Abstract
Background
Ependymomas account for up to 10% of childhood CNS tumors and have a high rate of tumor recurrence despite gross total resection. Recently, classification into molecular ependymoma subgroups has been established, but the mechanisms underlying the aggressiveness of certain subtypes remain widely enigmatic. The aim of this study was to dissect the clinical and biological role of telomerase reactivation, a frequent mechanism of cancer cells to evade cellular senescence, in pediatric ependymoma.
Methods
We determined telomerase enzymatic activity, hTERT mRNA expression, promoter methylation, and the rs2853669 single nucleotide polymorphism located in the hTERT promoter in a well-characterized cohort of pediatric intracranial ependymomas.
Results
In posterior fossa ependymoma group A (PF-EPN-A) tumors, telomerase activity varied and was significantly associated with dismal overall survival, whereas telomerase reactivation was present in all supratentorial RelA fusion-positive (ST-EPN-RELA) ependymomas. In silico analysis of methylation patterns showed that only these two subgroups harbor hypermethylated hTERT promoters suggesting telomerase reactivation via epigenetic mechanisms. Furthermore, chromosome 1q gain, a well-known negative prognostic factor, was strongly associated with telomerase reactivation in PF-EPN-A. Additional in silico analyses of gene expression data confirmed this finding and further showed enrichment of the E-twenty-six factor, Myc, and E2F target genes in 1q gained ependymomas. Additionally, 1q gained tumors showed elevated expression of ETV3, an E-twenty-six factor gene located on chromosome 1q.
Conclusion
Taken together we describe a subgroup-specific impact of telomerase reactivation on disease progression in pediatric ependymoma and provide preliminary evidence for the involved molecular mechanisms.
Keywords: chromosome 1q, ependymoma, promoter methylation, RelA fusion, telomerase
Importance of the study
Novel high-throughput technologies have revolutionized our understanding of pediatric ependymoma resulting in a new classification based on distinct molecular characteristics of the different subtypes. However, biomarkers for identification of highly aggressive tumors within these subgroups remain to be elucidated. Our study demonstrates that telomerase reactivation characterizes and is confined to the more aggressive ependymoma subtypes PF-EPN-A and ST-EPN-RELA. Moreover, telomerase activity represents a feasible biomarker for the identification of PF-EPN-A ependymoma with inferior clinical outcome. Detailed analyses revealed an association of telomerase reactivation with chromosome 1q gain, a well-described negative prognosticator in ependymoma. According to our in silico analyses, hTERT expression in 1q gained tumors might be driven by overexpression of ETV3, an E-twenty-six (Ets) transcription factor gene, located on chromosome 1q. Thus, telomerase reactivation represents one biological characteristic underlying the aggressive behavior of 1q gained ependymomas.
Ependymomas account for 5% to 10% of all CNS malignancies in childhood.1,2 Maximal safe surgery and subsequent radiotherapy are the mainstays of ependymoma treatment, and risk stratification is currently based on clinical parameters such as age and extent of resection.3–5 Despite being generally considered as standard care, the benefit of cytotoxic chemotherapy is not entirely clear, and therapeutic options for recurrent tumors remain limited.3,6,7 Tumors may recur even more than a decade after the primary disease, thus requiring long-term and continuous follow-up of ependymoma patients. The rarity of this tumor type requires cooperative multicenter studies conducted over a prolonged period of time to reach sufficient patient numbers. Yet, a disparate progress in neurosurgical techniques and neuroimaging over time might lead to a stratification bias influencing interpretation of results of patients treated in different centers. A meaningful evaluation of novel biomarkers, however, requires uniform treatment and homogeneous clinical documentation, such as an immediate postoperative MRI scan, thereby limiting the sample size within ependymoma patient cohorts.5
The World Health Organization (WHO) classification of CNS tumors distinguishes between subependymoma (SE) and myxopapillary ependymoma (MPE), both WHO grade I, as well as between classic (WHO grade II) and anaplastic ependymoma (WHO grade III).8 However, the prognostic impact of WHO grading has been discussed controversially.9,10 Recently, molecular high-throughput studies revealed that ependymomas in various CNS compartments comprise different molecular subtypes showing distinct biological and clinical behavior.11,12 According to this molecular classification, posterior fossa (PF) ependymomas (EPN) are divided into PF group A (PF-EPN-A) and PF group B (PF-EPN-B) tumors. While PF-EPN-A tumors rather occur in young children and are characterized by aggressive behavior, PF-EPN-B tumors are more frequent in older children and adults and have a better prognosis.3,11 In supratentorial (ST) ependymomas, oncogenic mutually exclusive gene fusions, involving either V-rel avian reticuloendotheliosis viral oncogene homolog A (RelA) or yes-associated protein 1 (Yap1), describe 2 distinct molecular subgroups.11,13,14 In contrast to Yap1 fusion-positive ependymomas (ST-EPN-YAP1), RelA fusion-positive ependymomas (ST-EPN-RELA) have been associated with a dismal prognosis.11 Moreover, chromosome 1q gain and homozygous deletion of the CDKN2A/B locus are associated with an adverse outcome.11,15–20 Furthermore, telomerase reactivation10,21–25 represents a significant predictor of poor survival and has been demonstrated to promote tumorigenicity in epen dymoma xenograft models.21
Telomerase is an enzymatic ribonucleoprotein capable of elongating shortened telomeres by adding the telomere-specific hexameric repeats (TTAGGG). The telomerase complex consists of the telomerase RNA component, which serves as a template, and the catalytic subunit termed human telomerase reverse transcriptase (hTERT). Via activation of telomerase, cancer cells circumvent cellular senescence caused by telomere shortening, thus obtaining the ability for repeated cell divisions.26 Transcription factors (TFs) involved in the regulation of hTERT expression include E-twenty-six (Ets) factors, RelA, Myc, and E2F1.27–30hTERT expression is additionally regulated by methylation within a cytosine-phosphate-guanine (CpG) island located in the hTERT promoter region, but the exact mechanisms remain unclear as promoter methylation either silences or enhances hTERT expression depending on the promoter region and tumor type.30,31 In pediatric brain tumors, hypermethylation of the hTERT promoter is linked to higher hTERT expression.25 Moreover, 2 somatic mutations within the promoter leading to novel binding sites for Ets factors and enhanced hTERT expression have been described in different tumor types, including glioblastoma and medulloblastoma, but could not be detected in ependymoma.21,32,33 Another Ets binding site within the hTERT promoter is disrupted by the rs2853669 single nucleotide polymorphism (SNP), which is known to influence telomerase activation in adult glioblastoma and other cancer types.32,34,35
Regarding ependymoma, previous studies suggested chromosomal gain of the hTERT locus and hTERT promoter hypermethylation as molecular alterations underlying telomerase reactivation.10,15,25 However, to date a comprehensive study analyzing the association of hTERT gene expression and enzymatic activity with promoter methylation patterns and chromosomal aberrations in ependymoma is not available. Hence, the aims of this study were to evaluate telomerase-associated parameters at DNA, RNA, and protein levels, to analyze their association with molecular and clinical parameters, and to explore potential mechanisms of telomerase activation.
Materials and Methods
Patient Samples and Clinical Data
Tumor tissue and clinical data were obtained from pediatric patients with intracranial ependymomas (WHO grade II or III) treated at the Medical University of Vienna between 1965 and 2015. The study was approved by the institutional review board of the Medical University of Vienna (EK190-2011). Fresh frozen tissue of 29 and formalin-fixed paraffin-embedded (FFPE) tissue of 72 primary or recurrent pediatric ependymomas were examined. For comparison, tumor material of spinal myxopapillary ependymoma (SP-MPE), spinal ependymoma (SP-EPN), PF-EPN-B, supratentorial subependymoma (ST-SE), and ST-EPN-YAP1 was also investigated. Epilepsy surgery brain and pilocytic astrocytoma were used as negative controls. Due to limited tissue availability, not all analyses could be performed for every case. A detailed description of tissue type and analyses performed is provided in Supplementary Table S1.
Survival analyses were restricted to 22 consecutive pediatric patients with intracranial ependymomas treated since 1992 in a uniform manner, including gross total resection (GTR) as assessed by a neurosurgeon and confirmed by immediate postoperative MRI. Within this cohort, all patients received focal radiotherapy and chemotherapy. Two additional patients fulfilling these criteria were excluded from survival analyses because of death of other cause. All 22 patients received follow-up examinations, including MRI on a regular basis for early detection of tumor recurrence. For validation we used a previously published cohort (Heidelberg, n = 112, Pajtler et al11) including all ependymoma subtypes.
Telomerase Activity
Telomerase activity was measured by the telomerase repeat amplifying protocol (TRAP) utilizing the TRAPeze Telomerase Detection Kit (Chemicon International) and analyzed according to the manufacturer’s instructions as previously published.36 Heat-inactivated samples of each specimen served as negative control and the glioblastoma cell line T98G (American Type Culture Collection) as positive control. Results were calculated as total product generated units. The value of negative samples was set as 0.
hTERT mRNA Expression
RNA expression of hTERT was determined by quantitative real-time PCR. Samples showing expression of hTERT after up to 40 cycles were considered positive for RNA expression. The expression level of negative samples was set as 0.
hTERT Promoter Methylation Analysis by Pyrosequencing
Following DNA isolation and bisulfite conversion, the specific region was amplified as previously published25 and analyzed on a PyroMark Q24 MDx (Qiagen). The cutoff of 10.4% for promoter hypermethylation was calculated as the mean of control samples (4.99%) plus 2.5 SD (SD = 2.17%).
hTERT Promoter Sequencing and Telomere Length Measurement
For analysis of the rs2853669 SNP and the described C250T, C229A, and C228T mutations,32,34 the region of interest was amplified by PCR and sequenced as previously published.32 Telomere lengths were analyzed by PCR and controlled by melting curve analysis as previously described.36 The osteosarcoma cell line SA-OS, known to exert alternative lengthening of telomeres (ALT), was used as the reference sample.36
DNA Methylation Profiling by 450k Methylation Array
DNA methylation profiling was performed at the DKFZ Genomics and Proteomics Core Facility (Heidelberg, Germany) utilizing the Illumina HumanMethylation450 BeadChip array (450k array) according to the manufacturer’s instructions. The resulting data were processed as previously described37 and samples were classified into molecular subgroups (PF-EPN-A, n = 19; PF-EPN-B, n = 1; ST-EPN-RELA, n = 5; ST-EPN-YAP1, n = 1).11 Additionally, copy number analyses were performed from 450k methylation array data using the conumee Bioconductor package. Individual aberrations, with special regard to gain of chromosome 1q, were manually assessed from each profile.37
Immunohistochemical Staining and Fluorescence In situ Hybridization
Staining and evaluation of the Ki-67 proliferation index were performed as previously described.38 Chromosome 1q gain was analyzed on FFPE sections by interphase fluorescence in situ hybridization as described previously.20
Analysis of Array Datasets
DNA methylation profiles and mRNA expression data were retrieved from our previously published datasets GSE65362 and GSE64415,11 publicly available at the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo, accessed March 4, 2017).
Statistical Analyses
Overall survival (OS) was defined as the interval between diagnosis by first imaging and death. Patients alive at the last follow-up were considered censored. Progression-free survival (PFS) was defined as the period from first diagnosis by imaging to disease progression or relapse. Statistical analyses were calculated with SPSS version 21.0 (IBM), GraphPad Prism version 5.0, and R version 3.2.3 (R Development Core Team, 2015).
A more detailed description of the methods is available in the Supplementary material.
Results
Telomerase-Associated Parameters, hTERT Promoter Methylation, and rs2853669 SNP Status Within Ependymoma Subgroups
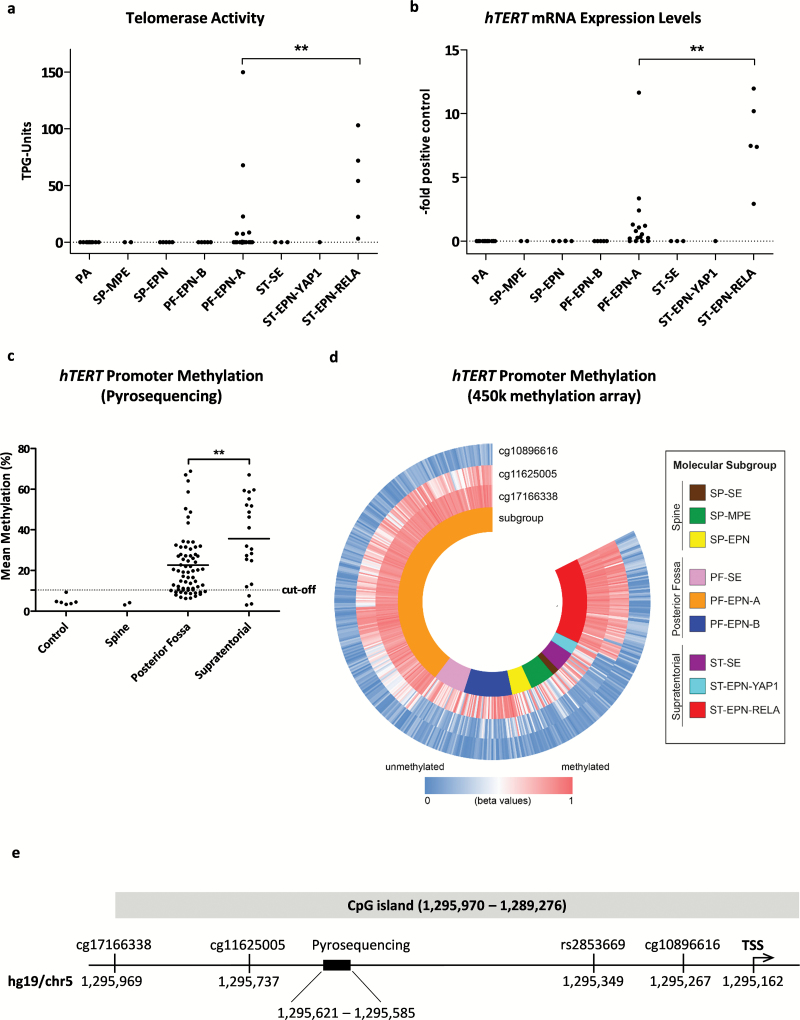
Telomerase activity was detected in 46% (11/24), hTERT gene expression in 71% (17/24), and hTERT promoter hypermethylation in 80% (53/66) of all intracranial primary ependymoma samples. Subgroup analyses revealed that ST-EPN-RELA tumors were characterized by significantly higher enzymatic activity (Fig. 1A) and hTERT mRNA expression (Fig. 1B) compared with PF-EPN-A. In contrast to these 2 subgroups, pilocytic astrocytomas (n = 11), SP-MPE (n = 2), SP-EPN (n = 5), PF-EPN-B (n = 5), ST-SE (n = 3), and ST-EPN-YAP1 (n = 1) completely lacked telomerase activity and hTERT mRNA expression. The gene was not or only marginally expressed in epilepsy surgery brain (n = 3, data not shown). With respect to the different CNS compartments, hTERT promoter methylation was absent in spinal ependymomas and was significantly elevated in ST compared to PF ependymoma (Fig. 1C). To corroborate these differences between molecular subgroups, the methylation levels of the hTERT promoter were additionally analyzed in our previously published 450k methylation array dataset of 500 ependymomas.11 Whereas the CpG site cg10896616, located adjacent to the transcription starting point, was predominantly hypomethylated, the more distal sites cg11625005 and cg17166338 exhibited distinct methylation differences between molecular subgroups (Fig. 1D, E). PF-EPN-A and ST-EPN-RELA were characterized by high but variable levels of methylation, whereas all other molecular ependymoma subgroups were widely hypomethylated within this promoter region. DNA sequencing showed no hTERT promoter mutations. Regarding the rs2853669 SNP, 63% (15/24) were noncarriers (TT), whereas within the carriers, 29% (7/24) harbored CT and 7% (2/24) CC alleles. No significant differences in allele distribution between the molecular ependymoma subgroups could be determined. Finally, analysis of telomere length revealed very long telomeres. Most ependymomas harbored markedly longer telomeres compared to the ALT-positive SA-OS osteosarcoma cell line (Supplementary Figure S1).
Fig. 1.
Telomerase activation in ependymoma. (A) Telomerase activity and (B) hTERT mRNA expression levels of pilocytic astrocytoma (PA), myxopapillary ependymoma (SP-MPE), spinal ependymoma WHO grade II (SP-EPN), posterior fossa group A (PF-EPN-A) and B (PF-EPN-B), supratentorial subependymoma (ST-SE), RelA fusion-positive (ST-EPN-RELA), and Yap1 fusion-positive (ST-EPN-YAP1) ependymomas. The dotted line at zero highlights negative samples. (C) hTERT promoter methylation stratified for tumor localization. (D) Methylation analysis of the hTERT promoter derived from 450k methylation array data, blue indicating low and red high DNA methylation and stratified for molecular subgroups (GSE65362, n = 500). (E) Map of the hTERT promoter showing the investigated regions. Sites are annotated according to hg19 (University of California Santa Cruz Genome Browser). Statistical analyses were performed by Mann–Whitney U test (**P < .01). The mean of each group is depicted as a solid line. TSS, transcription start site.
Telomerase Predicts Clinical Outcome of Patients with PF-EPN-A
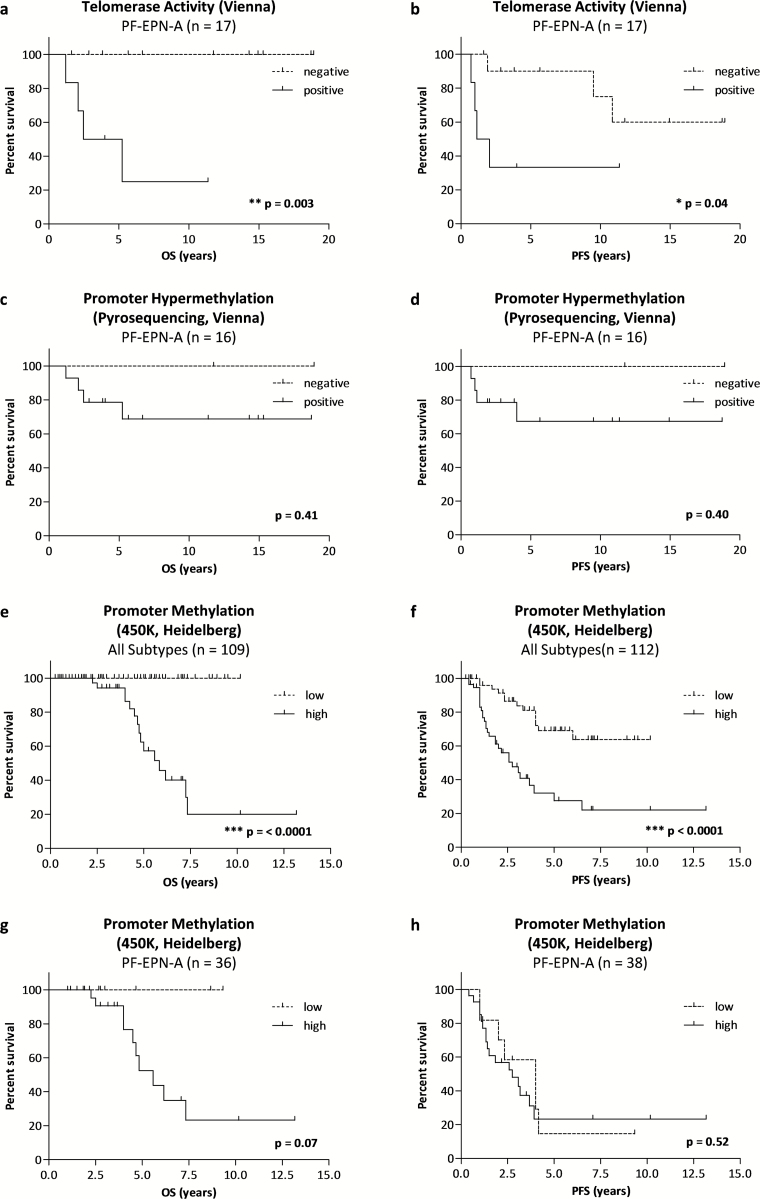
To determine the potential of telomerase-associated parameters as prognostic biomarkers, we tested the impact on survival probabilities in a cohort of 22 uniformly treated, closely monitored, and molecularly confirmed ependymoma cases. Fig. 2 provides an overview of clinical patient characteristics, outcome, and molecular parameters. No significant impact of sex, age, molecular subgroup, or tumor grade on OS and PFS was found in univariate analysis (Supplementary Table S2). Table 1 depicts 5-year and Supplementary Table S3 10-year survival analyses. Telomerase activity predicted both a significantly worse OS and PFS in the entire patient cohort. In contrast, the impact of hTERT mRNA expression on survival was not significant, although none of the 5 cases negative for hTERT mRNA expression relapsed within the 10-year observation period. The 2 cases without hTERT promoter hypermethylation showed excellent OS and PFS, yet association of this parameter with survival was not significant. hTERT promoter hypermethylation was investigated in an extended patient cohort of which FFPE material was available (n = 64, median follow-up 13.4 y, 1965–2015; Supplementary Figure S2) but was not significantly associated with shorter OS.
Fig. 2.
Summary of telomerase-associated markers and clinical and molecular parameters in a cohort of 22 uniformly treated primary pediatric ependymoma patients. RTX, radiotherapy; CTX, chemotherapy.
Table 1.
Impact of telomerase-associated parameters on survival probabilities*
| Parameter | OS | PFS | ||||||
|---|---|---|---|---|---|---|---|---|
| # | 5-y (95% CI) | Hazard Ratio (95% CI) | P | # | 5-y (95% CI) | Hazard Ratio (95% CI) | P | |
| Uniformly treated cohort (Vienna, n = 22) | ||||||||
| All subtypes | ||||||||
| Telomerase activity | .01 | .03 | ||||||
| Negative | 11 | 100% (63%–100%) | 11 | 90% (51%–100%) | ||||
| Positive | 11 | 58% (24%–89%) | 9.5 (1.6–56.3) | 11 | 36% (6%–69%) | 4.2 (1.1–16) | ||
| hTERT mRNA expression | .16 | .11 | ||||||
| Negative | 5 | 100% (40%–100%) | 5 | 100% (40%–100%) | ||||
| Positive | 16 | 74% (42%–92%) | 4.0 (0.6–28.6) | 16 | 47% (19%–74%) | 3.0 (0.8–11.2) | ||
| Promoter hypermethylationa | .42 | .17 | ||||||
| Negative | 2 | 100% (16%–100%) | 2 | 100% (16%–100%) | ||||
| Positive | 19 | 76% (50%–94%) | 3.1 (0.2 –48.8) | 19 | 58% (30%–80%) | 3.5 (0.6–20.1) | ||
| PF-EPN-A | ||||||||
| Telomerase activity | .003 | .04 | ||||||
| Negative | 11 | 100% (63%–100%) | 11 | 89% (47%–100%) | ||||
| Positive | 6 | 45% (8%–88%) | 32.2 (3.7–282.3) | 6 | 27% (1%–77%) | 6.1 (1.1–34.6) | ||
| hTERT mRNA expression | .13 | .16 | ||||||
| Negative | 5 | 100% (40%–100%) | 5 | 100% (40%–100%) | ||||
| Positive | 11 | 71% (33%–94%) | 4.8 (0.6–37.4) | 11 | 52% (18%–82%) | 3.0 (0.6–13.7) | ||
| Promoter hypermethylationa | .41 | .40 | ||||||
| Negative | 2 | 100% (16%–100%) | 2 | 100% (16%–100%) | ||||
| Positive | 14 | 79% (45%–95%) | 3.3 (0.2–51.2) | 14 | 67% (32%–91%) | 3.3 (0.2–51.2) | ||
| Validation cohort (Heidelberg, n = 112) | ||||||||
| All subtypes | ||||||||
| Promoter methylation (450kb) | < .0001 | < .0001 | ||||||
| Low | 55 | 100% (86%–100%) | 56 | 69% (50%–83%) | ||||
| High | 54 | 57% (34%–78%) | 16.4 (5.7–47.4) | 56 | 28% (12%–47%) | 4.1 (2.2–7.7) | ||
| PF-EPN-A | ||||||||
| Promoter methylation (450kb) | .07 | .52 | ||||||
| Low | 10 | 100% (16%–100%) | 11 | 15% (0.4%–57%) | ||||
| High | 26 | 52% (23%–79%) | 3.9 (0.9–16.9) | 27 | 23% (6%–49%) | 1.25 (0.5–3) | ||
*Survival probabilities in the uniformly treated cohort (n = 22, median follow-up 9.2 y) and the validation cohort (Heidelberg, n = 112, median follow-up 4.5 y).
a hTERT promoter hypermethylation (pyrosequencing).
b hTERT promoter methylation (450k, cg11625005).
As it is now accepted that molecular ependymoma subgroups represent distinct biological entities, we assessed survival probabilities separately for PF-EPN-A. Strikingly, telomerase activity was associated with a significant decrease in both OS and PFS (Fig. 3A and B, Supplementary Table S4). Interestingly, cases negative for hTERT mRNA expression or promoter hypermethylation showed excellent OS and PFS; however, this effect was not significant (Fig. 3C and D, Supplementary Table S4).
Fig. 3.
Prognostic value of telomerase-associated markers. Kaplan–Meier survival curves for OS (A, C, E, G) and PFS (B, D, F, H) within the respective patient subgroups. Survival curves for PF-EPN-A within the uniformly treated cohort (Vienna, n = 17) stratified for (A, B) telomerase activity and (C, D) hTERT promoter hypermethylation detected by pyrosequencing. Survival curves for PF-EPN-A within the validation cohort (Heidelberg, n = 112) stratified for methylation levels at the cg11625005 CpG site in all ependymoma subtypes (E, F), and restricted to posterior fossa group A (PF-EPN-A) (G, H). Tables of “numbers at risk” are summarized in Supplementary Table S4. P-values (log-rank test) are indicated.
Having determined subgroup-specific differences in telomerase reactivation and its prognostic value, we validated these findings in a previously published11 cohort (Heidelberg, n = 112) including all ependymoma subgroups. Therefore, we divided the cases into 2 groups according to the median methylation level at the cg11625005 CpG site. In line with our previous results, the more benign ependymoma subtypes were predominantly found in the low methylation subgroup, whereas all ST-EPN-RELA (22/22) but only 71% of PF-EPN-A (27/38) harbored high promoter methylation (Supplementary Table S5). Analysis across all subgroups confirmed the association of high hTERT promoter methylation and inferior clinical outcome (Fig. 3E and F, Supplementary Table S4). Also analysis of the PF-EPN-A subgroup showed no death of disease in cases harboring low promoter methylation (Table 1, Fig. 3C, Supplementary Table S4). No difference was observed with respect to PFS (Table 1, Fig. 3D, Supplementary Table S4).
Association Between Telomerase-Associated Parameters and Clinical and Histopathological Characteristics
Telomerase activity showed a strong correlation with both mRNA expression and promoter methylation (Supplementary Table S6). Furthermore, all parameters correlated with higher patient age (Supplementary Table S6), probably related to the higher age of ST-EPN-RELA patients (Fig. 2). Telomerase activity and hTERT mRNA expression showed an association with higher Ki-67 proliferation and anaplastic ependymoma (WHO grade III). Interestingly, gain of chromosome 1q, a well-described marker of dismal prognosis in pediatric ependymoma,10,17,20 significantly correlated with telomerase activity (Supplementary Table S6).
Telomerase Reactivation Is Associated with Chromosome 1q Gain in PF-EPN-A
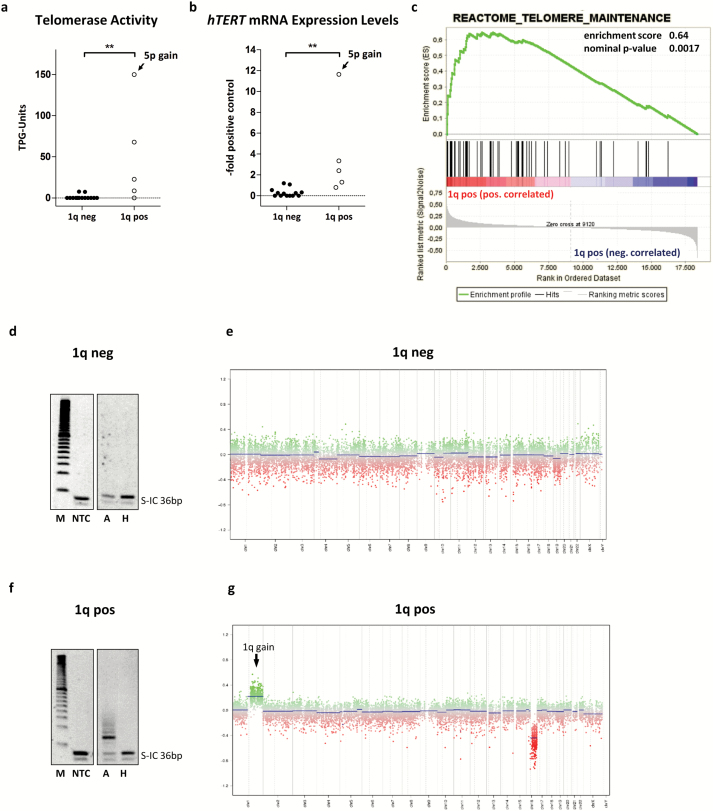
We investigated the association of 1q gain and telomerase reactivation with respect to molecular subgroups and found a strong association for PF-EPN-A (χ2, P = .03). On closer inspection, both telomerase activity and hTERT mRNA expression were significantly increased in PF-EPN-A tumors with chromosome 1q gain (Fig. 4A and B). Interestingly, one 1q gain–positive PF-EPN-A tumor additionally harbored a gain at the hTERT locus on chromosome 5p, but exclusion of this case did not change the level of significance (telomerase activity, P = .009; hTERT mRNA expression P = .007). To provide further evidence for the role of chromosome 1q gain in telomere stabilization, we performed gene set enrichment analysis (GSEA) in an expression dataset of 47 primary PF-EPN-A. Indeed, the curated gene set “REACTOME_TELOMERE_MAINTENANCE” was significantly enriched in PF-EPN-A ependymomas harboring chromosome 1q gain compared with samples without 1q gain (Fig. 4C, Supplementary Table S7). Representative examples of TRAP gels and copy number profiles for a PF-EPN-A tumor without and with chromosome 1q gain are shown (Fig. 4D–G). Due to the small number of ST-EPN-RELA tumors in our series, no statistical analyses could be performed for this subgroup and no enrichment was determined by GSEA (Supplementary Figure S3, Supplementary Table S7).
Fig. 4.
Association of chromosome 1q gain and telomerase reactivation in PF-EPN-A. (A) Telomerase activity and (B) hTERT mRNA expression levels segregated according to chromosome 1q gain. One single case with gain of chromosome 5p is indicated. The dotted line at zero highlights negative samples. (C) GSEA for the term “Reactome Telomere Maintenance.” TRAP gels and copy number profiles are shown for a 1q gain–negative (D, E) and a 1q gain–positive (F, G) tumor. Statistical analyses were performed by a Mann–Whitney U test (**P < .01); pos, positive; neg, negative; TPG, total product generated; M, marker; NTC, no template control; A, active sample; H, heat inactivated sample.
Myc, E2F, and Ets Transcription Factors Are Candidate Genes for Chromosome 1q Gain–Driven Telomerase Activation in Ependymoma
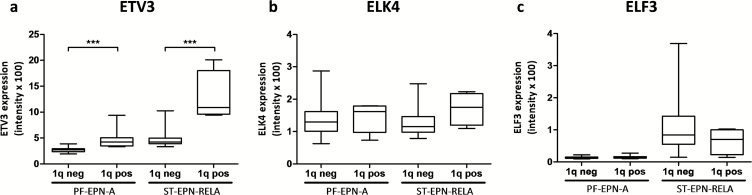
Next, we searched for differences in transcriptional activation patterns of 1q gain–positive versus –negative tumors by using GSEA within a dataset of cis-regulatory motifs in promoters and corresponding genes (“MSigDB C3: motif genesets: transcription factor targets v5”39). Among the top-ranked gene sets enriched in both PF-EPN-A and ST-EPN-RELA tumors with chromosome 1q gain were TF-binding motifs related to Myc (V$MAZ_Q6), E2F (V$E2F_Q2), and Ets TFs (V$ETF_Q6) (Supplementary Table S6). As Ets TFs are well-known activators of hTERT expression and, hence, telomerase activity,27 we further determined the expression of 3 Ets factors located on chromosome 1q—ie, ETV3, ELK4, and ELF3-in this dataset. Indeed, we found higher levels of ETV3 but not of the other Ets TFs in ependymomas harboring chromosome 1q gain (Fig. 5A–C).
Fig. 5.
Overexpression of ETV3 in ependymoma with gain of chromosome 1q. Messenger RNA expression levels of the 3 Ets transcription factors located on chromosome 1q: (A) ETV3, (B) ELK4, and (C) ELF3 stratified for 1q status and molecular subtype.
Discussion
In this study we uncovered that telomerase activity predicts dismal OS specifically in the PF-EPN-A subgroup, while telomerase-associated parameters are generally high in ST-EPN-RELA tumors. Several studies applying various detection methods have proposed telomerase as a prognostic marker for pediatric ependymoma without dissecting the association with this recently defined molecular subgroups.10,21–24 Moreover, the prognostic quality of telomere stabilizing parameters has not yet been validated in a uniformly treated patient cohort as performed in this study.
Within the entire cohort of pediatric intracranial epen dymomas, we detected telomerase activity in 46% of the cases, which was slightly lower than in previous studies ranging from 64% to 82%.21,23,24 This difference might be explained by the use of the gel-based TRAP assay, which is—due to the presence of an internal control—considered to be less sensitive but more reliable than the detection method based on enzyme-linked immunosorbent assay.40 Accordingly, hTERT mRNA expression was found in 71% of the tumors in our study in agreement with earlier reports. hTERT promoter hypermethylation was detected in 80% of cases using a cutoff of 10.4% determined by CNS tissue controls and not tissue of other organs as in previous studies.21,25 Frequencies of the different rs2853669 genotypes (TT, CT, and CC) were comparable to other patient populations.32,34 In accordance with earlier reports, our data confirm an association of telomerase reactivation with anaplastic ependymoma (WHO grade III) and a higher Ki-67 proliferation index.22,41 This supports a major contribution of telomerase activity to aggressiveness of ependymoma cases with biologically unfavorable characteristics.
Analysis of telomerase in ependymoma subgroups showed reactivation almost exclusively in intracranial ependymomas. All ST-EPN-RELA cases in our series harbored telomerase activation, whereas hTERT mRNA expression and telomerase activity were variable in PF-EPN-A tumors. These findings were corroborated by analysis of hTERT promoter methylation patterns by pyrosequencing in our cohort and additional in silico analyses of an independent 450k methylation dataset. Interestingly, the CpG site closest to the transcription start site (cg10896616) was hypomethylated across all ependymoma subgroups. In contrast, only the more aggressive PF-EPN-A and ST-EPN-RELA subtypes showed promoter hypermethylation in the more distal promoter region (cg11625005, cg17166338). These methylation patterns suggest a differential regulation within the CpG island resulting in an open chromatin structure close to the transcription start site and repression of TF binding in more distal parts of the hTERT promoter.30 The fact that the 450k array-based ependymoma subgroups are reflected in the methylation pattern of this promoter region might indicate a central function of the hTERT promoter in ependymoma biology.
Very recently, the requirement of GTR and radiotherapy for successful treatment of pediatric ependymoma has been confirmed.3,4 To rule out a treatment-derived bias in our survival analyses, we only included patients with GTR, who received uniform radio- and chemotherapy. This was further strengthened by the availability of long-term follow-up data. Thereby, we provide a unique cohort for validation of the prognostic impact of telomerase-associated markers on clinical outcome. Survival analyses in our series confirm the strong prognostic impact of telomerase activity on OS and PFS.21 In contrast to previous studies, we observed no significant associations for hTERT mRNA and hTERT promoter hypermethylation albeit cases negative for these parameters showed excellent OS.10,21,25 Stratification of our validation cohort according to cg11625005 methylation clearly segregated tumors with dismal OS, thus confirming the prognostic value of hTERT promoter methylation in collectives consisting of different molecular subgroups. Therefore, the significant effect observed in a previous study might be caused by a higher proportion of benign subtypes within the respective study collectives.25 Taken together, testing telomerase activity by TRAP seems to represent the most reliable method to detect aggressive tumors with telomerase reactivation. Although fresh frozen tissue is needed for testing telomerase activity, the facts that commercial kits are available, that only small amounts of tissue are needed, and that the enzymatic activity is more stable than mRNA make it a feasible marker for routine clinical practice.36,40
By stratification for molecular ependymoma subgroups we uncovered for the first time that telomerase activity is a feasible biomarker to detect PF-EPN-A tumors with dismal prognosis. Furthermore, our results suggest that apart from less aggressive molecular subtypes, hTERT promoter hypomethylation might also characterize PF-EPN-A tumors with a more favorable outcome. However, these observations require validation in larger patient collectives. PF-EPN-A represents the more common and more aggressive molecular subtype in the PF localization, but about half of the patients show long-term survival.3,11 The importance of telomerase in recurrent ependymoma has been previously described22 and could be further supported by our analyses (Supplementary Figure S4). Yet, the exact mechanisms underlying telomerase-mediated tumor aggressiveness and therapy resistance in pediatric ependymoma need to be further investigated. In neuroblastoma, for example, telomerase activity has been connected to resistance against radiotherapy.42
We have previously reported that chromosome 1q gain is an important prognostic factor for dismal outcome in PF ependymomas20 and show now an association between telomerase activity and chromosome 1q gain in PF-EPN-A tumors. On the basis of GSEA we provide additional bioinformatic evidence that expression of genes associated with telomerase maintenance is enriched in 1q-gained PF-EPN-A. Furthermore, the TF signatures enhanced in chromosome 1q–gained ependymoma top ranked Myc, E2F, and Ets family members. Interestingly, all these TFs are well known to activate hTERT expression.29,30,43 Of these TFs, the gene locus for ETV3 is indeed located on 1q21–q23, one of the most frequently gained subregions in pediatric ependymomas with 1q gain.17 Accordingly, we found higher expression of ETV3 in tumors with gain of chromosome 1q, an association also described in breast cancer.44 This points towards a central role of ETV3 as a driver of enhanced hTERT expression in PF-EPN-A tumors with 1q gain and might be one underlying biological process for the unfavorable behavior of this subtype. According to these results, testing for 1q gain and telomerase activity might detect the same highly aggressive tumors. Whereas the detection of 1q gain by fluorescence in situ hybridization can be carried out in most routine pathologic departments, TRAP is not yet widely available. However, as telomerase reactivation seems to be a central biological feature of aggressive PF-EPN-A, additional analysis of telomerase activity might help to detect tumors with dismal outcome—all the more because other mechanisms of telomerase reactivation, such as gains of the hTERT locus at chromosome 5p, might be present.
Our analyses further reveal that activation of telomerase is a characteristic feature of ST-EPN-RELA ependymomas. Accordingly, all tumors with C11orf95-RelA fusion showed telomerase activation in a previous study.21 Notably, also chromothripsis, characteristic for many ST-EPN-RELA genomes, has been linked to increased telomere lengths and hTERT expression in this tumor type.45
In summary, we describe telomerase activity as a potential biomarker characterizing a more aggressive subtype within PF-EPN-A tumors, show an association of telomerase reactivation with chromosome 1q gain and RelA fusion, and explore molecular mechanisms involved in telomerase activation in these ependymoma subtypes.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
This study was supported by the Jubiläumsfonds der Österreichischen Nationalbank (project # 15173 and #16152), the Medizinisch-Wissenschaftlicher Fonds des Bügermeisters der Bundeshauptstadt Wien (project # 14015), the Herzfelder’sche Familienstiftung, and the Forschungsgesellschaft fuer cerebrale Tumore. Additional support came from HIPO_036, the Illumina Medical Research Grant, and the German Childhood Cancer Foundation for MNP2.0.
Conflict of interest statement. The authors declare that they have no conflict of interest.
Supplementary Material
Acknowledgments
The authors thank Brigitta Hammer-Schmiedel and Tsuyoshi Araki for skillful technical assistance as well as Prof Harald Heinzl for excellent statistical advice.
References
- 1. Ostrom QT, Gittleman H, Liao P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. 2015;17 (supplOctober):iv1–iv62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ostrom QT, de Blank PM, Kruchko C, et al. Alex’s Lemonade Stand Foundation infant and childhood primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol. 2015;16Suppl 10:x1–x36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ramaswamy V, Hielscher T, Mack SC, et al. Therapeutic impact of cytoreductive surgery and irradiation of posterior fossa ependymoma in the molecular era: a retrospective multicohort analysis. J Clin Oncol. 2016;34(21):2468–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Massimino M, Miceli R, Giangaspero F, et al. Final results of the second prospective AIEOP protocol for pediatric intracranial ependymoma. Neuro Oncol. 2016;18(10):1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu J, Armstrong TS, Gilbert MR. Biology and management of ependymomas. Neuro Oncol. 2016;18(7):902–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grundy RG, Wilne SA, Weston CL, et al. ; Children’s Cancer and Leukaemia Group (formerly UKCCSG) Brain Tumour Committee. Primary postoperative chemotherapy without radiotherapy for intracranial ependymoma in children: the UKCCSG/SIOP prospective study. Lancet Oncol. 2007;8(8):696–705. [DOI] [PubMed] [Google Scholar]
- 7. Wright KD, Gajjar A. New chemotherapy strategies and biological agents in the treatment of childhood ependymoma. Childs Nerv Syst. 2009;25(10):1275–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 9. Ellison DW, Kocak M, Figarella-Branger D, et al. Histopathological grading of pediatric ependymoma: reproducibility and clinical relevance in European trial cohorts. J Negat Results Biomed. 2011;10:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Modena P, Buttarelli FR, Miceli R, et al. Predictors of outcome in an AIEOP series of childhood ependymomas: a multifactorial analysis. Neuro Oncol. 2012;14(11):1346–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pajtler KW, Witt H, Sill M, et al. Molecular classification of ependymal tumors across All CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27(5):728–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Witt H, Mack SC, Ryzhova M, et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell. 2011;20(2):143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pietsch T, Wohlers I, Goschzik T, et al. Supratentorial ependymomas of childhood carry C11orf95-RELA fusions leading to pathological activation of the NF-κB signaling pathway. Acta Neuropathol. 2014;127(4):609–611. [DOI] [PubMed] [Google Scholar]
- 14. Parker M, Mohankumar KM, Punchihewa C, et al. C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature. 2014;506(7489):451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mendrzyk F, Korshunov A, Benner A, et al. Identification of gains on 1q and epidermal growth factor receptor overexpression as independent prognostic markers in intracranial ependymoma. Clin Cancer Res. 2006;12(7 Pt 1):2070–2079. [DOI] [PubMed] [Google Scholar]
- 16. Korshunov A, Witt H, Hielscher T, et al. Molecular staging of intracranial ependymoma in children and adults. J Clin Oncol. 2010;28(19):3182–3190. [DOI] [PubMed] [Google Scholar]
- 17. Kilday JP, Mitra B, Domerg C, et al. Copy number gain of 1q25 predicts poor progression-free survival for pediatric intracranial ependymomas and enables patient risk stratification: a prospective European clinical trial cohort analysis on behalf of the Children’s Cancer Leukaemia Group (CCLG), Societe Francaise d’Oncologie Pediatrique (SFOP), and International Society for Pediatric Oncology (SIOP). Clin Cancer Res. 2012;18(7):2001–2011. [DOI] [PubMed] [Google Scholar]
- 18. Kilday JP, Rahman R, Dyer S, et al. Pediatric ependymoma: biological perspectives. Mol Cancer Res. 2009;7(6):765–786. [DOI] [PubMed] [Google Scholar]
- 19. Godfraind C, Kaczmarska JM, Kocak M, et al. Distinct disease-risk groups in pediatric supratentorial and posterior fossa ependymomas. Acta Neuropathol. 2012;124(2):247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Araki A, Chocholous M, Gojo J, et al. Chromosome 1q gain and tenascin-C expression are candidate markers to define different risk groups in pediatric posterior fossa ependymoma. Acta Neuropathol Commun. 2016;4(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barszczyk M, Buczkowicz P, Castelo-Branco P, et al. Telomerase inhibition abolishes the tumorigenicity of pediatric ependymoma tumor-initiating cells. Acta Neuropathol. 2014:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tabori U, Wong V, Ma J, et al. Telomere maintenance and dysfunction predict recurrence in paediatric ependymoma. Br J Cancer. 2008;99(7):1129–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tabori U, Ma J, Carter M, et al. Human telomere reverse transcriptase expression predicts progression and survival in pediatric intracranial ependymoma. J Clin Oncol. 2006;24(10):1522–1528. [DOI] [PubMed] [Google Scholar]
- 24. Ridley L, Rahman R, Brundler MA, et al. ; Children’s Cancer and Leukaemia Group Biological Studies Committee. Multifactorial analysis of predictors of outcome in pediatric intracranial ependymoma. Neuro Oncol. 2008;10(5):675–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Castelo-Branco P, Choufani S, Mack S, et al. Methylation of the TERT promoter and risk stratification of childhood brain tumours: an integrative genomic and molecular study. Lancet Oncol. 2013;14(6):534–542. [DOI] [PubMed] [Google Scholar]
- 26. Martínez P, Blasco MA. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat Rev Cancer. 2011;11(3):161–176. [DOI] [PubMed] [Google Scholar]
- 27. Xu D, Dwyer J, Li H, Duan W, Liu JP. Ets2 maintains hTERT gene expression and breast cancer cell proliferation by interacting with c-Myc. J Biol Chem. 2008;283(35):23567–23580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zuo QP, Liu SK, Li ZJ, et al. NF-kappaB p65 modulates the telomerase reverse transcriptase in the HepG2 hepatoma cell line. Eur J Pharmacol. 2011;672(1-3):113–120. [DOI] [PubMed] [Google Scholar]
- 29. Alonso MM, Fueyo J, Shay JW, et al. Expression of transcription factor E2F1 and telomerase in glioblastomas: mechanistic linkage and prognostic significance. J Natl Cancer Inst. 2005;97(21):1589–1600. [DOI] [PubMed] [Google Scholar]
- 30. Daniel M, Peek GW, Tollefsbol TO. Regulation of the human catalytic subunit of telomerase (hTERT). Gene. 2012;498(2):135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guilleret I, Yan P, Grange F, Braunschweig R, Bosman FT, Benhattar J. Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int J Cancer. 2002;101(4):335–341. [DOI] [PubMed] [Google Scholar]
- 32. Spiegl-Kreinecker S, Lötsch D, Ghanim B, et al. Prognostic quality of activating TERT promoter mutations in glioblastoma: interaction with the rs2853669 polymorphism and patient age at diagnosis. Neuro Oncol. 2015;17(9):1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110(15):6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rachakonda PS, Hosen I, de Verdier PJ, et al. TERT promoter mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism. Proc Natl Acad Sci U S A. 2013;110(43):17426–17431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Simon M, Hosen I, Gousias K, et al. TERT promoter mutations: a novel independent prognostic factor in primary glioblastomas. Neuro Oncol. 2015;17(1):45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lötsch D, Ghanim B, Laaber M, et al. Prognostic significance of telomerase-associated parameters in glioblastoma: effect of patient age. Neuro Oncol. 2013;15(4):423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hovestadt V, Jones DT, Picelli S, et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature. 2014;510(7506):537–541. [DOI] [PubMed] [Google Scholar]
- 38. Wolfsberger S, Fischer I, Höftberger R, et al. Ki-67 immunolabeling index is an accurate predictor of outcome in patients with intracranial ependymoma. Am J Surg Pathol. 2004;28(7):914–920. [DOI] [PubMed] [Google Scholar]
- 39. Xie X, Lu J, Kulbokas EJ, et al. Systematic discovery of regulatory motifs in human promoters and 3ʹ UTRs by comparison of several mammals. Nature. 2005;434(7031):338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saldanha SN, Andrews LG, Tollefsbol TO. Analysis of telomerase activity and detection of its catalytic subunit, hTERT. Anal Biochem. 2003;315(1):1–21. [DOI] [PubMed] [Google Scholar]
- 41. Rushing EJ, Yashima K, Brown DF, et al. Expression of telomerase RNA component correlates with the MIB-1 proliferation index in ependymomas. J Neuropathol Exp Neurol. 1997;56(10):1142–1146. [DOI] [PubMed] [Google Scholar]
- 42. Wesbuer S, Lanvers-Kaminsky C, Duran-Seuberth I, et al. Association of telomerase activity with radio- and chemosensitivity of neuroblastomas. Radiat Oncol. 2010;5:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu KJ, Grandori C, Amacker M, et al. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21(2):220–224. [DOI] [PubMed] [Google Scholar]
- 44. Mesquita B, Lopes P, Rodrigues A, et al. Frequent copy number gains at 1q21 and 1q32 are associated with overexpression of the ETS transcription factors ETV3 and ELF3 in breast cancer irrespective of molecular subtypes. Breast Cancer Res Treat. 2013;138(1):37–45. [DOI] [PubMed] [Google Scholar]
- 45. Ernst A, Jones DT, Maass KK, et al. Telomere dysfunction and chromothripsis. Int J Cancer. 2016;138(12):2905–2914. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.