.. Ten simple rules for developing usable software in Computational Biology. PLoS Comput. Biol. 2017; 13:e1005265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lorenz R., Bernhart S.H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bernhart S.H., Mückstein U., Hofacker I.L.. RNA Accessibility in cubic time. Algorithms fMol. Biol. 2011; 6:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alkan F., Wenzel A., Palasca O., Kerpedjiev P., Rudebeck A.F., Stadler P.F., Hofacker I.L., Gorodkin J.. RIsearch2: suffix array-based large-scale prediction of RNA–RNA interactions and siRNA off-targets. Nucleic Acids Res. 2017; doi:10.1093/nar/gkw1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lewis B.P., Shih I.-h., Jones-Rhoades M.W., Bartel D.P., Burge C.B.. Prediction of mammalian microRNA targets. Cell. 2003; 115:787–798. [DOI] [PubMed] [Google Scholar]
- 23. Rehmsmeier M., Steffen P., Höchsmann M., Giegerich R.. Fast and effective prediction of microRNA/target duplexes. RNA. 2004; 10:1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pain A., Ott A., Amine H., Rochat T., Bouloc P., Gautheret D.. An assessment of bacterial small RNA target prediction programs. RNA Biol. 2015; 12:509–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wright P.R., Richter A.S., Papenfort K., Mann M., Vogel J., Hess W.R., Backofen R., Georg J.. Comparative genomics boosts target prediction for bacterial small RNAs. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E3487–E3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cayrol B., Fortas E., Martret C., Cech G., Kloska A., Caulet S., Barbet M., Trepout S., Marco S., Taghbalout A. et al. Riboregulation of the bacterial actin-homolog MreB by DsrA small noncoding RNA. Integr. Biol. (Camb.). 2015; 7:128–141. [DOI] [PubMed] [Google Scholar]
- 27. Wang J., Rennie W., Liu C., Carmack C.S., Prevost K., Caron M.-P., Masse E., Ding Y., Wade J.T.. Identification of bacterial sRNA regulatory targets using ribosome profiling. Nucleic Acids Res. 2015; 43:10308–10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bos J., Duverger Y., Thouvenot B., Chiaruttini C., Branlant C., Springer M., Charpentier B., Barras F.. The sRNA RyhB regulates the synthesis of the Escherichia coli methionine sulfoxide reductase MsrB but not MsrA. PLoS One. 2013; 8:e63647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Benjamin J.-A.M., Masse E.. The iron-sensing aconitase B binds its own mRNA to prevent sRNA-induced mRNA cleavage. Nucleic Acids Res. 2014; 42:10023–10036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moon K., Six D.A., Lee H.-J., Raetz C. R.H., Gottesman S.. Complex transcriptional and post-transcriptional regulation of an enzyme for lipopolysaccharide modification. Mol. Microbiol. 2013; 89:52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. De Lay N., Gottesman S.. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol. Microbiol. 2012; 86:524–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Coornaert A., Chiaruttini C., Springer M., Guillier M.. Post-transcriptional control of the Escherichia coli PhoQ-PhoP two-component system by multiple sRNAs involves a novel pairing region of GcvB. PLoS Genet. 2013; 9:e1003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Beisel C.L., Updegrove T.B., Janson B.J., Storz G.. Multiple factors dictate target selection by Hfq-binding small RNAs. EMBO J. 2012; 31:1961–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen J., Gottesman S.. Spot 42 sRNA regulates arabinose-inducible araBAD promoter activity by repressing synthesis of the high-affinity low-capacity arabinose transporter. J. Bacteriol. 2016; doi:10.1128/JB.00691-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang Q., Figueroa-Bossi N., Bossi L.. Translation enhancing ACA motifs and their silencing by a bacterial small regulatory RNA. PLoS Genet. 2014; 10:e1004026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holmqvist E., Wright P.R., Li L., Bischler T., Barquist L., Reinhardt R., Backofen R., Vogel J.. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 2016; doi:10.15252/embj.201593360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lease R.A., Cusick M.E., Belfort M.. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:12456–12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Møller T., Franch T., Udesen C., Gerdes K., Valentin-Hansen P.. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 2002; 16:1696–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Beisel C.L., Storz G.. The base-pairing RNA Spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol. Cell. 2011; 41:286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wright P.R. Predicting small RNA targets in prokaryotes - a challenge beyond the barriers of thermodynamic models. 2016; Albert-Ludwigs-University Freiburg; PhD thesis doi:10.6094/UNIFR/11472. [Google Scholar]
- 41. Robledo M., Frage B., Wright P.R., Becker A.. A stress-induced small RNA modulates alpha-rhizobial cell cycle progression. PLoS Genet. 2015; 11:e1005153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Eberhard C. et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016; 44:W3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.